Abstract
High-throughput multiplexed proteomics of small-volume biospecimens will generate new opportunities in theranostics. Achieving parallel top-down and bottom-up mass spectrometry analyses of target proteins using a unified apparatus will improve proteome characterization. We have developed a novel silicon-based microfluidic device, multinozzle emitter array chip (MEA chip), as a new platform for small-volume proteomics using liquid chromatography-nanoelectrospray ionization mass spectrometry (LC-nanoESI-MS). We demonstrate parallel, on-chip, and on-line LC-MS analysis of hemoglobin and its tryptic digests directly from microliters of blood, achieving a detection limit of less than 5 red blood cells. Our MEA chip will enable clinical proteomics of small-volume samples.
Given that proteome reflects the physiological and pathological states of a patient, proteomics is a powerful tool for early diagnostics of diseases and monitoring of therapeutic responses. The majority of current protein assays in clinical settings are based on ELISA immunoassays, which require high-quality antibodies and are hard to achieve high multiplexing (>10) due to the cross-reactivity of antibodies. Mass spectrometry (MS) measures the mass-to-charge ratio of charged species, and has become an enabling technology for proteomics 1, 2. Aside from de novo identification of target proteins, MS has advantages over ELISA for detecting protein mutations, modification, truncations, and adductations etc. Once hyphenated with the LC, LC-MS enables separation, identification, characterization, and quantitation of complex mixtures of proteins and peptides. However, the penetration of MS into the in vitro diagnostics market particularly for clinical proteomics has remained low 3–5. Key challenges remain for the MS-based platform to achieve robustness, sensitivity, and throughput comparable to those of ELISA for analysis of small-volume biospecimens such as blood and urine 6–10.
In this letter, we report the design, fabrication, and proof-of-principle applications of 24-plex MEA chips for on-chip and on-line LC-nanoESI-MS analysis of low-volume whole blood samples. The chip is built on our recent breakthroughs in developing the silicon-based microfabricated monolithic multinozzle emitters (M3 emitters) and the first-generation MEA chips for nanoelectrospray MS 11, 12. These devices collectively offer a straightforward yet novel solution to the longstanding problem of the efficient coupling between silicon microfluidic chips and ESI-MS, and pave the way for large-scale integration on microfluidic chips for MS-based proteomics. Our previous MEA chips achieved both high-sensitivity (via multinozzle emitters) and high-throughput (via multi-channel) MS on a single silicon microfluidic chip. However, proteins and peptides were separated by off-chip cartridge-based LC columns, and the fluidic interface was not robust enough for automatic control of high-throughput multiplexed proteomics. Our next-generation MEA chips directly addressed these challenges.
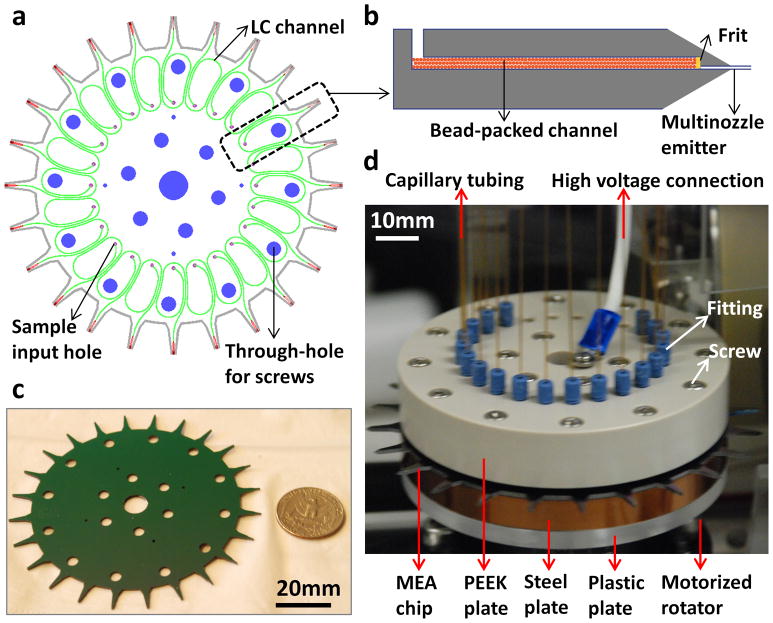
As shown in Figure 1, our MEA chip consists of 24 identical units uniformly distributed in a circular array format on a 4-inch silicon substrate (1a and 1c). Each unit (1b) contains three key functional modules: an input hole for sample injection, a LC channel (with a frit) packed with beads of desired coatings for analyte separation (or unpacked for direct infusion), and a multinozzle emitter for nanoESI-MS identification and quantitation of proteins and peptides. The LC channel has a cross section of 200 μm × 200 μm and a total length of 5 cm (with turns). The emitter has protruding nozzles with a cross-sectional area of 10 μm × 10 μm, a length of 150 μm, and a wall thickness of 0.5 μm. The emitters were constructed between the two silicon layers. The end of the emitter stem was sharpened with an angle of ~20o to enhance the electric fields at the nozzle tips 12. In addition, the chip contains through-holes designed for screws and alignment pins for connection to a fluidic manifold assembly (1d). The full assembly contains four layers including a PEEK plate, MEA chip, steel plate, and plastic plate. The top PEEK plate has 24 threaded ports for Upchurch fittings to provide leak-free connections with capillary tubings for delivering solvent gradients from a nanoflow source for LC separation (Suppl. Figure 1). High voltage for electrospray ionization is provided through a metal wire connected to the center of the MEA chip.
Figure 1.
The 24-plex MEA chip. (a) Schematic of the 2D layout. The chip consists of 24 identical units on a two-layer 4-inch silicon substrate. Each unit contains a sample input hole for sample injection (purple), a LC channel for separation (green), and a multinozzle emitter for MS (red). The through-holes (blue) are designed for filling screws during assembly. (b) Schematic of the cross-sectional view of a single unit (not to scale). Beads coated with desired chemistry are packed in the LC channel (length × width × depth: 5 cm × 200 μm × 200μm). Frits are used to retain beads inside the channel. All channels and emitters are embedded between the two silicon layers. (c) High-resolution photograph of a MEA chip with a US quarter for size comparison. (d) High-resolution photograph of a MEA chip assembly interfaced to MS. The assembly includes two clamping plates (made of PEEK and steel, respectively) to sandwich the MEA chip, a thin layer of gasket to prevent fluidic leakage, and a plastic adapter plate for attachment to a motorized rotator. The top clamping plate has 24 threaded ports for fittings to provide fluidic connections with capillary tubings. High voltage is provided through a metal wire connected to the center of the MEA chip.
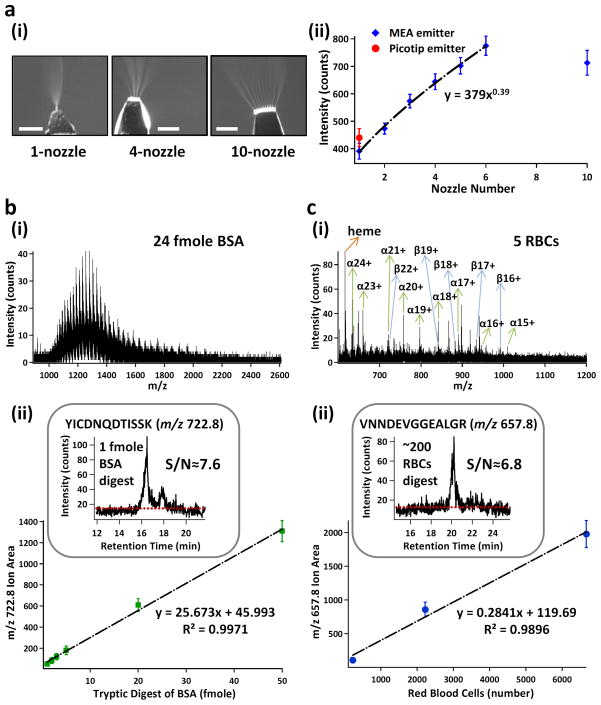
We first validated the performance of MEA emitters for MS analysis of standard proteins and peptides (Figure 2a). We demonstrated electrosprays from emitters with varying numbers of nozzles (2a(i)), and confirmed that the MS sensitivity using MEA emitters was approximately proportional to the square root of the number of the spraying nozzles (2a(ii)) 13. Importantly, multinozzle emitters achieved sensitivity significantly higher than that of the commercial Picotip capillary emitters. However, the 10-nozzle emitter was a clear outlier. This was probably due to two factors. First, the maximum voltage (5kV) provided by our current mass spectrometer might not generate electric fields high enough to produce cone-jet mode spray for all the 10 nozzles uniformly. Second, ion collection and transmission by the Z-spray sample cone of our current mass spectrometer was much less efficient for 10-nozzle emitters, because electrosprays were spread out significantly, resulting in a plume much larger in size than the MS cone inlet. Future implementation of a funnel-shaped sample cone for more efficient ion collection 14 may further increase MS sensitivity for multinozzle emitters.
Figure 2.
Proof-of-principle applications of the 24-pex MEA chip. (a) Dependence of ESI-MS sensitivity on the nozzle number of MEA emitters. (a–i) Representative images for electrospray of 1-, 4-, and 10-nozzle emitters, Scale bars are 500μm; and (a–ii) Sensitivity dependence on the nozzle numbers of MEA emitters (blue). A Picotip emitter was used as a comparison (red). MS sensitivity of MEA emitters using GFP (100 fmole/μL) illustrates a power-law relation to spraying nozzle numbers (1–6) with a power constant of 0.39. 10-nozzle ones were excluded from the curve fitting. (b) Representative mass spectrum of 24 fmole BSA (b–i) and calibration curve for quantifying tryptic digests of BSA (b–ii). Mass spectra were acquired by direct infusion of full-length BSA in 50/50 acetonitrile/H2O+0.1% formic acid. The correlation curve between the peak areas for a selected ion and the amount of BSA digests was obtained by LC-MS/MS analysis of BSA digests. The insert shows the extracted ion chromatogram of a selected tryptic peptide ion (m/z 722.8, YICDNQDTISSK) from 1 fmole BSA digest. The signal-to-noise ratio (S/N) was calculated to be ~ 7.6. (c) Representative mass spectrum of hemoglobin from ~5 RBC lysate (c–i) and calibration curve for quantifying tryptic digests of RBC lysate (c–ii). Mass spectra were acquired by direct infusion of RBC lysate in 50/50 acetonitrile/H2O+0.1% formic acid. The ions of the heme group, and multiply-charged α- and β-subunits of hemoglobin are designated. The correlation curve between the peak areas for a selected ion and the number of RBCs was obtained by LC-MS/MS analysis of trypsin digests of RBC lysate. The insert shows the extracted ion chromatogram of a selected tryptic peptide ion (m/z 657.8, VNNDEVGGEALGR) from the digest of ~200 RBC lysate. The signal-to-noise ratio (S/N) was calculated to be ~ 6.8. Error bars, s.d. (n≥6).
We next validated the performance of 24-plex MEA chips as a unified platform for parallel analysis of standard proteins and their tryptic peptides. We used one unpacked channel for direct infusion MS analysis of full-length proteins, and another channel packed with C18 beads for LC-MS analysis of their tryptic digests. We first confirmed the reproducibility of on-chip LC-MS analysis using Glu-fibrinopeptide B (GFP) standards (Suppl. Figure 2a). We then tested the separation efficiency and detection sensitivity using BSA and its tryptic digests (Figure 2b and Suppl. Figure 2b(i)). We achieved high-sensitivity detection of full-length BSA (~24 fmole) using direct infusion MS analysis, and a limit of detection (LOD, signal-to-noise ratio S/N=3) of ~400 attomole (i.e., 10−18 mole) BSA tryptic digests for the m/z 722.8 ion (YICDNQDTISSK) using LC-MS/MS. We observed a linear correlation between the LC-MS peak area and the amount of BSA tryptic digests (2b(ii)). The MS sensitivity is comparable to those of conventional cartridge-based LC-nanoESI-MS or HPLC-Chip/MS (Agilent), which are typically in the range of ~1 fmole for detecting BSA tryptic peptides.
We then evaluated the performance of our MEA chips for small-volume proteomic analysis of pooled human blood samples. We started with less than 10 μl of whole blood. By direct infusion MS of the blood lysate, we confidently detected the heme group and several multiply-charged species of hemoglobin (Hb)α and β subunits from as little as 5 red blood cells (RBCs) (containing ~ 2.3 fmole Hb) (Figure 2c(i)). This sensitivity is still lower than that of the current ELISA assay, which is typically in the range of 0.1–1.0 fmole (assuming 1–10 μl of 100 fmole/ml samples), depending on the characteristics of the antibody–antigen interaction. We achieved a LOD (S/N=3) of Hb tryptic digests from ~90 RBCs for the m/z 657.8 ion (VNNDEVGGEALGR) using LC-MS/MS (2c(ii) and Suppl. Figure 2b(ii)). We also observed a linear correlation between the LC-MS peak area and the number of lysed RBCs (2c(ii)). Therefore, one of the immediate clinical applications of our MEA chips may be low-cost and high-throughput detection of Hb variants using a pinprick of blood in newborn screening 15. With both top-down and bottom-up MS analyses of Hb using extremely-small volumes of blood on a single device, our MEA chip platform is expected to have distinct advantages in accuracy, resolution, and throughput for Hb analysis over existing methods, including matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS 16 and direct infusion electrospray MS 17.
In summary, we have developed a Si-based, scalable, and integrated MEA chip that has the potential to become an enabling platform for MS-based small-volume proteomics. We have demonstrated on-chip and on-line LC-MS analysis of microliters (e.g., pricks) of human blood. On our MEA chips, multinozzle emitters improve MS sensitivity, and on-chip and on-line multichannel LCs increase throughput. Detection of full-length proteins and their tryptic peptides on a unified platform facilitates protein identification, characterization, and quantitation. Our 24-plex MEA chip also opens up the possibility of multiplex analysis of multi-class analytes in parallel from a single sample on the same platform. For example, by packing separate LC channels using C4 and C18 beads, we can characterize the plasma proteome at both protein and peptide (endogenous or tryptic digests) levels. We can also use TiO2 beads for on-chip enrichment of phosphopeptides and phosphoproteins for phosphoproteomics. The separation efficiency of on-chip LC columns can be improved by increasing their length (e.g., to 15 cm) using more turns or larger wafers (e.g., 6 inch). In addition, implementation of sub-2μm particle columns may enable UHPLC capability and hence enhances chromatographic separation. To be comparable to immunoassays, the ESI-MS sensitivity of MEA emitters can be further improved by increasing their nozzle numbers and optimizing their geometry, configuration, and sharpening. The detection sensitivity of MEA chips can be further increased by interfacing them with the latest mass spectrometers such as Q-TOF SYNAPT® G2 MS (Waters Corp.), which has the sensitivity for peptide detection and quantitation in the low attomole range. The throughput of MEA chips can be further increased by integrating more channels (e.g., from 24 to 96), and by optimizing the multiplex LC operation to increase the MS duty cycle using the strategy of staggered parallel separations 18. If incorporated with functional modules for sample preparation such as cell lysis and on-chip protein digestion, plus automatic fluidic switching among on-chip LCs, and further interfaced with quantitative proteomics strategies such as multiple reaction monitoring (MRM)19 and “sequential window acquisition of all theoretical fragment-ion spectra” (SWATH) 20, our MEA chip will serve as a low-cost, fully-integrated, high-sensitivity, and high-throughput platform for multiplexed proteomics of small-volume biospecimens.
Supplementary Material
Acknowledgments
The authors acknowledge the financial support from the Director, Office of Science, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. P.M. and D.W. acknowledge the support under Award Number R43ES022360 from the National Institute of Environmental Health Sciences of the National Institutes of Health (to Newomics Inc.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Department of Energy. We thank UC-Berkeley Marvell Nanofabrication Laboratory for facility access.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Bodovitz S. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boja E, Hiltke T, Rivers R, Kinsinger C, Rahbar A, Mesri M, Rodriguez H. J Proteome Res. 2010;10:66–84. doi: 10.1021/pr100532g. [DOI] [PubMed] [Google Scholar]
- 4.Regnier FE, Skates SJ, Mesri M, Rodriguez H, Tezak Z, Kondratovich MV, Alterman MA, Levin JD, Roscoe D, Reilly E, Callaghan J, Kelm K, Brown D, Philip R, Carr SA, Liebler DC, Fisher SJ, Tempst P, Hiltke T, Kessler LG, Kinsinger CR, Ransohoff DF, Mansfield E, Anderson NL. Clin Chem. 2010;56:165–171. doi: 10.1373/clinchem.2009.140087. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez H, Tezak Z, Mesri M, Carr SA, Liebler DC, Fisher SJ, Tempst P, Hiltke T, Kessler LG, Kinsinger CR, Philip R, Ransohoff DF, Skates SJ, Regnier FE, Anderson NL, Mansfield E. Clin Chem. 2010;56:237–243. [Google Scholar]
- 6.Fan R, Vermesh O, Srivastava A, Yen BK, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liotta LA, Petricoin E. Cancer Cell. 2011;20:279–280. doi: 10.1016/j.ccr.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, Wong CH, Zhang Q, Chin A, Park KS, Goodman G, Gazdar AF, Sage J, Dinulescu DM, Kucherlapati R, Depinho RA, Kemp CJ, Varmus HE, Hanash SM. Cancer Cell. 2011;20:289–299. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. J Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Faca V, Hanash S. J Proteome Res. 2011;10:46–50. doi: 10.1021/pr101052y. [DOI] [PubMed] [Google Scholar]
- 11.Kim W, Guo M, Yang P, Wang D. Anal Chem. 2007;79:3703–3707. doi: 10.1021/ac070010j. [DOI] [PubMed] [Google Scholar]
- 12.Mao P, Wang HT, Yang P, Wang D. Anal Chem. 2011;83:6082–6089. doi: 10.1021/ac2011813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang K, Lin Y, Matson DW, Kim T, Smith RD. Anal Chem. 2001;73:1658–1663. doi: 10.1021/ac001191r. [DOI] [PubMed] [Google Scholar]
- 14.Kelly RT, Page JS, Zhao R, Qian WJ, Mottaz HM, Tang K, Smith RD. Anal Chem. 2008;80:143–149. doi: 10.1021/ac701647s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinert P, Schmid M, Zurbriggen K, Speer O, Schmugge M, Roschitzki B, Durka SS, Leopold U, Kuster T, Heizmann CW, Frischknecht H, Troxler H. Clin Chem. 2008;54:69–76. doi: 10.1373/clinchem.2007.089961. [DOI] [PubMed] [Google Scholar]
- 16.Kiernan UA, Black JA, Williams P, Nelson RW. Clin Chem. 2002;48:947–949. [PubMed] [Google Scholar]
- 17.Edwards RL, Creese AJ, Baumert M, Griffiths P, Bunch J, Cooper HJ. Anal Chem. 2011;83:2265–2270. doi: 10.1021/ac1030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. Anal Chem. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.