Abstract
Imbalances in GABA (γ-aminobutyric acid) homoeostasis underlie psychiatric and movement disorders. The ability of the 65 kDa isoform of GAD (glutamic acid decarboxylase), GAD65, to control synaptic GABA levels is influenced through its capacity to auto-inactivate. In contrast, the GAD67 isoform is constitutively active. Previous structural insights suggest that flexibility in the GAD65 catalytic loop drives enzyme inactivation. To test this idea, we constructed a panel of GAD65/67 chimaeras and compared the ability of these molecules to auto-inactivate. Together, our data reveal the important finding that the C-terminal domain of GAD plays a key role in controlling GAD65 auto-inactivation. In support of these findings, we determined the X-ray crystal structure of a GAD65/67 chimaera that reveals that the conformation of the catalytic loop is intimately linked to the C-terminal domain.
Keywords: auto-inactivation, catalytic loop, chimaera, C-terminal domain, glutamic acid decarboxylase, X-ray crystallography
Abbreviations: AET, 2-aminoethylisothiouronium bromide; GABA, γ-aminobutyric acid; GAD, glutamic acid decarboxylase; PLP, pyridoxal-5′-phosphate; PMP, pyridoxamine phosphate; SSA, succinic semialdehyde; TLS, translation libration screw-rotation; WT, wild-type
INTRODUCTION
GAD (glutamic acid decarboxylase) is a PLP (pyridoxal-5′-phosphate)-dependent enzyme that catalyses the α-decarboxylation of l-glutamate into the essential mammalian inhibitory neurotransmitter GABA (γ-aminobutyric acid) [1,2]. GABA is the most abundant neurotransmitter in the CNS (central nervous system) and is fundamental in processes such as movement, neurogenesis and tissue development [3–8]. Accordingly, defects in regulation of GABA homoeostasis have been implicated in neurological disorders such as epilepsy, Parkinson's disease, schizophrenia, anxiety disorders, autism, bi-polar disorder and PTSD (post-traumatic stress disorder) [9–15].
Two GAD isoforms, GAD67 and GAD65, are present in humans. These molecules share about 70% sequence identity and function synchronously to produce and regulate the levels of intracellular GABA. Together these two enzymes maintain the only physiological supply of GABA in mammals.
Despite extensive sequence similarity, the two GAD isoforms perform strikingly different biological roles. GAD67 is constitutively active in GABA production and remains bound to the co-factor PLP. In contrast, GAD65 exists predominantly in an inactive apo-form that can be rapidly activated via PLP binding. Further, in addition to producing GABA, GAD65 is also able to convert l-glutamate into SSA (succinic semialdehyde) via a side reaction, during which the co-factor is released [as PMP (pyridoxamine phosphate)] and GAD65 becomes an inactive apo-enzyme [16]. Current knowledge of the molecular mechanism of GAD65 auto-inactivation remains incomplete, hampering the development of therapeutic treatments aimed at stabilizing GAD65 activity in neurological disease states [16].
Both isoforms of GAD form an obligate dimer, with the two active sites formed at the dimer interface (Supplementary Figure S1A at http://www.bioscirep.org/bsr/033/bsr033e013add.htm). Structural studies revealed that the active site is made up of both monomers with the catalytic loop region contributed in trans from one monomer to the other (Supplementary Figure S1). In GAD65, the catalytic loop (His422–Tyr433) is mobile; this flexibility results in the transient absence of the catalytic residue Tyr425 from the active site. We proposed that, in the absence of Tyr425 from the GAD65 active site, the enzyme produces SSA (rather than GABA) through the side reaction, forming apo-GAD65 [16]. By contrast, our structural studies revealed that the same catalytic loop region in GAD67 (Cys431–Tyr442) is well ordered, and hence the side reaction and auto-inactivation is disfavoured. This observation is consistent with the fact that GAD67 catalyses the production of GABA constitutively under physiological conditions [17].
In support of these ideas, we have previously demonstrated that a GAD65 mutant (GAD65Phe283Tyr_67loop) containing the catalytic loop of GAD67, together with the mutation of a residue in the catalytic site (Phe283Tyr), had a significantly reduced ability to auto-inactivate. Consistent with these findings, the reverse mutation in GAD67 (i.e. GAD67Tyr292Phe_65loop) resulted in an enzyme that more closely resembled GAD65. However, we were unable to create a GAD65 variant that fully resembled GAD67 (and vice versa) [16].
In this study, we build on our initial observations and comprehensively investigate the structural basis for GAD65 auto-inactivation. We have created a panel of GAD65–67 chimaeras that covers three different regions of interest (Supplementary Figure 2A at http://www.bioscirep.org/bsr/033/bsr033e013add.htm): the catalytic loop (residues 422–435 and 431–444 in GAD65 and 67, respectively), the base of the catalytic site (residues Phe283 and Tyr292 in GAD65 and 67, respectively) and the C-terminal domain (residues 454–585 and 463–594 in GAD65 and 67, respectively). We have developed and applied a new NMR-based assay to compare the level of auto-inactivation by studying the residual activity across all mutant and wild type enzymes. Strikingly, our data reveal that the C-terminal domain of GAD plays a key role in controlling GAD65 auto-inactivation. We have also crystallized and determined the structure of the GAD6765loop mutant in order to gain further insights into the residue composition and mobility of the catalytic loop. In one of the monomers, we are able to ascertain the conformation of the GAD65 loop in the context of the active site. Analysis of the second monomer reveals that conformational change in the catalytic loop is intimately linked to the conformation of the C-terminal domain.
EXPERIMENTAL
GAD wild-type and GAD catalytic loop-swap mutants were generated as described [16]; additional mutants were made as follows. Coding sequences of human GAD65, GAD67 and GAD catalytic loop-swap enzymes (GAD67Tyr292Phe_65loop and GAD65Phe283Tyr_67loop) were cloned into pCR® blunt and used for mutation studies. GAD65Phe283Tyr_67loop_Leu435Val and GAD67Tyr292Phe_65loop_Val444Leu were made by PCR site-directed mutagenesis using the specific GAD loop swap/pCR® blunt clones and the following primers, 5′cagtatgacgtctcctatgacactggagacaaggcc and 3′gtcataggagacgtcatactgtttatctggctgaaag, 5′cattatgatctgtcctacgacaccggggacaag and 3′gtcgtaggacagatcataatgcttgtcttgctg, respectively. The C-terminal swap mutants were made by taking advantage of the DraI restriction enzyme site in GAD65, ten amino acids on the N-terminal side of the C-terminal domain boundary. A DraI restriction site was incorporated into GAD67/blunt and GAD67Tyr292Phe_65loop_Val444Leu/blunt constructs in the equivalent location to GAD65 using site-directed mutagenesis with the following primers: 5′gatatctttaaattctggctgatgtggaaagc and 3′cagccagaatttaaagatatccacgtcgcggcc. The resulting GAD67-DraI plasmids were subcloned into pAS-1N to generate the C-terminal swap mutants.
GAD enzymes were expressed in Saccharomyces cerevisiae and purified as previously described [16], using immobilized metal-affinity followed by size-exclusion chromatography methods; proteins were concentrated to 800 μg/ml and stored at −80°C in PBS (136 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2HPO4 pH 7.2), 1.0 mM 2-mercaptoethanol, 1.0 mM AET (2-aminoethylisothiouronium bromide), 20 μM PLP and 66% (v/v) glycerol. All experiments were performed within 2 weeks of purification.
Auto-inactivation of GAD occurs upon incubation with glutamate; in the absence of free PLP in the reaction, this process generates inactive apo-GAD. We measured the residual GAD activity by NMR, on samples pre-incubated with glutamate for 10 min and compared that with samples without the pre-incubation (0 min). The results are presented as percentage residual activities [see eqn (1)], as detailed in Supplementary Figure S3 (at http://www.bioscirep.org/bsr/033/bsr033e013add.htm). Briefly, auto-inactivation of GAD was performed by first incubating GAD samples with 20 μM PLP for 5 min at 37°C, excess PLP was removed by applying the GAD sample to a 5 ml HiTrap™ desalting column (GE Healthcare) in 100 mM K/NaHPO4 (pH 7.2), 1.0 mM 2-mercaptoethanol, 1.0 mM AET and 5% (v/v) glycerol. GAD was pre-warmed at 37°C for 5 min and then auto-inactivated by incubating with 10 mM glutamic acid, at 275 μg/ml and 37°C with shaking at 500 rev./min, for either 0 or 10 min. Auto-inactivation was stopped by applying the sample to a 5 ml HiTrap™ desalting column (GE Healthcare) in the above-mentioned buffer. After all desalting column experiments the protein concentration was determined by Bradford's assay using Bradford's reagent (BioRad) and BSA protein standards; concentrations were measured on a FluoStar Optima (BMG Labtech) at 595 nm. Importantly, the desalting column step repurified GAD from any carryover reagents from the auto-inactivation reactions, including glutamate, PMP and GABA, which can interfere with the metabolic activity of the enzymes or subsequent calculation of the final yield of GABA. GAD activity was determined by incubating pre-warmed (5 min at 37°C) 0 and 10 min GAD samples (50 μg/ml) with 4.0 mM glutamic acid in a final volume of 350 μl at 37°C with shaking at 500 rev./min, GAD activity was stopped by heating at 95°C for 5 min, and samples were made up to a final volume of 700 μl with milliQ water.
The measurement of glutamate turnover was carried out using NMR spectroscopy. Studies were performed on a Bruker 400 MHz spectrometer. The NMR samples were produced by adding 2H2O (70 μl) to each assay sample (600 μl). The samples were analysed using a water pre-saturation technique (128 scans) and interpreted using Topspin 2.1. Enzyme turnover was determined by measuring the amount of glutamic acid as a percentage of the total calculated from the integrations of the total amount of glutamic acid (2.3 p.p.m., triplet) and GABA (2.2 p.p.m., triplet). All experimental conditions were performed in quadruplicate unless otherwise stated.
| (1) |
Analysed data was plotted using Prism® GraphPad, statistical analysis was performed in Prism using an unpaired, two-tailed Student's t test. All Figures were created using Adobe® Illustrator.
Crystallization experiments used freshly prepared GAD6765loop concentrated to 10 mg/ml followed by addition of a 1/10 (v/v) solution of 4 mM chelidonic acid (GAD inhibitor) at room temperature. Crystals of GAD6765loop were obtained in a number of conditions, but the largest crystals were obtained using the hanging drop method in 25% (w/v) PEG 3350 and 100 mM Bis–Tris (pH 6.0). Crystals were grown at 22°C, appeared overnight and reached a size of approximately 0.5×0.3×0.2 mm in 2 days. Prior to data collection, crystals were transferred to a cryoprotectant solution containing 25% (w/v) of PEG3350, 100 mM Bis–Tris (pH 6.0), 10% (v/v) glycerol and 15% (v/v) MPD (2-methyl-2,4-pentanediol) for 5 s, and flash-cooled in liquid nitrogen. X-ray diffraction data sets were collected at the Australian synchrotron high-throughput macromolecular crystallography MX1 beamline. The crystals diffracted to 2.1 Å (1 Å=0.1 nM) resolution and the data were processed using the programs MOSFLM and SCALA [18]. The GAD6765loop crystals belong to space group P21 and have unit cell dimensions of a=86.00 Å, b=64.08 Å, c=108.14 Å, α=γ=90, β=108.14°, consistent with two molecules per asymmetric unit. Subsequent structural analysis was done using the CCP4i interface [19] to the CCP4 suite [20] and autoBuster [21]. Five percent of the dataset was flagged for calculation of the Rfree (free R factor) with neither a sigma, nor a low-resolution cut-off applied to the data. A summary of statistics is provided in Table 1.
Table 1. Data collection and refinement.
Values in parentheses are for highest-resolution shell. See Table 2 for a detailed description of GAD6765loop. R.M.S.D., root mean square deviation.
| Parameter | GAD6765loop |
|---|---|
| Data collection | |
| Space group | P21 |
| No. molecule/symmetric unit | 2 |
| Cell dimensions | |
| a, b, c (Å) α, β, γ (°) | 86.00, 64.08, 102.64 90.00, 108.14, 90.00 |
| Resolution (Å) | 54.8 (2.1) |
| Rmerge | 3.19 (16.3) |
| I / σI | 14.9 (4.0) |
| Completeness | 99.9 (99.9) |
| Redundancy | 3.7 (2.7) |
| Refinement | |
| Resolution (Å) | 2.1 |
| Numbe of reflections (work/free) | 62194/3149 |
| Rwork/Rfree | 21.7/25.3 |
| Number of atoms | |
| Protein | 7854 |
| Ligand | 36 |
| Water | 467 |
| R.M.S.D | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.14 |
| MolProbity analysis | |
| Ramachandran outliers | 0.10% |
| Ramachandran favoured | 97.36% |
| MolProbity score | 1.64 (95th percentile) |
The structure of the GAD6765loop mutant was solved by molecular replacement method using PHASER [22]. The search model was constructed using the crystal structure of wild-type GAD67 (2OKJ) trimmed to remove residues 228–242 from the catalytic loop. Structure refinement and model building proceeded using one dimer GAD6765loop in the asymmetric unit. Maximum-likelihood refinement using REFMAC [23], incorporating TLS (translation, libration screw-rotation) displacement refinement was carried out using a bulk solvent correction (Babinet model with mask) and autoBUSTER with TLS and the weighting fixed at 0.006 [21]. TLS groups for REFMAC refinement were made using the TLSMD server [24]. Throughout the refinement, tight NCS restraints were imposed on most residues, except in the region of high structural divergence between the monomers. All model building and structure validation were performed using COOT and MolProbity [25,26].
PyMOL [27] was used to produce structure figures. Structures were superimposed using the program MUSTANG [28].
The quality of the structure has been validated using MolProbity [29] (Table 1), and the co-ordinates for the GAD6765loop protein have been deposited in the Protein Data Bank with accession code 3VP6.
RESULTS
Design of GAD65–GAD67 chimaeras
The catalytic loop of GAD packs against the C-terminal domain of the enzyme. Analysis of structures of GAD65 and GAD67 reveals that the majority of interactions between the catalytic domain and the C-terminal domain are conserved in both isoforms (Supplementary Table S1 at http://www.bioscirep.org/bsr/033/bsr033e013add.htm). However, despite this, the C-terminal domain of GAD65 has shifted up to 4 Å (Supplementary Figure S1B) with respect to the corresponding domain in GAD67. Analysis of B-factors as well as electron density further reveals that the C-terminal domain of GAD65 is also substantially more mobile and contains a short region (residues 518–520) that is not visible in electron density (Supplementary Figure S1B). We therefore reasoned that mobility in both the catalytic loop and the C-terminal domain may together contribute to regulating the enzyme auto-inactivation. To test this idea, we generated chimaeric molecules in which the C-terminal domain was ‘swapped’ between the two isoforms. We investigated this change in the context of catalytic loop swaps, as well as an additional catalytic loop residue, as detailed below. All catalytic loop chimaeras had the additional mutation of the catalytic site residue Phe283Cys (GAD65) or Tyr292Phe (GAD67), previously shown to be important in loop mobility/stability [16].
In our previously published work, our GAD catalytic loop swap variants failed to take account of the C-terminal end of the catalytic loop (Asp434 and Leu435 in GAD65 and Asp443 and Val444 in GAD67). This may be important, since the crystal structure of GAD67 reveals that Val444 packs tightly against the catalytic loop (Supplementary Figure S1C) and we reasoned that substitution of this residue with a larger leucine residue may significantly contribute to destabilization of the overall catalytic loop structure. In order to test these hypotheses, we constructed GAD chimaeras that interchanged these important structural motifs from one isoform to the other. A full table of mutations investigated in this study is presented in Table 2 and Supplementary Figures S2(A) and S2(B).
Table 2. Residual activity and auto-inactivation of GAD chimaeras.
Residual activity±S.D. was calculated by dividing the glutamate turnover of the 10 min auto-inactivated sample by the untreated sample. All assays were performed at least four times. Auto-activation was calculated as 100%-% residual activity. Increase or decrease was calcualted as the fold difference in percentage auto-inactivation compared with respective WT (wild-type) calculated from the percentage residual activity.
| GAD chimaera | Residual activity (%) | Auto-inactivation (%) | Fold increase (↑) or decrease (↓) in auto-inactivation§ |
|---|---|---|---|
| GAD67WT | 101.8±2.4 | 0±2.4 | N/A |
| GAD67Tyr292Phe_65loop* | 84.3±4.8 | 15.7±4.8 | ↑ 1.21 |
| GAD67Tyr292Phe_65loop_Val444Leu | 51.1±6.2 | 48.9±6.2 | ↑ 1.99 |
| GAD6765CT† | 80.1±4.3 | 19.9±4.3 | ↑ 1.27 |
| GAD67Tyr292Phe_65loop_Val444Leu_65CT | 41.9±1.9 | 58.1±1.9 | ↑ 2.43 |
| GAD65WT | 45.2±3.6 | 54.8±3.6 | NA |
| GAD65Phe283Tyr_67loop‡ | 79.5±5.5 | 20.5±5.5 | ↓ 1.76 |
| GAD65Phe283Tyr_67loop_Leu435Val | 76.9±2.7 | 23.1±2.7 | ↓ 1.70 |
| GAD6567CT§ | 95.7±10.3 | 4.3±10.3 | ↓ 2.12 |
| GAD65Phe283Tyr_67loop_Leu435Val_67CT | 99.8±8.0 | 0.2±8.0 | ↓ 2.21 |
*65loop=GAD65 residues 422–433.
†65CT=GAD65 residues 454–585.
‡67loop=GAD67 residues 431–442.
§67CT=GAD67 residues 463–594.
Comparison of the residual activity of GAD chimaeras upon auto-inactivation
The measurement of GAD auto-inactivation is outlined in Supplementary Figure S3. We measured the enzyme activity of an untreated sample (t=0) and an auto-inactivated sample (t=10 min), and residual activity was calculated according to eqn (1). Conventionally, the residual activity is monitored through an end point radiolabelled 14CO2 trapping assay [16,17]. The auto-inactivation assay is of insufficient sensitivity to monitor subtle changes between populations of auto-inactivated GAD samples and the standard errors within an experiment are very high. Further, at the point where residual GAD activity of a sample is measured, the quantity of glutamic acid carried over from the auto-inactivation reaction is unknown, as is the amount of GABA generated. Both glutamic acid and GABA can impact upon the rate of enzyme activity, either positively or negatively, respectively [30]. We therefore re-engineered the endpoint assay by repurifying the auto-inactivated GAD samples with a desalting column, removing all substrates and products, and then performing the GAD assay in the presence of a known concentration of glutamate. For the current experimental set up (Supplementary. Figure S3), glutamate turnover is measured by NMR spectroscopy instead of radiolabelled 14CO2. This assay gave rise to highly reproducible results and, as a consequence, permits reliable monitoring of GAD auto-inactivation of the mutants.
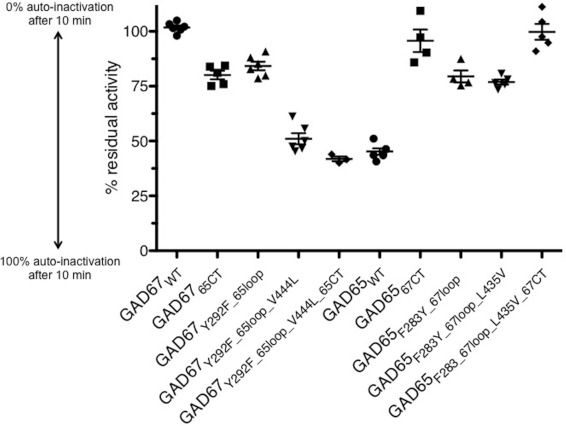
Using the NMR assay described above, GAD67WT (wild-type) was found to be fully active after pre-incubation with glutamate (100% residual activity), whereas GAD65WT retains 45% residual activity (Table 2). Among the GAD65 mutants studied, our data show that replacing the GAD65 catalytic loop with that of GAD67 results in a 1.76-fold decrease in auto-inactivation (79.5% residual activity), while mutation of the flanking residue (Leu435Val) has little additional effect (1.70-fold decrease in auto-inactivation, 76.9% residual activity) (Figure 1 and Table 2). However, both constructs are clearly auto-inactivated upon incubation with glutamate, and are significantly different to GAD67WT (Table 3). In contrast, when the C-terminal domain (alone) of GAD65 was replaced by that of GAD67, a 2.12-fold reduction in auto-inactivation (95.7% residual activity) resulted, which is indistinguishable from GAD67 (Table 3). No further increase in auto-inactivation was seen in the construct combining the catalytic loop with the C-terminal domain swap (GAD65Phe283Tyr_67loop_Leu435Val_67CT, 99.8% residual activity).
Figure 1. Residual activity of wild-type GAD compared with GAD chimaeras upon auto-inactivation.
Comparison of the auto-inactivation of GAD wild-type (●) to GAD loop-swap chimaeras with (▼) and without (▲) V444L/L435V, C-terminal domain swap chimaeras (■) and combination C-terminal domain and catalytic loop-swap chimaeras (◆). Y-axis shows The percentage of residual activity (% auto-inactivation in blue) is shown on the y-axis. Values are means±S.E.M.; significance of results can be found in Table 3. See Table 2 for a detailed description of the chimaeras.
Table 3. Significance of change (P values) in auto-inactivation of each mutation compared with all data.
P values was calculated using an unpaired two-tailed Student's t test. All values were calculated using PRISM® GraphPad. See Table 2 for a detailed description of the chimaeras.
| GAD67WT | GAD6765CT | GAD67Tyr292Phe_ 65loop | GAD67Tyr292Phe_65loop Val444Leu | GAD67Tyr292Phe_65loop_ Val444Leu_65CT | GAD65WT | GAD6567CT | GAD65Phe283Tyr_ 67loop | GAD65Phe283Tyr_ 67loop_Leu435Val | |
|---|---|---|---|---|---|---|---|---|---|
| GAD6765CT | <0.0001 | ||||||||
| GAD67Tyr292Phe_65loop | <0.0001 | NO | |||||||
| GAD67Tyr292Phe_65loop_Val444Leu | <0.0001 | <0.0001 | <0.0001 | ||||||
| GAD67Tyr292Phe_65loop_Val444Leu_65CT | <0.0001 | <0.0001 | <0.0001 | 0.0442 | |||||
| GAD65WT | <0.0001 | <0.0001 | <0.0001 | NO | NO | ||||
| GAD6567CT | NO | 0.017 | 0.0416 | <0.0001 | 0.0003 | <0.0001 | |||
| GAD65Phe283Tyr_67loop | <0.0001 | NO | NO | <0.0001 | <0.0001 | <0.0001 | 0.0317 | ||
| GAD65Phe283Tyr_ 67loop_Leu435Val | <0.0001 | NO | 0.0136 | <0.0001 | <0.0001 | <0.0001 | 0.0052 | NO | |
| GAD65Phe283Tyr_ 67loop_Leu435Val_ 67CT | NO | 0.0013 | 0.0031 | <0.0001 | <0.0001 | <0.0001 | NO | 0.0036 | 0.0003 |
The parallel experiments on GAD67 mutants reveal a different picture. Swapping the catalytic loop alone has a small but significant effect on auto-inactivation (1.21-fold increase, 84.3% residual activity). Similarly, swapping the C-terminal region of GAD65 into GAD67 had a modest influence on auto-inactivation (1.27-fold increase, 81% residual activity). Interestingly, however, including the flanking residue mutation Val444Leu with the catalytic loop swap results in an enzyme that closely resembles GAD65 (1.99-fold increase in auto-inactivation, 51.1% residual activity). Combining all three mutations (GAD67Tyr292Phe_65loop_Val444Leu _65CT) results in an enzyme indistinguishable from GAD65WT (Table 3; 41.9% residual activity).
X-ray crystal structure of GAD6765loop
We wanted to understand the structural basis for our observation that the catalytic loop and the C-terminal domain can both influence enzyme auto-inactivation. Crystallization of GAD mutants is challenging, as they do not readily yield diffraction quality crystals. We established trials on all of the different chimaeric proteins detailed in this study as well as another variant previously reported – the straight catalytic loop swap of GAD65 into GAD67 (GAD6765loop). Despite these setbacks, we were able to crystallize and determine the 2.1 Å structure of GAD6765loop; these structural data permitted a greater understanding of the influence of both the catalytic loop and the C-terminal domain on GAD function.
As with GAD67WT, the GAD6765loop enzyme forms an obligate dimeric assembly, with two monomers in the asymmetric unit [16]. The active sites of both monomers, A and B, reveal electron density consistent with the active site lysine (Lys405) covalently bound to the co-factor PLP (as an internal aldimine). These data confirm that the enzyme is in an active state. The higher resolution of the data also revealed electron density consistent with the presence of the GAD inhibitor chelidonic acid in the active site.
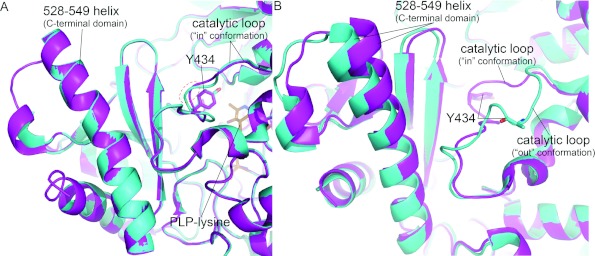
Comparison of the structure of GAD6765loop with the published GAD67WT structure (PDB ID 2OKJ) revealed certain interesting and important findings. In the active site of monomer B, the catalytic loop region contributed in trans from monomer A is structurally similar to the conformation of the catalytic loop in GAD67WT (Figure 2A). The catalytic tyrosine, Tyr434, is in the same position in both GAD structures. The minor differences in overall conformation of the GAD65WT catalytic loop can largely be attributed to the presence of a serine instead of a glycine at position 433. Gly433 in GAD67WT mediates a tight turn and is in a +/+ conformation (Figure 2A). Therefore the overall conformation of the two loops is similar. Further, analysis of the remainder of molecule B reveals no other major conformational changes – in particular the position of the C-terminal domain is the same.
Figure 2. Two conformations of the GAD6765loop catalytic loop.
(A) A cartoon representation of GAD6765loop (cyan, ‘in conformation’) and GAD67 (magenta) active site from monomer B, with the catalytic loop supplied in trans by monomer A. Although the catalytic loops have slightly different conformations, the catalytic tyrosine (Tyr434) is in the same position; a red dashed circle shows the region of most divergence, centred on GAD6765loop residue Ser433. In addition, the C-terminal domain α-helix (528–549) and PLP-lysine (Lys405, orange sticks) are shown. (B) A cartoon representation of GAD6765loop (cyan – out conformation) and GAD67 (magenta – in conformation) active site from monomer A, with the catalytic loop supplied in trans by monomer B. A large divergence between the two catalytic loops can be seen, along with the movement in the GAD6765loop α-helix 528–549. The side chain of GAD6765loop Tyr434 was not visible in the electron density and thus has been omitted from the structure; each catalytic tyrosine has been labelled (sticks).
Analysis of the GAD6765loop monomer A active site reveals a different picture. In active site A the first eight residues (431–439) of the catalytic loop (contributed by monomer B) have swung out of the active site. The effect of this change is that the catalytic tyrosine Tyr434 moves 13 Å out of the catalytic site (Figure 2B). Interestingly, the catalytic loop of GAD6765loop pivots out at positions His431 (Cys in GAD67) and Gln438 (Pro in GAD67), two of only four non-conserved residues in the catalytic loop. Most importantly, however, the C-terminal domain has also moved away from the catalytic site, presumably as a result of the conformational change in the catalytic loop. In particular, the N-terminal end of the 528–549 helix has shifted (by 3 Å at the top), together with the surrounding region. Note also, this region is substantially disordered in GAD65WT structure. Thus, it appears that proper packing of the catalytic loop into the GAD active site can influence the structure of the C-terminal domain. Conversely, these data, by implication, start to explain why the C-terminal domain can also impact upon the activity of the enzyme.
DISCUSSION
In the present study, our objective was to investigate the structural motifs critical for auto-inactivation via mutational studies on GAD65 hybrid enzymes containing GAD67 motifs (and vice versa). In order to achieve this, we have established a reliable assay for monitoring GAD auto-inactivation. This approach is highly reproducible, as evidenced by the low standard error in the results (Figure 2).
Our current observations challenge our original hypothesis [16] that residues within the catalytic loop alone influence overall enzyme function and auto-inactivation. We have shown that the C-terminal domain also plays a critical role in the auto-inactivation process.
We proposed originally that the higher residual activity upon auto-inactivation of GAD65 is due to the mobility of the catalytic loop [16]. Our current data suggest that the C-terminal domain also plays an important role in the rate of auto-inactivation, possibly via stabilizing the catalytic loop, such that substitution of the GAD67 C-terminal region alone into GAD65 reduces auto-inactivation substantially, thus increasing residual activity (from 45.2 to 95.7% residual activity). Interestingly, these data also suggest that the GAD65 C-terminal region alone in GAD6765CT did not have an equivalent impact on auto-inactivation, suggesting other structural motifs in GAD65 play a crucial role in the auto-inactivation process.
Indeed in GAD67, we showed that both GAD65 catalytic loop (431–441 and in particular Val444Leu) and the C-terminal region were required to produce a GAD65-like molecule. Simply swapping either the catalytic loop or the C-terminal domain (GAD6765CT) alone had only a modest influence on auto-inactivation. This observation further supports our hypothesis that the C-terminal domain of GAD plays an important role in the stability of the catalytic loop, which is reflected by the residual activity upon auto-inactivation.
Our structure of the GAD6765loop is consistent with such observations. Our data show that the GAD67 C-terminal domain is capable of stabilizing the GAD65 loop in a ‘GAD67-like’ conformation. The structure further reveals, however, that even in the context of the GAD67 C-terminal region, the mutations within the GAD65 catalytic loop sequence are sufficient to maintain conformational change whereby the catalytic loop can flip out of the active site. Interestingly, these rearrangements also impacted upon the conformation of the C-terminal domain, particularly around the 528–549 helix. Together, these data reveal that the structure and stability of the catalytic loop and the C-terminal region are intimately linked to GAD auto-inactivation.
AUTHOR CONTRIBUTION
Christopher Langendorf designed the study and the experiments, carried out most of the experiments and analysis and co-wrote the paper. Kellie Tuck and Trevor Key carried out the NMR experiments and analysis and provided critical review of the paper before submission. Gustavo Fenalti carried out X-ray crystallographic experiments and co-wrote the paper. Robert Pike carried out analysis and critical review of paper before submission. Carlos Rosado and Anders Wong carried out the experiments. Ashley Buckle carried out X-ray crystallographic analysis and provided critical review of the paper before submission. Ruby Law and James Whisstock designed and co-ordinated the study and co-wrote the paper. All authors read and approved the paper.
ACKNOWLEDGEMENTS
We thank the Monash NMR, Monash Crystalmation high-throughput crystallization platform and protein production platforms for technical assistance. We also thank the Australian Synchrotron MX1 beamline staff for technical assistance, especially Dr Tom Caradoc-Davies.
FUNDING
This study was supported by the National Health and Medical Research Council (NHMRC) of Australia [grant number NHMRC 436740], as well as the Australian Research Council (ARC). C.G.L. is a NHMRC Postgraduate Research Scholar, and J.C.W. is an ARC Federation Fellow and honorary NHMRC principal research fellow.
References
- 1.Capitani G., De Biase D., Aurizi C., Gut H., Bossa F., Grütter M. G. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 2003;22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gut H., Pennacchietti E., John R. A., Bossa F., Capitani G., De Biase D., Grutter M. G. Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. EMBO J. 2006;25:2643–2651. doi: 10.1038/sj.emboj.7601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asada H., Kawamura Y., Maruyama K., Kume H., Ding R-G., Kanbara N., Kuzume H., Sanbo M., Yagi T., Obata K. Cleft palate and decreased brain γ-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6496–6499. doi: 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erlander M. G., Tobin A. J. The structural and functional heterogeneity of glutamic acid decarboxylase: a review. Neurochem. Res. 1991;16:215–226. doi: 10.1007/BF00966084. [DOI] [PubMed] [Google Scholar]
- 5.Fagiolini M., Fritschy J-M., Löw K., Möhler H., Rudolph U., Hensch T. K. Specific GABAA circuits for visual cortical plasticity. Science. 2004;303:1681–1683. doi: 10.1126/science.1091032. [DOI] [PubMed] [Google Scholar]
- 6.Ge S., Goh E. L. K., Sailor K. A., Kitabatake Y., Ming G-L., Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439:589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kash S., Johnston R. S., Tecott L. H., Noebels J. L., Mayfield R. D., Hanahan D., Baekkeskov S. Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14060–14065. doi: 10.1073/pnas.94.25.14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatsu Y., Tyndale R. F., DeLorey T. M., Durham-Pierre D., Gardner J. M., McDanel H. J., Nguyen Q., Wagstaff J., Lalando M., Sikela J. M., et al. A cluster of three GABAA receptor subunit genes is deleted in a neurological mutant of the mouse p locus. Nature. 1993;364:448–450. doi: 10.1038/364448a0. [DOI] [PubMed] [Google Scholar]
- 9.Akbarian S., Huang H-S. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res. Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Kaplitt M. G., Feigin A., Tang C., Fitzsimons H. L., Mattis P., Lawlor P. A., Bland R. J., Young D., Strybing K., Eidelberg D., During M. J. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 11.Lanoue A. C., Dumitriu A., Myers R. H., Soghomonian J-J. Decreased glutamic acid decarboxylase mRNA expression in prefrontal cortex in Parkinson's disease. Exp. Neurol. 2010;226:207–217. doi: 10.1016/j.expneurol.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry T. L., Buchanan J., Kish S. J., Hansen S. γ-Aminobutyric acid deficiency in brain of schizophrenic. Lancet. 1979;313:237–239. doi: 10.1016/s0140-6736(79)90767-0. [DOI] [PubMed] [Google Scholar]
- 13.Thompson M., Weickert C. S., Wyatt E., Webster M. J. Decreased glutamic acid decarboxylase67 mRNA expression in multiple brain areas of patients with schizophrenia and mood disorders. J. Psychiatr. Res. 2009;43:970–977. doi: 10.1016/j.jpsychires.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Treiman D. M. GABAergic Mechanisms in Epilepsy. Epilepsia. 2001;42:8–12. doi: 10.1046/j.1528-1157.2001.042suppl.3008.x. [DOI] [PubMed] [Google Scholar]
- 15.Vaiva G., Thomas P., Ducrocq F., Fontaine M., Boss V., Devos P., Rascle C., Cottencin O., Brunet A., Laffrargue P., Goudemand M. Low posttrauma GABA plasma levels as a predictive factor in the development of acute posttraumatic stress disorder. Biol. Psychiatry. 2004;55:250–254. doi: 10.1016/j.biopsych.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Fenalti G., Law R. H. P., Buckle A. M., Langendorf C., Tuck K., Rosado C. J., Faux N. G., Mahmood K., Hampe C. S., Banga J. P., et al. GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop. Nat. Struct. Mol. Biol. 2007;14:280–286. doi: 10.1038/nsmb1228. [DOI] [PubMed] [Google Scholar]
- 17.Battaglioli G., Liu H., Martin D. L. Kinetic differences between the isoforms of glutamate decarboxylase: implications for the regulation of GABA synthesis. J. Neurochem. 2003;86:879–887. doi: 10.1046/j.1471-4159.2003.01910.x. [DOI] [PubMed] [Google Scholar]
- 18.Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 19.Potterton E., Briggs P., Turkenburg M., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. BUSTER. 2.9 Ed. Cambridge, U.K.: Global Phasing Ltd; 2011. [Google Scholar]
- 22.McCoy A. J., Storoni L. C., Read R. J. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:1220–1228. doi: 10.1107/S0907444904009990. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Painter J., Merritt E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLano W. L. The PyMOL User's Manual (2002) DeLano Scientific. San Carlos, CA: 2002. [Google Scholar]
- 28.Konagurthu A. S., Whisstock J. C., Stuckey P. J., Lesk A. M. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 29.Chen V. B., Arendall W. B., III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin D. L., Rimvall K. Regulation of γ-aminobutyric acid synthesis in the brain. J. Neurochem. 1993;60:395–407. doi: 10.1111/j.1471-4159.1993.tb03165.x. [DOI] [PubMed] [Google Scholar]