Abstract
Traumatic brain injuries are the leading cause of disability each year in the US. The most common and devastating consequence is the stretching of axons caused by shear deformation that occurs during rotational acceleration of the brain during injury. The injury effects on axonal molecular and functional events are not fully characterized. We have developed a strain injury model that maintains the three dimensional cell architecture and neuronal networks found in vivo with the ability to visualize individual axons and their response to a mechanical injury. The advantage of this model is that it can apply uniaxial strains to axons that make functional connections between two organotypic slices and injury responses can be observed in real-time and over long term. This uniaxial strain model was designed to be capable of applying an array of mechanical strains at various rates of strain, thus replicating a range of modes of axonal injury. Long term culture, preservation of slice and cell orientation, and slice-slice connection on the device was demonstrated. The device has the ability to strain either individual axons or bundles of axons through the control of microchannel dimensions. The fidelity of the model was verified by observing characteristic responses to various strain injuries which included axonal beading, delayed elastic effects and breakdown in microtubules. Microtubule breakdown was shown to be dependent on the degree of the applied strain field, where maximal breakdown was observed at peak strain and minimal breakdown is observed at low strain. This strain injury model could be a powerful tool in assessing strain injury effects on functional axonal connections.
Introduction
Traumatic brain injury (TBI) is the leading cause of death and disability in the United States (US).1 Inertial forces that occur during acceleration/deceleration of the brain can lead to the generation of shearing or straining forces exerted on axons.2 The consequence of these physical forces is that axons are injured and the resultant damage is commonly referred to as diffuse axonal injury (DAI). DAI is the most common type of pathology in TBI occurring in approximately 40–50% of reported cases.3, 4 After the initial mechanical trauma, secondary injury cascades are initiated which include among others, cytoskeletal damage, calcium influx, neurotransmitter release, and mitochondrial dysfunction.5 There is currently no “standard treatment” protocol for these secondary effects of TBI and with almost one third of all injury related deaths in the US attributed to a TBI related incident, finding relevant therapies is of utmost importance.1
In order to develop appropriate interventions, reliable and accurate models that can simulate an applied injury need to be developed. These models must be capable of applying physiological levels of injury and assessing the extent of injury. Numerous strain induced injury models have been developed both in vivo and in vitro.6 Animal models are unfortunately both costly and inherently variable, and even though injuries can be precisely applied, it is particularly difficult to know how these forces and strains are translated from the point of application to the cellular level. In vitro models vary from using dissociated cells to organotypic tissue slices and from applying uniaxial to biaxial strains. While organotypic slices maintain the cellular cytoarchitecture and functional connections, knowing exactly how the applied strain is translated through the thickness of the slice and imaging injury effects in real time limits the imaging access of this approach.7 Dissociated cells however, are easily cultured and precise strains can be applied, but they unfortunately lack the natural connectivity and interaction with surrounding extracellular matrix (ECM) and Glial cells.8, 9 Both biaxial and uniaxial applied strains are equally important since both are physically found to occur during TBI events.10
A model that can incorporate the benefits of both organotypic and cellular level cultures will thus be able to overcome multiple issues. It will maintain the tissue structure found in vivo while being able to, in real time, monitor cellular level changes following injury. This is accomplished by incorporating microfluidics with organotypic slice cultures. Two organotypic slices are placed in close proximity to each other but separated by micron sized channels. These microchannels allow for axons that extend from the periphery of the slice to be guided from one slice to the other, thereby allowing functional connections to be made.11, 12 Depending on the slices placed in culture, specific physiologically relevant networks can be tested. Hippocampus slices are the ideal organotypic tissue to use in TBI injury models since they are frequently injured during TBI events and maintain their structure and functional networks in culture over extended periods of time.11, 13, 14
We have thus developed and characterized a uniaxial strain device that can apply precise levels of strain to individual and/or bundles of axons that extend from an organotypic hippocampus slice and connect to an adjacent organotypic slice. These organotypic slices are cultured on an elastic substrate manufactured from Polydimethylsiloxane (PDMS) and remain healthy for over three weeks in culture. The strain injury is applied to the connecting axons by pressurizing a cavity beneath the microchannels. The relationship between degree of strain and microtubule integrity following injury and responses typically seen in vivo are observed using this device.
Materials and Methods
Making/Assembling device
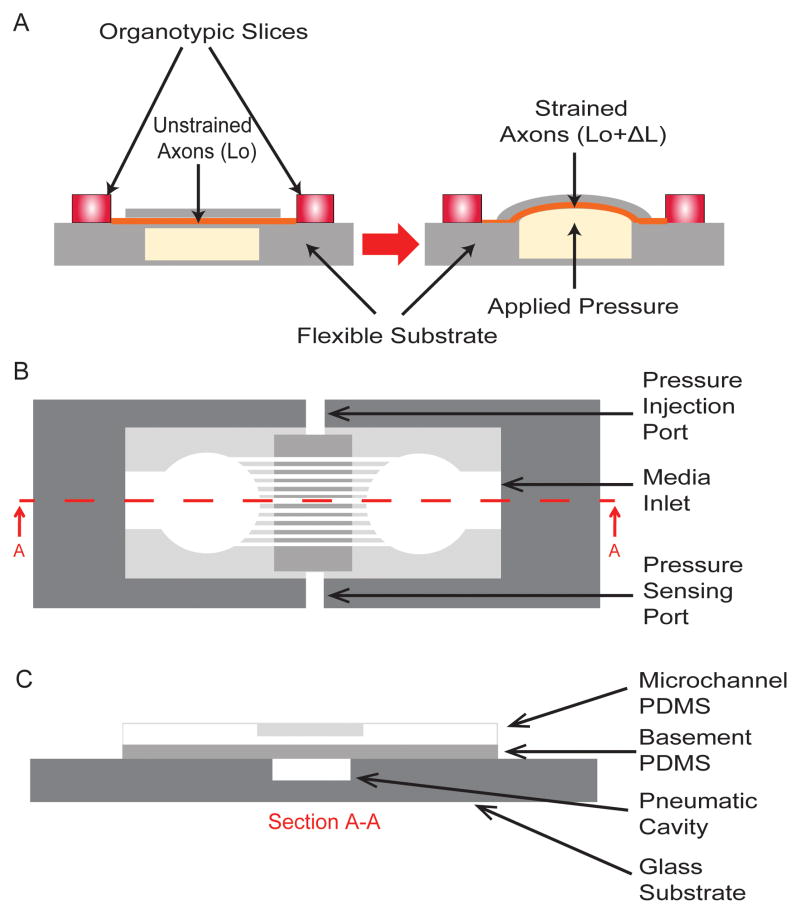
Multiple uniaxial strain devices were manufactured using cross-linked PDMS (Sylgard 184, Dow Corning Corporation, Auburn, MI, USA). Two PDMS layers were fabricated using soft-lithography methods. The first layer (referred to as the basement PDMS layer) is a smooth PDMS surface on which organotypic slices are placed and axons extend, and the second layer (referred to as the microchannel layer) is where extending axons are guided to grow between two organotypic slices. A schematic of the device is shown in Figure 1A and a more detailed top and section view is shown in Figures 1B and C. Both PDMS layers were fabricated by mixing PDMS to its crosslinker in a 10:1 ratio followed by 40mins of degassing in a desiccant chamber. The basement PDMS layer was produced by spincoating PDMS onto a blank silicon wafer (El-Cat Inc, Waldwick, NJ, USA) at 1000rpm for 30s resulting in a thickness of 75±5μm. The microchannel PDMS layer was manufactured using silicon mold masters that were prepared using a negative chrome mask (Fine Line Imaging, Colorado Springs, CO, USA) and SU-8 (2002) photoresist (Microchem, Newton, MA, USA). Two SU-8 patterns were produced: 50μm (microchannel width) × 6μm (microchannel height) × 50μm (microchannel spacing) and 25μm (microchannel width) × 3μm (microchannel height) × 50μm (microchannel spacing). These dimensions were confirmed using a profilometer (Dektak 3030, Veeco, Plainview, NY, USA). The microchannel PDMS layer was produced by spincoating PDMS onto a SU-8 master at 1000rpm for 30s resulting in a thickness of 71±5μm. Both layers of PDMS were cross-linked at 65°C overnight. The manufacturing protocol for both PDMS layers was strictly followed so as to ensure that the material thicknesses and resulting material properties (which were tested periodically) were consistent from batch to batch. The cured PDMS was then cut into appropriate shapes. The basement PDMS layers were cut into strips of 40mm × 45mm. The microchannel PDMS was cut into 22mm diameter circles with 3mm mini-wells punched at a spacing of 2mm from each other with a media access channel leading to each of the mini- wells (critical for allowing sufficient nutrient exchange for long term slice cultures). The small mini-well that the organotypic slice is placed in provides a buffering against shear forces that occur during device movement and media changes to both the organotypic slice as well as to delicate axons extending from the slice.11 Before irreversibly bonding PDMS to either glass (basement PDMS layer) or PDMS (microchannel PDMS to basement PDMS), all surfaces were cleaned. This was important since these layers experience relatively large pressures and may be susceptible to delamination. The glass substrate was cleaned by boiling in 0.5M Hydrochloric Acid (HCL) for 10mins, rinsed in deionized water (diH2O) for 10mins and then dehydrated by placing in an oven at 65°C overnight. The basement and microchannel PDMS were cleaned by first applying scotch tape to both surfaces to remove any large adherent debris, placed in methanol and sonicated for 10mins, boiled in 0.5M HCL for 10mins, rinsed in diH2O for 10mins and then dehydrated by placing in an oven at 65°C overnight. The basement PDMS layer was first bonded to the glass substrate. Both surfaces were oxygen plasma treated (ENI, Rochester, NY, USA) at 100W for 30s at 60% O2. The exposed surfaces were immediately placed into contact with each other and then placed on a hotplate at 100°C for 30mins with a 0.5kg weight applied on top. The microchannel PDMS was bonded to the basement PDMS layer by treating both surfaces with oxygen plasma at 100W for 30s at 60% O2. The exposed surfaces were immediately placed into contact with each other. To contain the media for the organotypic slices, a PDMS well was manufactured with an outer diameter of 38.1mm and inner diameter of 31.8mm. This was bonded to the basement PDMS by exposing both surfaces to oxygen plasma at 100W for 30s at 60% O2. To ensure that this outer well was leak tight, it was epoxy sealed using EPO-TEK 353ND (Epoxy Technology Inc, Billerica, MA, USA). Media evaporation was reduced by placing a semi-permeable membrane, i.e., permeable to O2 and CO2 but not permeable to H2O, (ALA Scientific Instruments, Farmingdale, NY, USA) over the PDMS outer well and sealed with an O-ring.
Fig. 1.
Organotypic uniaxial axonal strain device. A) Schematic of device before and after strain application. B) Detailed top view of assembled device. C) Detailed sectioned view (A–A) of assembled device. All figures are not to scale.
FEA – Abaqus
The commercially available finite element analysis software Abaqus 6.10 – CAE/Standard (Dassault Systemes, Waltham, MA, USA) was used to assess the degree of PDMS strain that occurs when applying a pressure and to ensure that uniaxial strain is applied across all relevant microchannels. PDMS material properties obtained from tensile material tests were input into the software. These tensile tests were done according to ASTM D 412 standards using a Bose/EnduraTEC ELF 3200 mechanical testing machine (EnduraTEC, Eden Prairie, MN, USA). The size of the test specimens were scaled down to one fourth of those specified within the standard.15 A hourglass shaped die was cast using rapid prototyping (Viper, 3D Systems, Overview Drive, SC, USA) and used to cut out samples of PDMS for testing (8.25mm-length by 2.5mm-width). These samples were securely placed in the jaws of the mechanical testing machine and tensile strained at a rate of 3.75mm/s. The force and displacement readings were recorded at a rate of 80HZ. Markings were applied to the PDMS around the jaws to assess for slippage during loading. Given the highly elastic nature of PDMS, a second order Ogden hyperelastic material model was used to calculate strains. A three-dimensional model (8-node linear brick (C3D8), hybrid, constant pressure model was used with a seed size of 0.015mm) was created that included three components, namely the microchannel PDMS, basement PDMS and glass substrate. The glass substrate was fixed and since all three components were irreversibly bonded to one another no slippage between the PDMS and glass substrate or between the PDMS layers was assumed, i.e. no translation or rotation boundary conditions were applied. With respect to the bond between the glass substrate and the basement PDMS, if the bonding wasn’t absolutely ideal, with the range of pressures applied, delamination occurred. The bonding between the two PDMS layers was tested before and after strain and no slippage was ever observed. The pressure loading was applied to the under surface of the basement PDMS and resulting PDMS displacements and strains (Exx, Eyy) were obtained.
Applying Strain
In order to produce a uniaxial strain field a pneumatic pressure is applied to a cavity beneath the microchannels (Fig 1A). This pneumatic pressure is produced by coupling to a pressure injection system that consists of a linear actuator (PS01-23x80, Lin Mot Inc, Elkhorn, WI, USA), controller (E100-MT -Lin Mot Inc, Elkhorn, WI, USA), a pressure sensor (Honeywell International, Morristown, NJ, USA) and a high speed sampling system (USB-5132, National Instruments Corporation, Austin, TX, USA). The linear actuator and controller are coupled to the inlet port on the device and controlled by a visual basic program. A pressure sensor is coupled to the pneumatic cavity and attached to a high speed digitizer that is controlled by a Labview program (National Instruments Corporation, Austin, TX, USA). The injected volume and rate of injection are varied to produce a variety of strains (11%, 25% and 42%) and rate of strain (260ms, 50ms and 22ms to peak deflection). Uniaxial strains were applied 14days after initially plating hippocampal slices on the strain injury device. To verify that the Abaqus model accurately describes the system, PDMS deflections were monitored at various cavity pressures. This was done by slowly increasing the cavity pressure using a syringe pump (PHD 2000 – Harvard Apparatus, Holliston, MA, USA) and measuring the corresponding deflection of the PDMS at the center of the pressure cavity using an Olympus IX81 DSU microscope(Olympus, Center Valley, PA, USA). This microscope has a high-resolution Z-axis motor capable of 0.01μm step sizes.
Organotypic Slice Isolation
The brains of Sprague-Dawley rat pups (Taconic, Hudson, NY, USA) between the ages of 4 to 6 days old were removed and placed in ice cold Gey’s Balanced Salt Solution (Sigma-Aldrich Corp, MO) supplemented with 10mM D-glucose (Sigma-Aldrich Corp, St. Louis, MO, USA) and 3μM Kynurenic Acid (Sigma-Aldrich Corp, St. Louis, MO, USA). In our hands, we have found that axon extension from the periphery of organotypic hippocampal slices is very dependent on the age of the rat pup they have been excised from with axon extension decreasing significantly after day 6 (data not shown). The hippocampi are separated from the surrounding cortex and sliced into 400μm thick slices using a McIllwain Tissue Chopper (Stoelting Co, Wood Dale IL, USA). The slices were carefully placed into PDMS (Sylgard 184, Fischer Scientific, Pittsburgh, PA, USA) mini-wells and orientated such that the dentate gyrus (DG) region was facing the CA1 region of the adjacent slice, i.e., these regions were facing the microchannels. The PDMS was precoated with poly-d-lysine (1mg/ml, Sigma-Aldrich Corp, St. Louis, MO, USA) and laminin (25μg/ml, Sigma-Aldrich Corp, St. Louis, MO, USA). The device was filled with 450μl of serum containing media (1:1:2 of heat inactivated horse serum, Hanks Balanced Salt Solution, Basal Medium Eagle, supplemented with 0.5mM L-Glutamine, 30μg/ml gentamycin and 10mM HEPES, all from Invitrogen, Carlsbad, CA, USA), i.e., enough media to just cover the exposed basement PDMS area. The cultures were placed on a rocker (~1 revolution/60s) in a humidified 5% CO2 incubator. Glial cell proliferation was reduced by changing from serum containing media to a serum free media after 24hrs (Neurobasal A, 1X B27, 0.5mM L-glutamine, 30μg/ml gentamycin and 10mM HEPES, all from Invitrogen, Carlsbad, CA, USA). Thereafter, half of the media was changed every 48hrs. All animal experiments were approved by the Animal Care and Use Committee of Rutgers, The State University of New Jersey.
DiI Staining
Axons traversing the distance from one organotypic slice and connecting to the adjacent slice was visualized by applying the lipophilic carbocyanine dye, DiI (1,1′,di-octadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate – Invitrogen, Carlsbad, CA, USA) to one organotypic slice. DiI is a nontoxic fluorescent probe that allows for the tracing of axons through the uptake of the dye in the cell body and via transport/diffusion, labels the axons through to its axon terminal. One small DiI crystal was carefully placed on top of one slice (in the center). Within 24–48hrs later the culture was imaged using a 540nm fluorescent filter on an Olympus IX81 DSU microscope. DiI labelling proved to be quite challenging due to the fact that the device is an open system, i.e., the media that supports one slice is the same that supports the adjacent slice (thus there is a potential for it to diffuse to the adjacent slice through the media thereby bypassing the connecting axons). For this reason the cultures were carefully handled making sure to not dislodge the crystal.
Immunohistochemical staining
Cultures were fixed before immunofluorescent staining for specific protein markers. This was done by washing three times in phosphate buffered saline (PBS) followed by incubating in 4% paraformaldehyde (Sigma Aldrich Corp, MO) for 25mins at room temperature (RT). The cultures were again washed three times in PBS for 5mins each at RT. For primary conjugated MAP2 and Tau staining, the following protocol was followed: non-specific binding of antibodies to proteins on/within the cells was reduced by first blocking for 45mins at RT by incubating with Tris Buffered Saline (TBS), 10% mouse serum (Invitrogen, Carlsbad, CA, USA), 1% Bovine Serum Albumin (BSA) (Sigma–Aldrich Corp, St. Louis, MO, USA) and 0.1% Triton X-100 (Sigma-Aldrich Corp, St. Louis, MO, USA). The cultures were then incubated for 45mins with the respective primary conjugated antibody in TBS and 1% mouse serum. The antibodies used were: 1.2 μg/ml mouse anti-MAP2B IgG1 conjugated with Alexa Fluor 647 (BD Pharmingen, San Diego, CA, USA) and 1.5 μg/ml mouse anti-Tau IgG2b conjugated with Alexa Fluor 488 (BD Pharmingen, San Diego, CA, USA). To account for non-specific background binding, isotype controls were used for each antibody: mouse IgG1 conjugated with Alexa Fluor 647 (BD Pharmingen, San Diego, CA, USA), and mouse IgG2b conjugated with Alexa Fluor 488. The cultures were washed three times for 15mins in PBS. Beta III tubulin was stained using indirect methods and the following protocol was followed: non-specific binding was reduced using the same as above. The cultures were incubated overnight at 4°C with 2μg/ml beta III tubulin rabbit IgG (Abcam, Cambridge, MA, USA) in TBS and 1% goat serum. To account for non-specific background binding, isotype control cultures were incubated at 4°C overnight with 2μg/ml rabbit IgG (BD Pharmingen, San Diego, CA, USA) in TBS and 1% goat serum. Both beta III tubulin and isotype control cultures were then incubated for 1hr with 2μg/ml Alexa Fluor 647 goat-anti-rabbit IgG (Invitrogen, Carlsbad, CA, USA) secondary antibody. The cultures were then washed three times for 15mins in PBS. Fluorescent images were acquired using a computer interfaced inverted Olympus IX81 DSU microscope. Specimens were excited using either 488nm or 647nm filters. Fluorescent intensity values were determined using Slidebook™software (Olympus, Center Valley, PA, USA) and control values were subtracted from experimental values.
Results
Device – PDMS material properties
In order to apply a uniaxial strain to axons, they needed to be cultured on a flexible elastic substrate.16 The substrate chosen was PDMS since it is transparent, permeable to gases, non-toxic to cells, highly elastic, can be bonded to other surfaces, and can rapidly produce micron-sized structures using soft lithography methods.17 The PDMS layers were manufactured in-house since both the thickness and the stiffness could be controlled in order to obtain the desired strains. The required PDMS thicknesses for the basement and microchannel layers were achieved by spincoating at 1000rpm for 30s. This speed resulted in material thicknesses of 75±5μm and 71±5μm for the basement and microchannel PDMS layers respectively. In order to accurately predict the strain applied at the axon surface, PDMS material properties were determined. The stress/strain properties were periodically obtained through tensile testing (Supplementary Fig S1, Supplementary Methods). The resultant PDMS stress/strain behaviour was fit to a 2nd order Ogden hyperelastic material model (μ1=107.6 kPa, α1=6.1, μ2=535.2 kPa, α2=−3.96) with a root mean square error of 0.993. (inset Supplementary Fig 1).
Applied strain predictions
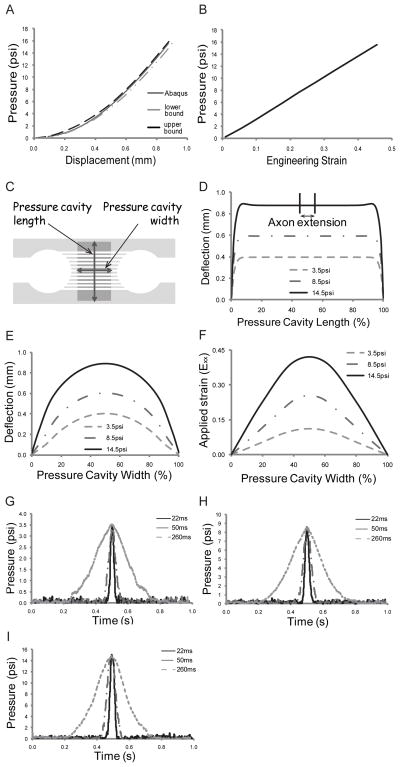
The finite element software Abaqus was used to calculate: 1) the strain axons will experience when attached to the PDMS after application of specific pressures to the device, and 2) confirmation that the area in which the axons extend and connect with the adjacent slice experiences uniform uniaxial strain. The fidelity of the Abaqus simulations was confirmed by initially applying discrete static pressures to the device and the amount of PDMS deflection measured and compared to simulation results (Fig 2A). As the graph indicates, the simulations correlate well to experimental deflections staying within the upper and lower bounds of deflection. The corresponding strain experienced by the PDMS as calculated by Abaqus is shown in Figure 2B. These strains have an almost linear relationship with respect to the applied pressure (R2=0.999).
Fig. 2.
PDMS characterization. Strain device displacements and Abaqus strain predictions. A) PDMS deflections: Experimental vs. Abaqus. B) Strain vs. applied pressure as determined from Abaqus. PDMS deflection profiles. C) Definition of deflection profiles. D) PDMS deflection profile along the pressure cavity length. Area in which axons extend is indicated on the top curve (between 45%–55%). E) PDMS deflection profile along the pressure cavity width. F) Strain (Exx) profiles across the pressure cavity width. Inertial effects within the system. Representative graphs of the pressures measured within the device for various times to peak deflection (22ms, 50ms, and 260ms). G) 3.5psi applied pressure, H) 8.5psi applied pressure, and I) 14.5psi applied pressure.
The strain injury device was characterized using three applied cavity pressures selected on the basis of a range of resulting strains, namely: 3.5psi, 8.5psi, and 14.5psi, which equate to ±11%, ±25%, and ±42% strain respectively. These strains span the range believed to cause damage during TBI events, i.e., 10%–50% strain.18–20 Deflection profiles across the length and width of the pressure cavity were determined (Fig 2C–F). When a width to length ratio of 1:16 was used (ratios above 8 have been shown to produce uniaxial strains),21 a uniform deflection profile occurred in the region in which axons traverse the microchannels (Fig 2D shows cavity length deflection profile). A uniform deflection along the length indicates a uniform strain field along the width of the cavity. We observe that in the region in which the axons extend (indicated on the 14.5psi deflection profile curve, i.e., between 45% and 55% along the length) the deflection is uniform for all applied pressures. Biaxial strain (strain along the length of the pressure cavity), was verified using Abaqus. At an applied pressure of 15.5psi (i.e., above our maximum applied pressure), there is a maximum of 0.11μm deflection along the length of the channel (i.e., 90° to the applied strain) equating to a biaxial strain of 0.006% (essentially zero). The PDMS deflection profile across the width of the pressure cavity can be seen in Figure 2E showing the relative shape of the respective deflections. As expected, the maximal deflection occurs at the midpoint of the cavity and is consistent for all applied pressures. The corresponding strain (Exx) profile across the pressure cavity width for the three applied pressures is shown in Figure 2F.
Dynamic pressure effects
This device is capable of applying strains at various strain rates. Figure 2G–I show three different times to reach peak deflection, i.e., 260ms, 50ms, and 22ms, for the three different applied pressures. Table 1 reflects the strain rates that equate to the three different deflection times. These strain rates fall within the range shown to produce damage during TBI. In Figure 2G all three curves have a maximum pressure of 3.5psi showing no dynamic effects on the applied strain. In Figure 2H and I, it may be difficult to observe, but at the highest applied rate, the maximum pressure is approximately 0.9% and 1.2% higher respectively, than at the lowest applied rate. These dynamic effects equate to an increase in <0.5% strain.
Table 1.
Strain rates that correspond to chosen peak deflection times.
| Time to peak deflection | 11% strain | 25% strain | 42% strain |
|---|---|---|---|
| 260ms | 0.4s−1 | 1.0s−1 | 1.6s−1 |
| 50ms | 2.2s−1 | 5.0s−1 | 8.4s−1 |
| 22ms | 5.0s−1 | 11.4s−1 | 19.1s−1 |
Monitoring hippocampus slice health on the device over time
Cell death within the organotypic slices was assessed to confirm that the slices remain healthy throughout the course of the expected experiment. This was done using the fluorescent molecule Propidium Iodide (PI). By Day 11 in vitro, cell death due to the initial slicing procedure had been cleared and remained around zero until Day 22 in vitro (Supplementary Fig S2, Supplementary Methods). This time progression of cell death is typical of other organotypic slice cultures on elastic substrates.7
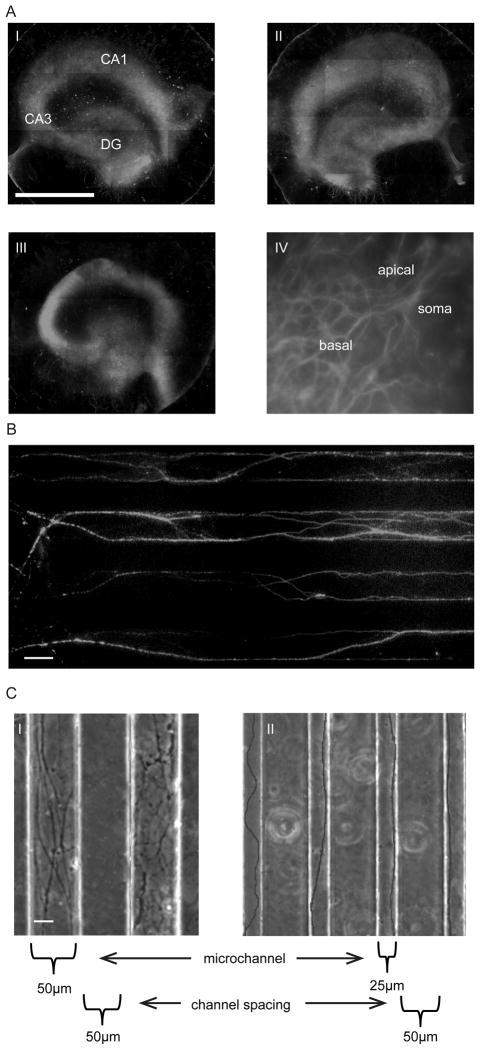
Organotypic slice health was not only assessed by cell death but also by observing the integrity of the different neuronal cell layers that are so distinct to the hippocampus over time (Fig 3A). Hippocampal slices stained with the neuronal cell marker MAP2 (microtubule associated protein) on Day 7 (Fig 3A i), 14 (Fig 3A ii) and 22 (Fig 3A iii) on the device clearly show the three distinct neuronal cell layers of the dentate gyrus (DG), CA3 and CA1. Not only are the distinct regions of the hippocampus maintained over time, but the specific cell orientation within the slice is maintained over the same time period (Fig 3A iv), where the characteristic apical and basal branching of a CA1 pyramidal neuron can be seen.
Fig. 3.
Organotypic and axonal culture on the injury device. A) MAP-2 staining of organotypic slices on the strain injury device. i) Day 7, ii) Day 14, iii) Day22, (DG – dentate gyrus), and iv) CA1 neuron on Day22, showing apical and basal branching. Scale, 1mm. B) Axon extension within microchannels (50μm × 6μm). Confirmation that extension is indeed axons by immunostaining for Tau (axonal marker). Scale, 50μm. C) PDMS microchannel dimensions control the number of entering axons, i) 50μm wide microchannels = large number of entering axons, and ii) 25μm wide microchannels = single axons/small number of entering axons. Scale, 20μm.
Axon extension within microchannels
Axons are observed to extend from the periphery of hippocampal slices in early postnatal slices (P4–P6). These extending axons generally enter microchannels that are closest to them. The axons extend down the length of the microchannels and connect with the adjacent slice. The extensions within the microchannels were confirmed to be axons and not dendrites by immunostaining for the axon specific marker Tau (stabilizes microtubules in axons) (Fig 3B). In this particular example, we see both individual as well as bundles of axons within the channels (thicker more intense stained axons).
The microchannel height was optimized to approximately 6μm in order to minimize cell migration down the channel (Supplementary Fig 3, Supplementary Methods). This was important since some Glial cells migrate out from the organotypic slice and if they enter a microchannel, may obstruct the progression of axons and interfere with the way in which axons experience the applied strain. The number of axons entering a given microchannel can be manipulated by changing its dimensions. Reducing the microchannel width not only controls the number of axons that enter a microchannel, but also reduces the tortuous path that the axon may take. By reducing the microchannel width from 50μm (Fig 3C i) to 25μm (Fig 3C ii) we were able to reduce the number of axons entering a given microchannel. Thus the system has the capability of investigating the effect of uniaxial strains on individual axons.
Slice-slice connection
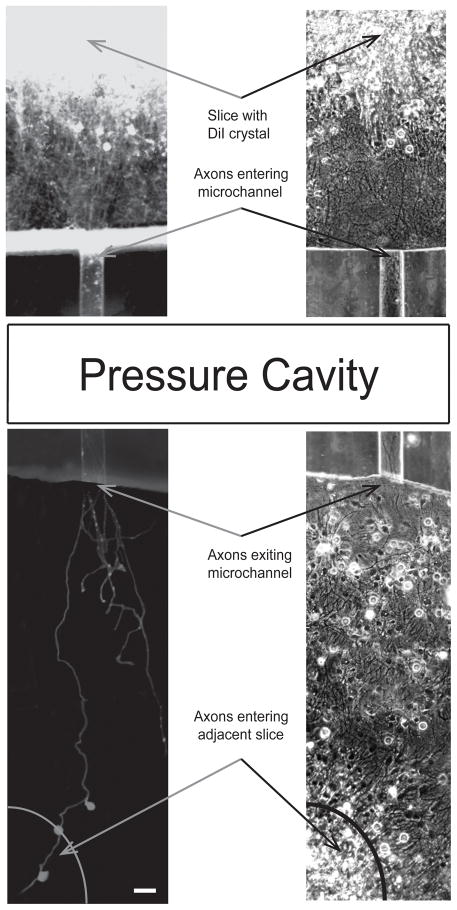
In order to confirm that axons from one organotypic slice do indeed extend the full distance and connect to the adjacent slice, axons were traced using the lipophilic plasma membrane dye, DiI (Fig 4). The slice on which the DiI crystal was placed (top slice) is very brightly fluorescent due to the very high concentration of DiI on that slice. The DiI has diffused along the plasma membrane of axons that extend from the periphery of the slice. This diffusion proceeds along their length where we observe axons entering a particular microchannel and extending down the full length (the section over the pressure cavity is not shown for conciseness) and emerging from the other end. In this particular example, one axon that has exited the microchannel clearly makes its way to the adjacent slice. Along the way it has transferred the dye to three connecting cells. A few axons that exit the microchannel have not quite made it to the adjacent slice by the time these images were taken.
Fig. 4.
Confirmation of slice-to-slice connection. DiI staining of axons extending from one slice (top), entering a microchannel (50μm × 6μm), and connecting to adjacent slice (bottom). Scale, 50μm.
Application of strain injury
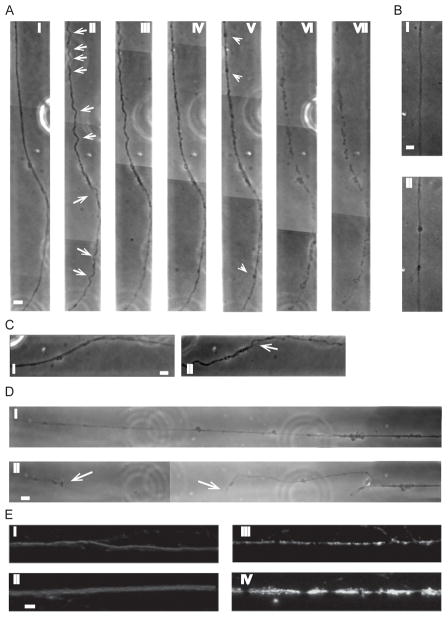
One way to confirm that our device does indeed simulate what is experienced in vivo, is to look for characteristic responses. Two of the more common axonal responses to strain injuries include axonal beading (due to transport interruptions) and delayed elastic effects (due to microtubule damage).22–24 When a uniaxial strain is applied to axons on our device we observe these typical responses, thus demonstrating that our device is proficient in applying uniaxial strains. Figure 5A shows an example of a typical response to a 42% strain, 19s−1 strain rate, applied to axons connecting hippocampal slices. This example shows a time progression of an axon bundle after a uniaxial strain. Immediately following injury, distinct delayed elastic effects can be seen along the axon bundle length in the form of waves/undulations (arrows) (Fig 5A ii). This effect typically disappears within 50minutes after injury (Fig 5A iv). In this particular example, partial degradation of the axon has occurred by 210minutes (Fig 5A vi) after injury and complete degradation by 24 hrs (Fig 5A vii). The results observed in Figure 5A for the applied strain of 42% and 19−1 were also observed at the lower applied strains (however only at the highest strain rate, i.e. strain duration of 22ms) just to lesser degrees, i.e. not as many undulations and smaller undulation amplitudes.
Fig. 5.
Application of uniaxial strain to axons connecting two organotypic hippocampal slices together (50μm × 6μm microchannels). A) Time progression of delayed elastic effect on axon bundle after application of 42% strain, 19s−1 strain rate, i) before injury, ii) immediately after injury, iii) 20mins after injury, iv) 50mins after injury, v) 75mins after injury, vi) 210mins after injury, vii) 24hrs after injury (arrows (ii) show individual “waves/undulations”, arrow heads (v) show beading). B) Axonal beading, i) before injury, ii) after injury. C) Example of axonal bundle unravelling after application of a 42% strain, 19s−1 strain rate, i) before injury, ii) after injury (arrow shows point of unravelling). D) Example of primary axotomy after application of 42% strain, 19s−1 strain rate, i) before injury, ii) after injury (arrows show free ends of axon). E) Effect of applied strain profile across the pressure cavity on axonal microtubules 4hrs after 42% strain injury and 19s−1 strain rate. β-tubulin immunostaining of axons at pressure cavity walls, i.e., regions of low applied strain, i) small diameter axon, ii) larger diameter bundle, and at the center of the pressure cavity, i.e., region of peak applied strain, iii) small diameter axon, iv) larger diameter bundle. Scale, 10μm.
Another characteristic response to axonal strain injury is multiple beads forming along the length of an axon due to a pathological build up in axonal transport proteins (Fig 5B i,ii).25 These beads appear to form at random positions along the length of the axon, however a larger number of axonal beads appear to form in areas that have experienced delayed elasticity, i.e. displayed temporary undulations (Fig 5A v – arrowheads). Once the axon has returned to its original length, is when axonal beading starts to be visible.
Not all axons within a particular bundle experience this delayed elastic effect demonstrating that not all axons have the same adhesion to the rest of the bundle. We observe on occasion at the higher applied strain and strain rate (42% applied strain and 19s−1 strain rate), one or more axons may locally return to its original length (as soon as the applied strain is removed) while the rest of the axons within the bundle undergo the temporary increase in length associated with delayed elastic effects (Fig 5C). Another effect that can be observed is primary axotomy of an axon or axon bundle following strain injury (Fig 5D). Again, this tends to occur at the higher applied strain and strain rate, and typically occurs in larger diameter bundles.
Cytoskeletal effects following strain injury
Microtubules are the stiffest structural components within the axon and when excessive rates of strain are experienced, they can mechanically break.24, 26 Figure 2F showed the PDMS strain profile across the pressure cavity width, thus axons within the microchannels will experience a peak strain in the center decreasing towards the two pressure cavity walls. An example of how this varying applied strain affects microtubules within the axon can be seen in Figure 5E. The figure shows axons that have been subjected to a 42% strain injury at a strain rate of 19s−1 immunostained for β-tubulin (microtubule marker). In this figure we see that in regions of low applied strain, i.e., at the pressure cavity walls (Fig 5E i, ii), there is a continuous flow of microtubules as seen by uninterrupted β-tubulin staining in both small and large diameter axon bundles. However, at the center of the cavity, i.e., at peak applied strain (Fig 5E iii, iv), we see a breakdown in microtubules (gaps in staining) occurring in both smaller and larger diameter axon bundles. We also observe brighter fluorescence intensity with punctuate staining at specific regions along the length, denoting areas of axonal beading on single axons within the bundle. The results observed in Figure 5E for the applied strain of 42% and 19−1 were also observed at the lower applied strains and strain rates, just to lesser extents.
Discussion
We successfully designed and evaluated a device that can apply uniaxial strains to an elastic substrate on which axons extend between two organotypic hippocampal slices. These axons are guided between slices by small PDMS microchannels that allow for the most direct route between the slices. The dimensions of these microchannels can be manipulated to control the number of axons entering a given channel. The application of various strains and strain rates was accurate, reproducible, and believed to be within those experienced during TBI and resulted in characteristic axonal injury responses. These responses vary according to the applied strain field and demonstrate device fidelity.
In vivo models of TBI have the advantage of being able to measure electrophysiological and behavioural outcomes that are based on quantitative and semi-quantitative observations.27 It is however, very difficult to translate applied stresses and strains to the cellular level given the complex geometry, boundary conditions and material properties of brain tissue. On the other hand, in vitro models offer precise control over the environment, amount of injury that is imposed and allow for dynamic measurements of injury responses.28, 29 Various dissociated cell culture injury models have been developed over the years that have provided very useful information on neuronal cell and axonal injury responses.8, 9, 28, 30–32 However dissociated cell culture models lack the three dimensional nature of in vivo tissue where cell morphology, viability, ECM synthesis and cytokine production can affect injury responses. Thus the use of organotypic slices is an ideal compromise between in vivo and in vitro models.33 Organotypic slices retain their tissue and cellular architecture that are specific to that brain region, as demonstrated in this study and others where the layers and relative arrangement of pyramidal and granule neurons in hippocampal slices are maintained for relatively long periods.7, 11 Applying TBI injuries to organotypic slices is valuable for monitoring overall slice health and electrophysiological changes.34 However it is difficult to observe changes at the axonal level, since they are not easily accessible to microscopic techniques unless injection methods are used which prohibits the long-term study of axonal changes following injury. Organotypic slices extend axons when placed on an extension permissive substrate, and our lab has shown that these axons make functional connections to an adjacent organotypic slice.11, 12 This demonstrates that these extending axons are healthy and fully functional and may represent a more physiologically relevant DAI model than pure dissociated cell culture models. Having a fully functional connection between two organotypic slices before injury will allow researchers to investigate the effects of strain injury on slice-to-slice communication. For the purpose of this study, i.e., characterizing the strain injury device, we chose hippocampus-hippocampus connections (DG-CA1), however by incorporating different organotypic slices, the effects of strain injury could be tested on various functional pathways found to be affected during TBI events in vivo. Some other pathways that could be investigated are the entorhinal cortex-hippocampal pathway,35 septo-hippocampal projections,36 and thalamocortical pathways.37 Thus to combine the advantages of an organotypic slice culture system that maintains the in vivo tissue structure with the capability of monitoring single axon molecular and functional responses to strain injury in real-time could prove to be a powerful tool for studying TBI responses and developing potential therapeutics.
DAI has been shown to occur in up to 50% of TBI cases and been shown to occur across the spectrum of TBI cases from mild through severe.4, 38 It is believed that during non-contact inertial TBI events, brain tissue is subjected to strains in the order of 10–50% and at strain rates as high as 50s−1.18, 19 These wide ranging boundaries are most likely one of the reasons why we see DAI being manifested at all levels of TBI. We have designed our device such that our applied strains span those believed to produce damage during these events, i.e., a low (11%), an intermediate (25%) and a high applied strain (42%). The strain rates applied in this study fall within the lower half of the range shown to produce damage. However, in a comprehensive study conducted by Morrison and colleagues that applied varying strains (5–50%) and strain rates (0.1–50s−1) to organotypic hippocampus slices, they showed that injury response was more dependent on strain than on strain rate.39
One critical comment of other uniaxial strain models is the verification of adhesion of cells and axonal processes to the substrate. This in part, is due to the fact that some models apply a downward deflection to the substrate on which the axons have extended.8, 19, 30 Our device applies a positive pressure to a pressure cavity located beneath the basement and microchannel PDMS layers, resulting in the PDMS being deflected upwards. We do not observe any movement of axons due to the application of a strain (apart from the obvious “delayed elastic effects” observed), i.e., the axon is in the same exact position after injury as it was before injury implying that they are firmly attached to the PDMS substrate. Thus axons that are adhered to the top surface of the PDMS basement layer will be forced upwards and experience similar strains as that of the PDMS on which they have adhered.
The mechanical stiffness/properties of the PDMS are of utmost importance due to the fact that this is what dictates the applied. Thus the manufacture of the PDMS basement and microchannel layers is a very important and controlled step in the assembly of the strain device. In our hands, it was very important to be as consistent and accurate as possible with every step involved making the respective PDMS layers.
In the longitudinal deflection profile seen in Figure 2D small ridges are seen in the two opposing ends of the 14.5psi profile. These are edge effects due to the radius at the end of the pressure cavity, however they only reflect a 1.6% increase in deflection over the central stabilized portion of the device and within 7% of the length of the pressure cavity has stabilized to the maximum deflection observed in the region in which the axons extend. These edge effects are only observed at the higher pressures.
The device utilizes the inherent capability of organotypic hippocampal slices to extend axons from the periphery of the slice.11 This extension is likely to be due to the spontaneous response to the elimination of axonal projections during the dissection procedure. The axons that extend from the periphery of these organotypic slices are unmyelinated, and in vivo studies using a fluid percussion injury model have shown that unmyelinated axons are even more susceptible to TBI, than myelinated axons.40 Axonal response to strain injury also differs depending on the caliber of the axon.40 Our strain injury device has the capability of investigating injury effects on various caliber axons due to the fact that we have the ability to control the dimensions of the microchannels and thus control the number of axons entering a particular channel. The number of axons entering a channel correlates to the resulting axonal diameters within the channel since axons tend to aggregate together and form bundles. Although the system has the capability of investigating individual axon response to injury we chose to focus on a mix of individual axons and bundles of axons both of which are commonly found in vivo.41 Thus the majority of the figures shown in the manuscript were using 50μm wide × 6μm high microchannels.
The conventional methods for culturing hippocampal slices involve maintaining tissue slices of 400–600μm thickness at the air-liquid interface in order to improve oxygen levels in the tissue and avoid hypoxia and necrosis. In the interface method developed by Stoppini et al, slices are placed on top of porous membranes with culture medium below and only a thin layer of medium covering the tissue that allows for ample oxygen flow to the cells.14 An older method developed by Gahwiler et al calls for attaching organotypic slices to a glass cover slip using a blood clot, which is then placed in a roller-tube and rotated at slow speed to alternatively dip the slice into culture medium or bring it into the air.42 However, since the interface method has medium beneath the cultures this prohibits the possibility of applying a strain to the substrate through the application of a pressure and since we are relying on the ability of axons to extend from the periphery of the organotypic slice, this precludes the use of blood clots for slice attachment. More recently, our lab and others have used a combination of the two methods where hippocampal slices are cultured on glass substrates11 or on elastic substrates such as PDMS.19, 7, 43 Our method employs enough media to just cover the substrate on which they are cultured with the addition of placing our strain device on a rocker in order to ensure adequate oxygen supply. Using this method, we demonstrated that organotypic slices remain healthy over a period of 22days in vitro. Our device is consistent with other slice injury models that wait until slices are cultured for approximately 14 days in vitro and then either mechanically or chemically injure the slices.7, 44 Over this initial 14 day period, the slice not only stabilizes, i.e., we observe a decrease in cell death, but also matures. This 14 day slice stabilizing period is convenient since it takes between 7–10 days for the axons to traverse the distance from one slice to the other.
In contrast to other uniaxial strain models, we observe primary axotomy occurring at peak strains of 42%. Smith and colleagues report that on their strain injury device they observe primary axotomy occurring at strains only above 65%.8 This disparity in results could be attributed to a number of different reasons, one of them being the fact that the Smith model stretches their membrane downwards which may result in their axons not experiencing the full applied strain since they have the potential to detach from the substrate. Our model however applies a positive pressure beneath the axons thus stretching the axons to the full extent of the substrate strain. Another reason could be due to the fact that our device utilizes axons that extend from organotypic slices whereas their model uses dissociated cells from the NTera-2 cell line. The neuronal cell bodies from which our axons originate have a fairly rigid anchoring within the tissue being firmly embedded in extracellular matrices and surrounded by other cell bodies. Once axons have traversed the full distance and connected to the adjacent slice, a few of them are seen to slightly straighten along their length. This straightening reduces the tortuous path axons can take along the length of the microchannel thus taking up the “slack” along the length which would result in the axon experiencing larger stresses and strains during injury. Organotypic slices have been shown to mature in vitro,45 and with the fact that CNS tissue has also been shown to become stiffer with increasing age,46 what we may be observing is a slight stiffening of axons extending from the organotypic slice.45 If the axons are slightly more stiff when a strain injury is applied, slightly higher forces would be experienced that might result in primary axotomy occurring at lower applied strains. This may demonstrate that some of the axons extending within this model may be physiologically closer to those found in vivo.
Axons are normally elastic and thus return to their original length and shape after a temporary strain has been applied.8 It is only at high rates of strain that delayed elastic effects are observed. These effects are observed on our device similar to what has been observed in other in vitro uniaxial strain models.8 This response is due to a breakdown in microtubules following high rates of strain injury that lead to misalignment of individual microtubules within the axon resulting in them not being able to slide freely over one another.24 This delay in the axon returning to its original length once the applied strain has been removed results in the undulations observed along the axon length. This strain induced microtubule damage correlates to the observation of an increase in the number of axonal beads occurring in areas of delayed elasticity (microtubule damage leading to transport interruptions). This increase in axonal beading also correlates to more rapid axonal degradation occurring in these areas as seen by the amount of degradation occurring within 4hrs (Fig 5A vi). The return to the original axonal length within 50minutes is comparable to that seen in other in vitro models.8, 24
Kilinc and colleagues have suggested that DAI and axonal beading following a TBI event is due to mechanoporation (increase in axonal plasma membrane permeability).47 In their model they observe significant amounts of Lucifer Yellow (480Da) entering neurons following injury. They however use a different mechanism to induce injury, i.e. fluid shear stress which could account for the permeability observed. In our model, we observe no axonal plasma membrane permeability to molecules 622Da (calcein) and larger following injury (data not shown). We demonstrate that the initial formation of axonal beading is probably due more to primary microtubule damage than to mechanoporation.
The effect of strain injury on microtubules appears to be dependent on the degree of strain that is applied. In the region in which peak strain is experienced, i.e., in the central portion of the pressure cavity, β-tubulin staining showed significant microtubule breakdown as compared to regions of low strain, i.e., at the edges of the pressure cavity, where little to no microtubule breakdown is seen. In low strain regions we observe constant intensity fluorescent staining along the length of both single axons and larger axon bundles. Whereas in regions of peak strain we observe sections of either no fluorescence, indicating a breakdown in microtubules, or pockets of high fluorescence, indicating an accumulation of microtubules typically found in axonal beads. In regions where delayed elasticity occurs, we observe some unravelling of axon bundles at specific locations along the bundle. In these sections one or more axons return to their original length while surrounding axons undergo delayed elasticity. It appears that axonal bundles that form either do not completely bind along the full length of the bundle or the elastic recoil force inherent to the axon exceeds the binding force of the axon to the rest of the bundle. Typically there is a slight delay in degradation of these axons, although by 24 hours following injury, even these axons have degraded (as did the rest of the bundle). This unravelling is however only observed at higher applied strains.
One potential application of this model is to assess how various strain injuries affect action potential propagation between two organotypic slices. This could be accomplished through the incorporation of multiple electrode arrays into the device.11, 48 This strain injury device could prove to be particularly useful in elucidating mechanisms involved in TBI induced alterations in axonal action potential propagation, since TBI induced epilepsy is the most common form of acquired epilepsy with almost half of people experiencing a severe TBI event succumbing to bouts of epilepsy.49
Supplementary Material
Acknowledgments
This work was supported by the New Jersey commission for Brain Injury Research (BIR-1.08.016) and NIH Biotechnology Training Program (T32 GM008339).
Footnotes
Electronic Supplementary Information (ESI) available: [Supplementary methods and results; supplementary Figures 1, 2 and 3].
Contributor Information
Jean-Pierre Dollé, Email: jpdolle@rutgers.edu.
Barclay Morrison, III, Email: bm2119@columbia.edu.
Rene R. Schloss, Email: schloss@soemail.rutgers.edu.
Martin L. Yarmush, Email: yarmush@rci.rutgers.edu.
References
- 1.Coronado VG, Xu L, Basavaraju SV, McGuire LC, Wald MM, Faul MD, Guzman BR, Hemphill JD. MMWR Surveillance summaries: Morbidity and mortality weekly report Surveillance summaries/CDC. 2011;60:1–32. [PubMed] [Google Scholar]
- 2.Smith DH, Meaney DF, Shull WH. The Journal of head trauma rehabilitation. 2003;18:307–316. doi: 10.1097/00001199-200307000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Iwata A, Stys PK, Wolf JA, Chen XH, Taylor AG, Meaney DF, Smith DH. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:4605–4613. doi: 10.1523/JNEUROSCI.0515-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meythaler JM, Peduzzi JD, Eleftheriou E, Novack TA. Archives of physical medicine and rehabilitation. 2001;82:1461–1471. doi: 10.1053/apmr.2001.25137. [DOI] [PubMed] [Google Scholar]
- 5.Andriessen TM, Jacobs B, Vos PE. Journal of cellular and molecular medicine. 2010;14:2381–2392. doi: 10.1111/j.1582-4934.2010.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison B, 3rd, Elkin BS, Dolle JP, Yarmush ML. Annual review of biomedical engineering. 2011;13:91–126. doi: 10.1146/annurev-bioeng-071910-124706. [DOI] [PubMed] [Google Scholar]
- 7.Morrison B, 3rd, Cater HL, Benham CD, Sundstrom LE. Journal of neuroscience methods. 2006;150:192–201. doi: 10.1016/j.jneumeth.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Smith DH, Wolf JA, Lusardi TA, Lee VM, Meaney DF. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1999;19:4263–4269. doi: 10.1523/JNEUROSCI.19-11-04263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfister BJ, Weihs TP, Betenbaugh M, Bao G. Annals of biomedical engineering. 2003;31:589–598. doi: 10.1114/1.1566445. [DOI] [PubMed] [Google Scholar]
- 10.Strich SJ. Langmuir. 1961;1:443–448. [Google Scholar]
- 11.Berdichevsky Y, Sabolek H, Levine JB, Staley KJ, Yarmush ML. Journal of neuroscience methods. 2009;178:59–64. doi: 10.1016/j.jneumeth.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berdichevsky Y, Staley KJ, Yarmush ML. Lab on a chip. 2010;10:999–1004. doi: 10.1039/b922365g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tate DF, Bigler ED. Learn Mem. 2000;7:442–446. doi: 10.1101/lm.33000. [DOI] [PubMed] [Google Scholar]
- 14.Stoppini L, Buchs PA, Muller D. Journal of neuroscience methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 15.Mata A, Fleischman AJ, Roy S. Biomed Microdevices. 2005;7:281–293. doi: 10.1007/s10544-005-6070-2. [DOI] [PubMed] [Google Scholar]
- 16.Choi KM, Rogers JA. Journal of the American Chemical Society. 2003;125:4060–4061. doi: 10.1021/ja029973k. [DOI] [PubMed] [Google Scholar]
- 17.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 18.Margulies SS, Thibault LE, Gennarelli TA. Journal of biomechanics. 1990;23:823–836. doi: 10.1016/0021-9290(90)90029-3. [DOI] [PubMed] [Google Scholar]
- 19.Morrison B, 3rd, Meaney DF, McIntosh TK. Annals of biomedical engineering. 1998;26:381–390. doi: 10.1114/1.61. [DOI] [PubMed] [Google Scholar]
- 20.Meaney DF, Thibault LE. International Conference on the Biomechanic of Impacts Lyon, France: IRCOBI. 1990:215–224. [Google Scholar]
- 21.Lusardi TA, Rangan J, Sun D, Smith DH, Meaney DF. Annals of biomedical engineering. 2004;32:1546–1558. doi: 10.1114/b:abme.0000049038.75368.75. [DOI] [PubMed] [Google Scholar]
- 22.Gennarelli TA, Thibault LE, Tipperman R, Tomei G, Sergot R, Brown M, Maxwell WL, Graham DI, Adams JH, Irvine A, et al. Journal of neurosurgery. 1989;71:244–253. doi: 10.3171/jns.1989.71.2.0244. [DOI] [PubMed] [Google Scholar]
- 23.Tomei G, Spagnoli D, Ducati A, Landi A, Villani R, Fumagalli G, Sala C, Gennarelli T. Acta neuropathologica. 1990;80:506–513. doi: 10.1007/BF00294611. [DOI] [PubMed] [Google Scholar]
- 24.Tang-Schomer MD, Patel AR, Baas PW, Smith DH. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:1401–1410. doi: 10.1096/fj.09-142844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang-Schomer MD, Johnson VE, Baas PW, Stewart W, Smith DH. Experimental neurology. 2012;233:364–372. doi: 10.1016/j.expneurol.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gittes F, Mickey B, Nettleton J, Howard J. The Journal of cell biology. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cernak I. NeuroRx: the journal of the American Society for Experimental NeuroTherapeutics. 2005;2:410–422. doi: 10.1602/neurorx.2.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf JA, Stys PK, Lusardi T, Meaney D, Smith DH. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2001;21:1923–1930. doi: 10.1523/JNEUROSCI.21-06-01923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison B, 3rd, Saatman KE, Meaney DF, McIntosh TK. Journal of neurotrauma. 1998;15:911–928. doi: 10.1089/neu.1998.15.911. [DOI] [PubMed] [Google Scholar]
- 30.Ellis EF, McKinney JS, Willoughby KA, Liang S, Povlishock JT. Journal of neurotrauma. 1995;12:325–339. doi: 10.1089/neu.1995.12.325. [DOI] [PubMed] [Google Scholar]
- 31.Cargill RS, 2nd, Thibault LE. Journal of neurotrauma. 1996;13:395–407. doi: 10.1089/neu.1996.13.395. [DOI] [PubMed] [Google Scholar]
- 32.Geddes DM, Cargill RS., 2nd Journal of biomechanical engineering. 2001;123:247–255. doi: 10.1115/1.1374201. [DOI] [PubMed] [Google Scholar]
- 33.Sundstrom L, Morrison B, 3rd, Bradley M, Pringle A. Drug discovery today. 2005;10:993–1000. doi: 10.1016/S1359-6446(05)03502-6. [DOI] [PubMed] [Google Scholar]
- 34.Cater HL, Gitterman D, Davis SM, Benham CD, Morrison B, 3rd, Sundstrom LE. Journal of neurochemistry. 2007;101:434–447. doi: 10.1111/j.1471-4159.2006.04379.x. [DOI] [PubMed] [Google Scholar]
- 35.Del Turco D, Deller T. Methods Mol Biol. 2007;399:55–66. doi: 10.1007/978-1-59745-504-6_5. [DOI] [PubMed] [Google Scholar]
- 36.Fischer Y, Gahwiler BH, Thompson SM. The Journal of physiology. 1999;519(Pt 2):405–413. doi: 10.1111/j.1469-7793.1999.0405m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molnar Z, Blakemore C. Experimental neurology. 1999;156:363–393. doi: 10.1006/exnr.1999.7032. [DOI] [PubMed] [Google Scholar]
- 38.Smith DH, Meaney DF. Neuroscientist. 2000;6:483–495. [Google Scholar]
- 39.Cater HL, Sundstrom LE, Morrison B., 3rd Journal of biomechanics. 2006;39:2810–2818. doi: 10.1016/j.jbiomech.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Reeves TM, Phillips LL, Povlishock JT. Experimental neurology. 2005;196:126–137. doi: 10.1016/j.expneurol.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Wyss JM, Swanson LW, Cowan WM. Anatomy and embryology. 1980;158:303–316. doi: 10.1007/BF00301819. [DOI] [PubMed] [Google Scholar]
- 42.Gahwiler BH. Experientia. 1984;40:235–243. doi: 10.1007/BF01947561. [DOI] [PubMed] [Google Scholar]
- 43.Bottlang M, Sommers MB, Lusardi TA, Miesch JJ, Simon RP, Xiong ZG. Journal of neurotrauma. 2007;24:1068–1077. doi: 10.1089/neu.2006.3772. [DOI] [PubMed] [Google Scholar]
- 44.Kawalec M, Kowalczyk JE, Beresewicz M, Lipkowski AW, Zablocka B. Neurochemical research. 2011;36:2091–2095. doi: 10.1007/s11064-011-0568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buchs PA, Stoppini L, Muller D. Brain research Developmental brain research. 1993;71:81–91. doi: 10.1016/0165-3806(93)90108-m. [DOI] [PubMed] [Google Scholar]
- 46.Thibault KL, Margulies SS. Journal of biomechanics. 1998;31:1119–1126. doi: 10.1016/s0021-9290(98)00122-5. [DOI] [PubMed] [Google Scholar]
- 47.Kilinc D, Gallo G, Barbee KA. Experimental neurology. 2008;212:422–430. doi: 10.1016/j.expneurol.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 48.Yu Z, Graudejus O, Tsay C, Lacour SP, Wagner S, Morrison B., 3rd Journal of neurotrauma. 2009;26:1135–1145. doi: 10.1089/neu.2008.0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowenstein DH. Epilepsia. 2009;50(Suppl 2):4–9. doi: 10.1111/j.1528-1167.2008.02004.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.