Abstract
The proposed macroscopic optical histopathology includes a broad-band light source which is selected to illuminate the tissue glass slide of suspicious pathology, and a hyperspectral camera that captures all wavelength bands from 450 to 950 nm. The system has been trained to classify each histologic slide based on predetermined pathology with light having a wavelength within a predetermined range of wavelengths. This technology is able to capture both the spatial and spectral data of tissue. Highly metastatic human head and neck cancer cells were transplanted to nude mice. After 2–3 weeks, the mice were euthanized and the lymph nodes and lung tissues were sent to pathology. The metastatic cancer is studied in lymph nodes and lungs. The pathological slides were imaged using the hyperspectral camera. The results of the proposed method were compared to the pathologic report. Using hyperspectral images, a library of spectral signatures for different tissues was created. The high-dimensional data were classified using a support vector machine (SVM). The spectra are extracted in cancerous and non-cancerous tissues in lymph nodes and lung tissues. The spectral dimension is used as the input of SVM. Twelve glasses are employed for training and evaluation. The leave-one-out cross-validation method is used in the study. After training, the proposed SVM method can detect the metastatic cancer in lung histologic slides with the specificity of 97.7% and the sensitivity of 92.6%, and in lymph node slides with the specificity of 98.3% and the sensitivity of 96.2%. This method may be able to help pathologists to evaluate many histologic slides in a short time.
Keywords: Cancer detection, hyperspectral imaging, Head and neck cancer, Optical imaging, Infrared imaging, Support vector machine
1. INTRODUCTION
The current gold standard for diagnosis of cancer is biopsy, which involves the removal of tissue samples for examination. Laboratory results for the determination of histopathology of a suspected tumor may generally take several days [1]. Hyperspectral image data provide a powerful tool for non-invasive tissue analyses. This technology is capable of capturing both the spatial and spectral information of an organ in one snapshot. In fact, a hyperspectral imaging system produces several narrow band images at different wavelengths. Compared to conventional color cameras and other filter-based imaging systems, this system produces full neighboring spectral data with spectral and spatial information [2].
Metastatic tumor or a metastasis is one of the most important concerns in cancer. Hyperspectral imaging (HSI) can extend the human vision to near-infrared and infrared wavelength regions. HSI has already been applied in the medical field [1, 3–14]. HSI is applied to provide quantitative data about the tissue oxygen saturation in patients with peripheral vascular disease, to detect ischemic regions of the intestine during surgery, to predict and follow healing in foot ulcers of diabetic patients, and to diagnose hemorrhagic shock [15]. The hyperspectral imaging that operates as a tunable optical band pass filter has been utilized to discriminate between non-cancerous and cancerous tissue [16]. Recently, Fourier transform infrared (FTIR) spectroscopy has widely been used as a cancer diagnostic technique that can only be applied to point measurements [17–22]. The hyperspectral imaging was evaluated for cytologic diagnosis of cancer cells. HSI was employed to obtain hyperspectrum from a normal human fibroblast, as well as its telomerase-immortalized and SV40-transformed derivatives. Novel algorithms were developed to differentiate among these cell models based on spectral and spatial differences [23]. The high-resolution hyperspectral imaging microscopy was evaluated to detect abnormalities in skin tissue using Hematoxylin and Eosin stained preparations of normal and abnormal skin, benign nevi and melanomas [6]. The capability of this method was demonstrated by observing gastric tumors in 10 human subjects. The spectral signatures of the gastric cancer and non-cancerous stomach tissue were created in infrared wavelengths. The hyperspectral images were processed via the standard deviation of the spectral diagram, support vector machine, and first derivatives and integral of in spectral diagram were proposed to enhance and detect the cancerous regions [24]. So far, none of the research has used hyperspectral imaging for cancer detection in pathological slides. The goal of this study is to test the potential of using hyperspectral imaging and advance image analysis methods for cancer detection.
2. METHODS
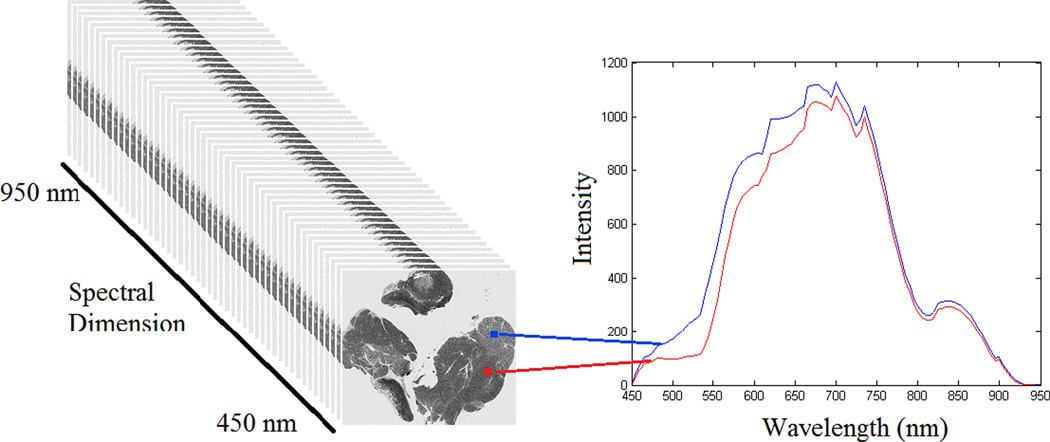
The hyperspectral images were captured of three normal and three pathologic slides for each organ, i.e. lung and lymph node, using a hyperspectral camera. Each pixel in a hyperspectral image has different intensities at different wavelengths. These intensities make the spectral diagram or the spectral signature for each pixel. Figure 1 shows a schematic view of the hyperspectral image.
Figure 1.
A schematic view of hyperspectral images (left). Right: The spectral graphs of two pixels from the histologic slide of lymph node tissue. The graph depicts the intensities for each pixel in that wavelength.
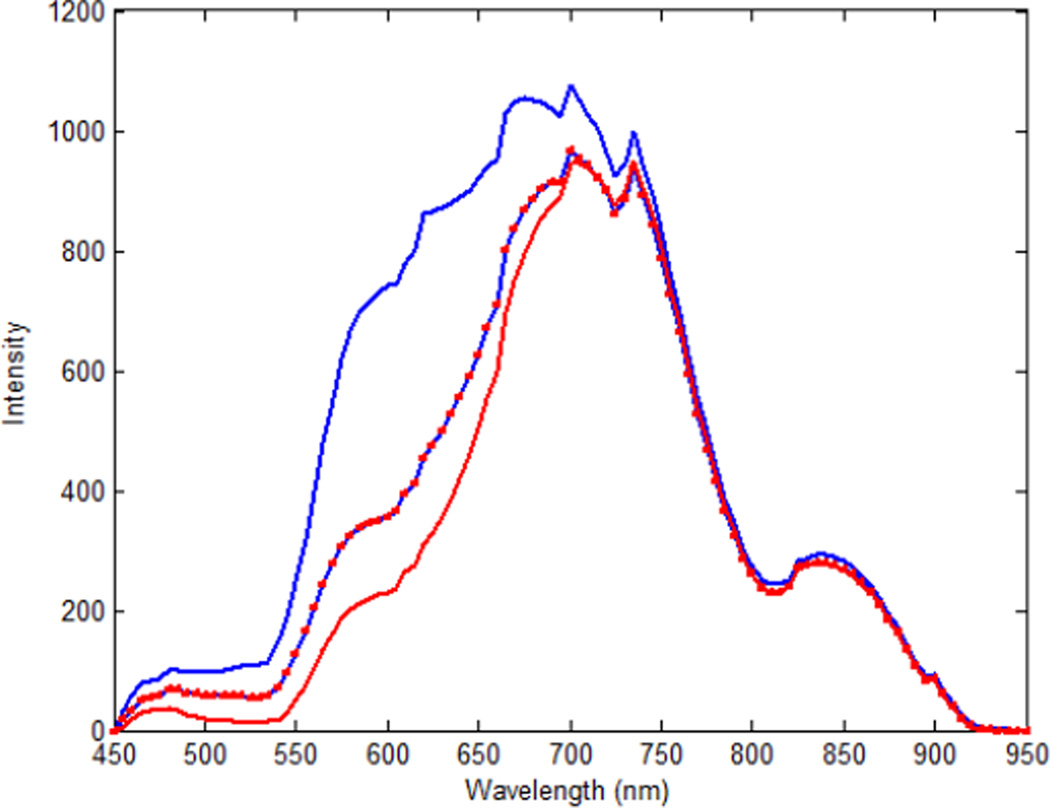
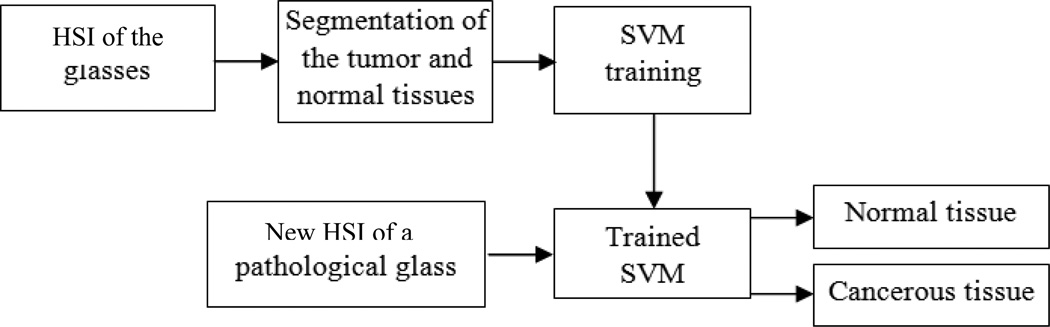
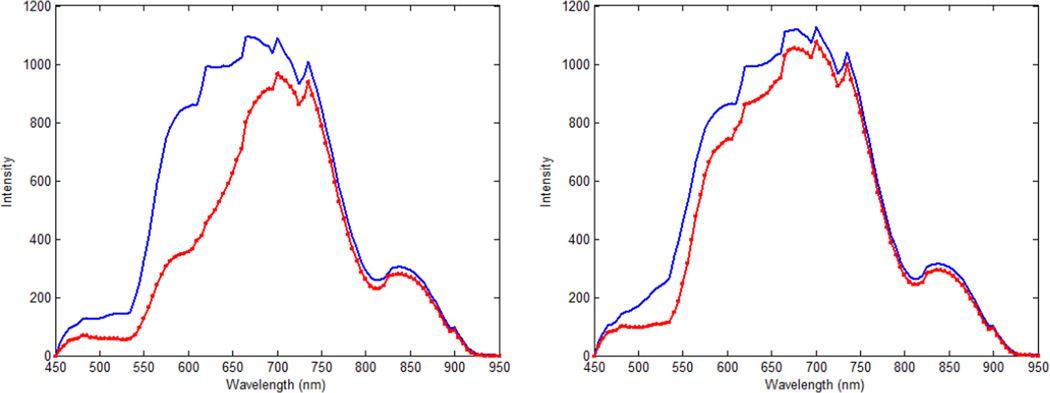
We made a spectra library by capturing spectral diagrams of the original cancerous tissue and the metastatic tumor in the lungs and lymph nodes. Figure 2 shows the intensity spectra of the head and neck tumor and its metastasis to the lungs and the lymph nodes. The defined regions were utilized to train the support vector machine (SVM) to classify the normal and malignant regions. Using the trained SVM, each tissue slide was analyzed and classified. The SVM was trained and evaluated with the leave-one-out cross-validation method [25, 26]. Figure 3 shows the flowchart of the method.
Figure 2.
The mean of spectral signatures of the original cancerous tissues and the metastatic tumor in mice. The horizontal axis shows different wavelengths in nanometers. The vertical axis shows the intensities. The original tumor pixels are shown with the red line; the metastatic tumor in the lymph nodes are shown in the blue line with red squares; and the metastatic tumor in the lungs are shown in the blue line.
Figure 3.
The flowchart of the method.
The head and neck tumor was transplanted into 19 nude mice. The cell line that was used in this study was 686LN-M4e. This cell line is a highly metastatic, human head and neck cancer cell line. After 2–3 weeks, the mice were euthanized and the lymph nodes and lung tissues were sent to the pathology facility at our institution. The metastatic cancer is studied in lymph nodes and lungs. The pathological slides were imaged using the hyperspectral camera. To capture the hyperspectral image data, a CRi imaging system system (Caliper, Hopkinton, MA) was used.
A successful approach to HSI data classification is based on the use of multilayer perceptrons (MLP) and radial basis function neural networks (RBFNNs). However, these approaches are not effective when dealing with a high number of spectral bands, since they are highly sensitive to the Hughes phenomenon. In the recent years, support vector machines (SVMs) have been successfully used for HSI data classification. The SVMs can efficiently handle large input spaces or noisy samples, and produce sparse solutions [27]. Therefore, to classify the compressed data and to segment the hyperspectral image, support vector machine (SVM) is chosen for this study.
2.1 CRi Maestro systems
CRi Maestro systems can acquire full hyperspectral data with down to 25 micron/pixel resolution in only a few seconds. Inside the Maestro system is a liquid crystal tunable filter that utilizes a solid-state liquid-crystal design that blocks unwanted wavelengths and transmits the required wavelength. It changes wavelengths in milliseconds without noise or vibration. The CRi's spectral imaging system generates x-number of images, where×is user-defined. Each image set contains the measurement of the spectrum of all the points which comprise it. The wavelength range of interest was defined between 450–950 nm with 5 nm increments. The series or spectral cube contains spectra from 450 to 950 nm, with each image containing 1.4 million data points. The excitation light was set on the white illumination plus the infrared internal illumination and no emission filter was used. Each pixel in the hyperspectral image has a sequence of reflectance at different spectral wavelengths that can display the spectral signature of that pixel.
2.2 Least Squares SVMs
In Support Vector Machines (SVMs), a convex quadratic programming (QP) solves the classification problem. Suykens and Vandewalle proposed a new version of SVM classifiers, which was named Least Squares SVMs (LS-SVMs) [28]. In LS-SVMs instead of inequality constraints, a two-norm with equality are applied. Therefore, instead of a QP problem in dual space, a linear set of equations are obtained. The SVM tries to find a large margin for classification. However, the LS-SVM that is used in this paper looks for a ridge regression for classification with binary targets. This method overcomes some disadvantages of SVM. For example the selection of hyperparameters is not as problematic. The size of the matrix involved in the QP problem is also directly proportional to the number of training points [29]. Suykens and Vandewalle modified the Vapnik’s optimization function of the SVM as follows [28]:
| (1) |
subject to the equality constraints
| (2) |
where w is the weighting vector, b is the bias term, e is for misclassifications, and γ is the tuning parameter. This constrained optimization problem can be solved by determining the saddle points in the Lagrange functional as,
| (3) |
where αi ∈ R are Lagrange multipliers that can be positive or negative in the LS-SVM formulation.
In SVMs, it is possible to choose several types of kernel functions including linear, polynomial, radial basis function (RBF), multilayer perceptron (MLP) with one hidden layer and splines. The RBF kernel is used in this paper as follows:
| (4) |
where σ is constant.
Spectral data of each pixel in the hyperspectral image was employed as the input of the SVM, and the output is either cancerous or non-cancerous pixel. In each slide, the regions that are definitely normal or malignant were selected and used for SVM training and evaluation. These regions were defined based on pathological reports. The leave-one-out cross-validation method is utilized in this study. Here we have input vectors of 101 elements in 5 nm spectral resolution images.
2.3 Evaluation Criteria
The method was evaluated based on pixel detection. Sensitivity and specificity were used as statistical measures of the performance of the binary classification method [30–33]. Sensitivity measures the proportion of actual positives which are correctly identified as positive, i.e. the percentage of tumor pixels which are correctly identified as tumor tissue. The sensitivity expresses as follows:
| (5) |
where TP and FN are true positive and false negative, respectively. When a pixel was not detected as a tumor pixel, the detection was considered as a false negative if the pixel was indeed a tumor pixel in the manually created map. When a pixel was detected as a tumor tissue, the detection was a false positive if the pixel was not tumor tissue.
Specificity measures the proportion of negatives which are correctly identified, i.e., the percentage of healthy tissue correctly identified as not having cancer. The following equation shows the specificity calculation:
| (6) |
where TN and FP are true negative and false positive, respectively.
The FNR was defined as the number of false negative pixels divided by the total number of the tumor pixel. When a pixel was detected as tumor tissue, the detection was false positive if the pixel was not a tumor. The FPR was defined as the number of false positive pixels divided by the total number of normal tissue. In each pathological slide, the region that is definitely malignant or normal tissue was chosen. The evaluation criteria were calculated based on these regions.
3. RESULTS
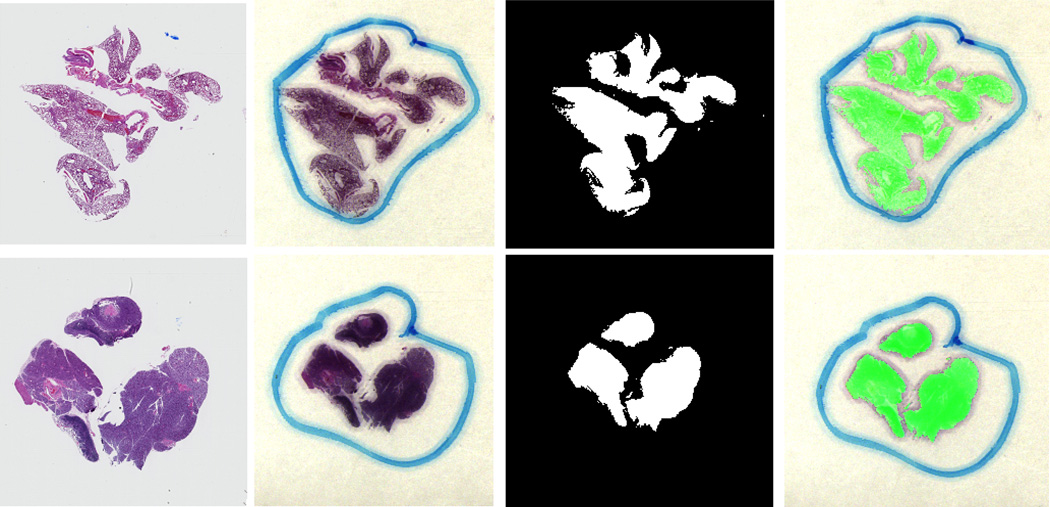
The method was evaluated by data sets of three slides of normal lung tissue, three slides of cancerous lung tissue, three slides of normal lymph node tissue, and three slides of cancerous lymph node tissue. Fig. 5 shows cancer detection results in the lung and at the lymph node in the pathological slides. The detection performance was evaluated with respect to the pathological report. The numerical results of the sensitivity and specificity are shown in Table 1.
Figure 5.
Cancer detection results in pathological slides for lung tissue (top) and lymph node tissue (bottom). From left to right, digital images by a microscope; RGB images that are made by using three bands of the hyperspectral image; the binary results, and the overlaid images where the green regions show the tumor.
Table 1.
Quantitative evaluation results of the pixel-by-pixel classification.
| Specificity (%) | Sensitivity (%) | FPR (%) | FNR (%) | |
|---|---|---|---|---|
| Lymph nodes | 98.3 | 96.2 | 1.7 | 3.8 |
| Lungs | 97.7 | 92.6 | 2.3 | 7.4 |
4. DISCUSSION AND CONCLUSION
Hyperspectral imaging and advance image analysis methods are proposed to aid the detection of cancer in pathologic slides. Hyperspectral image can help pathologist to detect the cancer without consuming long time for each slide and can be used not only for diagnosis but also for determining the tumor metastasis after biopsy. This technology expands the vision from the three RGB spectral bands to more than a hundred spectral bands. The large number of data in hyperspectral images can be processed to broaden the spectral range and can supply useful information for medical doctors. The proposed method detects a cancerous tissue pixel by pixel. The detection of one pixel as a cancerous pixel does not depend on adjacent pixels. Therefore, the cancer size or shape would not cause a problem. This method could detect the cancerous tissue in the lung and the lymph node with high specificity and sensitivity. Hyperspectral imaging can be used as a supporting tool for pathologist to evaluate a large number of slides in a short time. Histopathology is the current gold standard for diagnosis of cancer. However, even this method has problems such as invasiveness, great dependence on the judgment of pathologists, and needs time for results preparing. Moreover, the biopsy specimens can only be captured from a few points. A simple, noninvasive, and reliable technique that enables rapid detection of cancer would aid many physicians.
Figure 4.
The mean spectral signatures of the cancerous and normal tissue in mice. The horizontal axis shows different wavelengths in nanometers, and the vertical axis shows the intensity. Tumor pixels are shown in the red-squared line, and the normal tissue pixels are shown in the blue line. Left panel shows the lymph node; and the right panel shows the lung tissue.
ACKNOWLEDGEMENT
This research is supported in part by NIH grant R01CA156775 (PI: Fei), Coulter Translational Research Grant (PIs: Fei and Hu), Georgia Cancer Coalition Distinguished Clinicians and Scientists Award (PI: Fei), Emory Molecular and Translational Imaging Center (NIH P50CA128301), SPORE in Head and Neck Cancer (NIH P50CA128613), and Atlanta Clinical and Translational Science Institute (ACTSI) that is supported by PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program.
Footnotes
Copyright 2012 Society of Photo-Optical Instrumentation Engineers (SPIE). One print or electronic copy may be made for personal use only. Systematic reproduction and distribution, duplication of any material in this paper for a fee or for commercial purposes, or modification of the content of the paper are prohibited.
REFERENCES
- 1.Kong SG, Martin EM, Vo-Dinh T. Hyperspectral Fluorescence Imaging for Mouse Skin Tumor Detection. ETRI Journal. 2006;28(6):770–776. [Google Scholar]
- 2.Akbari H, Kosugi Y, Kojima K, et al. Detection and analysis of the intestinal ischemia using visible and invisible hyperspectral imaging. IEEE Trans Biomed.Eng. 2010;57(8):2011–2017. doi: 10.1109/TBME.2010.2049110. [DOI] [PubMed] [Google Scholar]
- 3.Cancio LC, Batchinsky AI, Mansfield JR, et al. Hyperspectral imaging: A new approach to the diagnosis of hemorrhagic shock. Journal of Trauma-Injury Infection and Critical Care. 2006;60(5):1087–1095. doi: 10.1097/01.ta.0000217357.10617.3d. [DOI] [PubMed] [Google Scholar]
- 4.Best SL, Thapa A, Holzer MS, et al. Assessment of Renal Oxygenation During Partial Nephrectomy Using DLP Hyperspectral Imaging. In: Douglass MR, Oden PI, editors. Book Emerging Digital Micromirror Device Based Systems and Applications Iii. Vol. 7932 2011. [Google Scholar]
- 5.Crow M, Marinakos S, Chilkoti A, et al. Simultaneous Molecular Imaging of EGFR and HER2 Using Hyperspectral Darkfield Microscopy and Immunotargeted Nanoparticles. Conference on Plasmonics in Biology and Medicine VI. 2009;7192 [Google Scholar]
- 6.Dicker DT, Lerner J, Van Belle P, et al. Differentiation of normal skin and melanoma using high resolution hyperspectral imaging. Cancer Biology & Therapy. 2006;5(8):1033–1038. doi: 10.4161/cbt.5.8.3261. [DOI] [PubMed] [Google Scholar]
- 7.Dolloff NG, Ma XH, Dicker DT, et al. Spectral imaging-based methods for quantifying autophagy and apoptosis. Cancer Biology & Therapy. 2011;12(4):349–356. doi: 10.4161/cbt.12.4.17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erives H, Targhetta NB. Implementation of a 3-D Hyperspectral Instrument for Skin Imaging Applications. IEEE Transactions on Instrumentation and Measurement. 2009;58(3):631–638. [Google Scholar]
- 9.Ishihara M, Sato M, Matsumura K, et al. Development of the hyperspectral cellular imaging system to apply to regenerative medicine. Optics in Tissue Engineering and Regenerative Medicine Iv. 2010;7566 [Google Scholar]
- 10.Jolivot R, Nugroho H, Vabres P, et al. Validation of a 2D multispectral camera: application to dermatology/cosmetology on a population covering five skin phototypes. Conference on Clinical and Biomedical Spectroscopy and Imaging II. 2011;8087 [Google Scholar]
- 11.Kashani AH, Kirkman E, Martin G, et al. Hyperspectral Computed Tomographic Imaging Spectroscopy of Vascular Oxygen Gradients in the Rabbit Retina In Vivo. Plos One. 2011;6(9) doi: 10.1371/journal.pone.0024482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellicut DC, Weiswasser JM, Arora S, et al. Emerging Technology: Hyperspectral Imaging. Perspectives in Vascular Surgery and Endovascular Therapy. 2004;16(1):53–57. [Google Scholar]
- 13.Kester RT, Gao L, Bedard N, et al. Real-Time Hyperspectral Endoscope for Early Cancer Diagnostics. Advanced Biomedical and Clinical Diagnostic Systems Viii. 2010;7555 [Google Scholar]
- 14.Kong SG, Du Z, Martin M, et al. Hyperspectral fluorescence image analysis for use in medical diagnostics. Advanced Biomedical and Clinical Diagnostic Systems III. 2005;5692 [Google Scholar]
- 15.Akbari h, Kosugi Y, Kojima K, et al. Hyperspectral Image Segmentation and its Application in Abdominal Surgery. International Journal of Functional Informatics and Personalised Medicine. 2009;2(2):201–216. [Google Scholar]
- 16.Martin ME, Wabuyele MB, Chen K, et al. Development of an advanced hyperspectral imaging (HSI) system with applications for cancer detection. Annals of Biomedical Engineering. 2006;34(6):1061–1068. doi: 10.1007/s10439-006-9121-9. [DOI] [PubMed] [Google Scholar]
- 17.Fujioka N, Morimoto Y, Arai T, et al. Discrimination between normal and malignant human gastric tissues by Fourier transform infrared spectroscopy. Cancer Detection and Prevention. 2004;28(1):32–36. doi: 10.1016/j.cdp.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Tannapfel A, Schmelzer S, Benicke M, et al. Expression of the p53 homologues p63 and p73 in multiple simultaneous gastric cancer. Journal of Pathology. 2001;195(2):163–170. doi: 10.1002/path.947. [DOI] [PubMed] [Google Scholar]
- 19.Cohenford MA, Godwin TA, Cahn F, et al. Infrared Spectroscopy of Normal and Abnormal Cervical Smears: Evaluation by Principal Component Analysis. Gynecologic Oncology. 1997;66(1):59–65. doi: 10.1006/gyno.1997.4627. [DOI] [PubMed] [Google Scholar]
- 20.Wong PTT, Goldstein SM, Grekin RC, et al. Distinct Infrared Spectroscopic Patterns Of Human Basal-Cell Carcinoma Of The Skin. Cancer Research. 1993;53(4):762–765. [PubMed] [Google Scholar]
- 21.Rigas B, Wong PTT. Human Colon Adenocarcinoma Cell-Lines Display Infrared Spectroscopic Features Of Malignant Colon Tissues. Cancer Research. 1992;52(1):84–88. [PubMed] [Google Scholar]
- 22.McIntosh LM, Jackson M, Mantsch HH, et al. Infrared spectra of basal cell carcinomas are distinct from non-tumor-bearing skin components. Journal of Investigative Dermatology. 1999;112(6):951–956. doi: 10.1046/j.1523-1747.1999.00612.x. [DOI] [PubMed] [Google Scholar]
- 23.Siddiqi AM, Li H, Faruque F, et al. Use of hyperspectral imaging to distinguish normal, precancerous, and cancerous cells. Cancer Cytopathology. 2008;114(1):13–21. doi: 10.1002/cncr.23286. [DOI] [PubMed] [Google Scholar]
- 24.Akbari H, Uto K, Kosugi Y, et al. Cancer detection using infrared hyperspectral imaging. Cancer Science. 2011;102(4):852–857. doi: 10.1111/j.1349-7006.2011.01849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XF, Schuster D, Master V, Nieh P, Fenster A, Fei BW. Automatic 3D Segmentation of Ultrasound Images Using Atlas Registration and Statistical Texture Prior. Proceedings of SPIE Medical Imaging: Visualization Image-Guided Procedures and Modeling. 2011;7964 doi: 10.1117/12.877888. 796432K-1~8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akbari H, Yang X, Halig L, Fei BW. 3D Segmentation of Prostate Ultrasound Images using Wavelet Transform. Proceedings of SPIE Medical Imaging - Image Processing. 2011;7962 doi: 10.1117/12.878072. 79622K-1~8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camps-Valls G, Bruzzone L. Kernel-based methods for hyperspectral image classification. IEEE Transactions on Geoscience and Remote Sensing. 2005;43(6):1351–1362. [Google Scholar]
- 28.Suykens JAK, Vandewalle J. Least Squares Support Vector Machine Classifiers. Neural Processing Letters. 1999;9(3):293–300. [Google Scholar]
- 29.van Gestel T, Suykens JAK, Baesens B, et al. Benchmarking least squares support vector machine classifiers. Machine Learning. 2004;54(1):5–32. [Google Scholar]
- 30.Wang H, Feyes D, Mulvihil J, et al. Multiscale fuzzy C-means image classification for multiple weighted MR images for the assessment of photodynamic therapy in mice. Proceedings of SPIE Medical Imaging: Image Processing. 2007;6512 doi: 10.1117/12.710188. 65122W-1~9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang HS, Fei BW. A modified fuzzy C-means classification method using a multiscale diffusion filtering scheme. Medical Image Analysis. 2009;13(2):193–202. doi: 10.1016/j.media.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XF, Fei BW. A multiscale and multiblock fuzzy C-means classification method for brain MR images. Medical Physics. 2011;38(6):2879–2891. doi: 10.1118/1.3584199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogie K, Wang X, Fei B, et al. New technique for real-time interface pressure analysis: getting more out of large image data sets. J Rehabil Res Dev. 2008;45(4):523–535. doi: 10.1682/JRRD.2007.03.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]