Abstract
Nonsteroidal anti-inflammatory drugs (NSAIDs) have been widely reported to inhibit tumor growth by a COX-independent mechanism, although alternative targets have not been well defined or used to develop improved drugs for cancer chemoprevention. Here, we characterize a novel sulindac derivative referred to as sulindac benzylamine (SBA) that does not inhibit COX-1 or COX-2, yet potently inhibits the growth and induces the apoptosis of human colon tumor cells. The basis for this activity appears to involve cyclic guanosine 3′,5′,-monophosphate phosphodiesterase (cGMP PDE) inhibition as evident by its ability to inhibit cGMP hydrolysis in colon tumor cell lysates and purified cGMP-specific PDE5, increase intracellular cGMP levels, and activate cGMP-dependent protein kinase G at concentrations that suppress tumor cell growth. PDE5 was found to be essential for colon tumor cell growth as determined by siRNA knockdown studies, elevated in colon tumor cells as compared with normal colonocytes, and associated with the tumor selectivity of SBA. SBA activation of PKG may suppress the oncogenic activity of β-catenin as evident by its ability to reduce β-catenin nuclear levels, Tcf (T-cell factor) transcriptional activity, and survivin levels. These events preceded apoptosis induction and appear to result from a rapid elevation of intracellular cGMP levels following cGMP PDE inhibition. We conclude that PDE5 and possibly other cGMP degrading isozymes can be targeted to develop safer and more efficacious NSAID derivatives for colorectal cancer chemoprevention.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world that accounts for approximately 600,000 deaths per year. While colonoscopy allows for the early detection of disease and the identification of individuals who are at high risk of disease progression, the mortality rate from CRC has decreased only marginally in the last 2 decades (1). In addition, certain lesions such as flat adenomas cannot be readily detected by colonoscopy (2) and surgical management of adenomas in high-risk individuals, such as with familial adenomatous polyposis (FAP) often requires complete or segmental removal of the colon (3). Given the slow progression of carcinogenesis and the limitations of colonoscopy, much research has focused on cancer chemoprevention to reduce the development and progression of CRC.
One class of drugs that has shown promise for chemoprevention is the nonsteroidal anti-inflammatory drugs (NSAIDs), a chemically diverse family of drugs commonly used for the treatment of pain, fever, and inflammation. Epidemiologic studies have shown that long-term use of NSAIDs such as aspirin can significantly reduce the incidence and risk of death from CRC (4). In addition, certain prescription strength NSAIDs, such as sulindac can cause the regression and prevent recurrence of adenomas in individuals with FAP (5). The antineoplastic activity of NSAIDs is widely attributed to their COX inhibitory activity because prostaglandins are elevated in colon tumors (6) and a significant percentage of colon tumors express high levels of the inducible COX-2 isozyme (7). However, there is evidence that alternative mechanisms either contribute to or fully account for the CRC chemopreventive activity of NSAIDs (8–10). For example, the non-COX inhibitory sulfone metabolite of sulindac has been reported to inhibit the growth and induce apoptosis of colon tumor cells in vitro (11, 12) and suppress colon tumorigenesis in animal models (13–15). Sulindac sulfone (exisulind) was also shown to suppress adenoma formation in individuals with FAP or sporadic adenomas (16, 17) but did not receive U.S. Food and Drug Administration (FDA) approval due to hepatotoxicity. Nonetheless, because the use of NSAIDs is associated with gastrointestinal, renal, and cardiovascular toxicities from suppressing prostaglandin synthesis (18, 19), the investigation of COX-independent mechanisms may provide insight that could lead to new drug candidates that are potentially safer and more efficacious for CRC chemoprevention.
Previous studies have suggested that there is a close association between the antineoplastic activity of NSAIDs and their ability to suppress Wnt/β-catenin signaling in colon tumor cells. For example, studies have shown that certain NSAIDs can decrease nuclear levels of β-catenin to inhibit the transcription of genes (e.g., cyclin D, survivin) that provide a survival advantage to allow for clonal expansion of neoplastic cells (20–22). Several groups have reported that sulindac sulfone can also induce proteosomal degradation of oncogenic β-catenin, which suggests that the underlying biochemical mechanism by which NSAIDs suppress β-catenin signaling may not require COX inhibition (22–24).
The mechanism responsible for the antineoplastic activity of sulindac sulfone has been previously reported to involve cyclic guanosine 3′,5′,-monophosphate (cGMP) phosphodiesterase (PDE) inhibition, although the specific isozymes involved were not identified (23, 25). More recently, we reported that the COX inhibitory sulfide metabolite of sulindac and certain other NSAIDs also inhibit cGMP PDE and that this activity is closely associated with PDE5 inhibition and their tumor cell growth inhibitory and apoptosis-inducing properties (26–28). Cyclic nucleotide PDEs are a large superfamily of enzymes responsible for regulating second messenger signaling by hydrolyzing the 3′,5′-phosphodiester bond in cGMP and/or cAMP. There are at least 11 PDE isozyme family members having different substrate specificity, regulatory properties, tissue localization, and inhibitor sensitivity (29). PDE1, 2, 3, 10, and 11 are dual substrate-degrading isozymes, whereas PDE5, 6, and 9 are selective for cGMP and PDE4, 7, and 8 are cAMP selective. In addition, each isozyme family contains multiple isoforms or splice variants. Depending on the PDE isozyme content of the target cell population and inhibitor selectivity, PDE inhibitors can increase the magnitude and/ or the duration of the cAMP and/or cGMP intracellular signal(s). Increasing cyclic nucleotide levels can induce specific signaling pathways, which, in the case of cGMP, can activate protein kinase G (PKG) to regulate cellular activity (30).
Here, we characterize the anticancer activity of a novel benzylamine derivative of sulindac that does not inhibit COX-1 or COX-2, yet can potently inhibit the growth of colon tumor cells by inhibiting proliferation and inducing apoptosis. The underlying biochemical mechanism appears to involve cGMP PDE inhibition as evident by its ability to selectivity inhibit cGMP hydrolysis in whole-cell lysates, as well as purified PDE5. Moreover, treatment of colon tumor cells with sulindac benzylamine (SBA) increased intracellular cGMP levels and activated cGMP-dependent PKG in colon tumor cells at concentrations that paralleled those required for inhibiting cGMP PDE/PDE5 and colon tumor cell growth. PKG activation by SBA was also found to be associated with decreased nuclear levels of β-catenin, T-cell factor (Tcf) transcriptional activity, and the suppression of the apoptosis regulatory protein, survivin; all of which preceded apoptosis induction.
Materials and Methods
Drugs and reagents
Sulindac sulfide was purchased from Sigma-Aldrich. Recombinant PDE isozymes were purchased from BPS Biosciences. Isozyme-specific PDE antibodies were purchased from GeneTex, whereas vasoactive stimulatory protein (VASP) antibodies were purchased from BD Transduction Laboratories. All other antibodies were purchased from Cell Signaling Technologies. Nontargeting control siRNA, PDE5-specific siRNAs, and the SureFECT transfection reagent were purchased from SA Biosciences. All other reagents were purchased from Sigma unless otherwise specified.
Sulindac benzylamine synthesis
SBA or [(Z)-N-benzyl-2-(5-fluoro-2-methyl-1-(4-(methylsulfinyl) benzylidene)-1H-inden-3-yl)ethanamine] was synthesized by converting sulindac to sulindac methyl ester. Refluxing sulindac acid in methanol in the presence of concentrated sulfuric acid, followed by recrystallization from ethyl ether gave the methyl ester as a yellow solid in quantitative yield. The ester was dissolved in methylene chloride and then treated with diisobutylaluminum hydride (2 mol/L in toluene) at –77° C. After stirring for 4 hours, the reaction was quenched by methanol. The temperature was slowly raised to 0° C, and the reaction solution was washed with aqueous solution of acetic acid. The organic layer was concentrated and dried in vacuum overnight to give the aldehyde as a yellow syrup. The aldehyde solution in ethanol was treated with benzylamine at –77° C in for 3 hours, followed bysodiumborohydride for 30 minutes. Methanol was added, and the temperature was slowly brought up to –40° C and stirred for 2 hours. Acetic acid was added slowly to quenchthe reaction. The reaction mixture was concentrated, purified with a silica gel column, and recrystallized from chloroform/acetone/ether 3 times, which resulted in a yellow solid. The final product was characterized by high resolution mass, nuclear magnetic resonance, and elemental analysis.
Cell culture
The human colon cancer cell lines, HT-29, SW480, and HCT 116 and normal fetal human colonocytes (FHC) were purchased from the American Type Culture Collection. HT-29, SW480, and HCT 116 cells were maintained in RPMI-1640 + 2.0 g/L glucose pH 7.4 + 5% FBS + 4 mmol/L glutamine (complete growth medium), incubated at 37° C in 5% CO2, and passaged at subconfluent density. The human fetal colon FHC cells were cultured in DMEM:Ham's F-12 medium supplemented with 10% FBS, cholera toxin (10 ng/mL), 5 μg/ml transferrin, 5 μg/mL insulin, and 100 ng/mL hydrocortisone. The passage number was routinely limited to approximately 20 and morphology monitored with each passage, but no additional authentication of the cell lines was conducted.
COX activity
COX-1 and COX-2 activities were measured using purified ovine COX-1 and COX-2 with colorimetric assay kits obtained from Cayman Chemical Company as previously reported (31). The activities of COX-1 and COX-2 were measured after the addition of arachidonic acid and incubation at 25° C for 5 minutes by absorbance at 590 nm as specified by the manufacturer.
PGE2 assay
U937 promonocytic cells (32) were differentiated into macrophage-like adherent cells by culture in 10 nmol/L phorbol 12-myristate 13-acetate (PMA) for 48 hours. Differentiated cells were washed with fresh media and plated at a density of 1.0 × 106 cells per well in 96-well half-area plates. Cells were allowed to adhere overnight and stimulated with 10 μg/mL lipopolysaccharide (LPS). Cells were treated with sulindac sulfide or SBA for 24 hours. Prostaglandin PGE2 levels in supernatants were measured using the HTRF PGE2 assay from Cisbio. This detection method uses the binding of exogenous PGE2 to disrupt the fluorescence resonance energy transfer (FRET) between a PGE2–d2 conjugate (acceptor) and a PGE2 antibody (donor).
PDE activity
PDE activity was measured by the IMAP fluorescence polarization assay (Molecular Devices) in which binding of hydrolyzed cyclic nucleotide substrate to immobilized metal coordination complexes increases fluorescence polarization as previously described (26). For studies involving tumor cell lysates, human colon tumor cells were harvested and lysed with PDE activity buffer (20 mmol/L Tris-acetate, 5 mmol/L magnesium acetate, 1 mmol/L EGTA, 1.0% Triton X-100, 50 mmol/L NaF, and protease inhibitor cocktail at pH 7.4). Tetramethylrhodamine (TAMRA)-cGMP and fluorescein-cAMP were used as substrates, each at final concentration of 50 nmol/L. Cell lysates were titrated to identify a suitable protein concentration that was in the mid portion of a concentration versus PDE activity curve. The PDE assay was done according to the manufacturer's specifications using either whole-cell lysates or recombinant enzymes. Fluorescence polarization was measured at excitation, emission wavelengths of either 530,590 nm for TAMRA-cGMP or 485,530 nm for fluorescein-cAMP using a Synergy4 (Biotek) microplate reader.
Growth assays
Cells were plated in 96-well microtiter plates at a density of 5,000 cells per well and allowed to adhere overnight before treatment. Cells were treated with a 2-log concentration range of either sulindac sulfide or SBA and incubated for an additional 72 hours. For siRNA assays, cells were transfected in OptiMEM media with 0.5% SureFECT transfection reagent and 200 nmol/L of either negative control or PDE5 siRNA and incubated at 37° C for 24 hours before treatment. At the end of the incubation period, relative cell viability was compared with vehicle controls using the CellTiter-Glo Assay (Promega), which measures viable cells based on ATP content, according to the manufacturer's specifications.
Proliferation assays
The antiproliferative activity of sulindac sulfide and SBA was determined by measuring EdU (5-ethynyl-2′-deoxyuridine) incorporation during DNA synthesis. Cells were seeded at a density of 1.5 × 106 cells per 10-cm tissue culture dish and incubated overnight at 37° C in 5% CO2. After growing the cells in serum-free medium for 24 hours, the cultures were treated with sulindac sulfide, SBA, or vehicle [0.1% dimethyl sulfoxide (DMSO)] in RPMI-1640 media with 5% serum for 4 hours. A final concentration of 16 μmol/L EdU was added to each dish and incubated for an additional 18 hours. Cells were harvested and analyzed using the Click-iT EdU Alexa Fluor 488 flow cytometry assay kit (Invitrogen) according to the manufacturer's specifications. Proliferating cells were quantified using a Guava EasyCyte Plus flow cytometer. A minimum of 5,000 events were collected in triplicate for each treatment group.
Apoptosis assays
HCT 116 cells were seeded at a density of 1 × 106 cells per 10-cm tissue culture dish, incubated for 48 hours, and treated with the specified compound or vehicle control. After 24 hours of treatment, cells were harvested from the treatment media and dish and fixed with 10% neutral buffered formalin (4% formaldehyde) on ice for 15 minutes. Samples were stained for DNA strand breaks using the APO-BrdU terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) assay (Invitrogen), which labels bromodeoxyuridine (BrdUrd) incorporation into DNA strand breaks with AlexaFluor-488. The assay was conducted according to the manufacturer's specifications. The percentage of TUNEL-positive cells was quantified using a Guava EasyCyte Plus flow cytometer. A minimum of 5,000 events were collected in triplicate for each treatment group with minimal electronic compensation. Data were analyzed with CytoSoft 5.0 Software (Guava Technologies).
Intracellular cGMP levels
Intracellular cGMP levels were measured using the Promega GloSensor cAMP Assay modified to use a firefly luciferase fused to the human PDE5 GAF-A cGMP binding domain (GloSensor cGMP-40F plasmid kindly provided by Promega Corporation). HEK293 cells were plated in 96-well white, clear bottom plates at a density of 25,000 cells per well and incubated overnight at 37° C, 5% CO2, 95% relative humidity. For each well, a final concentration of 100 ng of DNA and 0.25 μL PLUS reagent were added to 19.75 μL of Opti-MEM media and incubated at room temperature for 5 minutes; then 0.35 μL of Lipofectamine LTX reagent was added to the solution, mixed gently, and incubated for 30 minutes at room temperature. To each well 20 μL of DNA-Lipofectamine LTX reagent was added and incubated overnight under standard cell culture conditions. After incubation, the media was replaced with 100 μL of equilibration solution (88% CO2-independent media, 10% FBS, 2% GloSensor cAMP reagent) and incubated for 2 hours at room temperature protected from light. Background measurements were obtained 15 minutes before the end of the equilibration incubation by reading on a PerkinElmer Victor 3 luminometer. After the background measurements, sodium nitroprusside (SNP) or SNP plus SBA was added to the wells in 10 μL of CO2-independent media. The final concentration of SNP was 50 μmol/L. Plates were read every 2 minutes for 1 hour on the Victor 3 luminometer.
β-Catenin–mediated Tcf transcriptional activity
Experiments to determine the effects of SBA on β-catenin–mediated Tcf transcriptional activity were carried out using the TOP/FOP-flash Tcf reporter constructs as described previously (33). The TOP-flash plasmid contains Tcf binding sites for β-catenin, whereas the FOP-flash plasmid has mutated Tcf binding sites, which serves as a control for measuring nonspecific activation of the reporter. In brief, HCT 116 cells were plated in 24-well plates and cotransfected with 0.1 μg TOP-flash or FOP-flash plasmids and 0.1 μg β-galactosidase–expressing vector. Following treatment with SBA for 24 hours, luciferase activity was determined and normalized with activity of β-galactosidase. These data are expressed as the mean and SEM of triplicate values of the normalized TOP-flash and FOP-flash activity.
Western blotting
For Western blotting, cells were lysed in either ice-cold membrane lysis buffer (1.0% Triton X-100, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 1 mmol/L DTT, 10 mmol/L HEPES, 50 mmol/L NaF, and protease inhibitor cocktail), or nuclear lysis buffer (20 mmol/L HEPES, 1.5 mmol/L MgCl2, 200 μmol/L EDTA, 1 mmol/L DTT, 400 mmol/L NaCl, 25% glycerol, and protease inhibitor cocktail). For nuclear fractionation, cell lysate was maintained at 4° C and vortexed for 30 seconds every 5 minutes for 1 hour, followed by centrifugation for 15 minutes at 15,000 rpm. Protein concentrations were determined by the Lowry method. Proteins were separated by SDS-PAGE on 12% PAGE before transfer to nitrocellulose membranes. Membranes were blocked for 1 hour in 5% bovine serum albumin, 0.05% Tween 20, and incubated with primary antibodies overnight at 4° C. Membranes were incubated with horseradish peroxidase–conjugated secondary antibody for 1 hour followed by incubation with Super Signal West Pico Enhanced Chemiluminescence Reagent (Pierce). Protein bands were visualized on Hyblot CL (Denville Scientific) autoradiography film.
Molecular modeling
Molecular modeling was conducted using Schrödinger Suite 2010 (Schrödinger, LLC). The structural model of PDE5 catalytic domain was derived from the crystal structure of the PDE5-GMP complex from the protein databank (PDB ID: 1T9S). The induced fit docking (IFD) protocol, which takes into consideration the ligand-induced receptor conformational change, was used for all docking studies. Specifically, residues within 6 é from ligands were allowed to be flexible; docking results were scored using the extra-precision (XP) mode of Glide version 5.6 (Schrödinger, LLC). IDF docking protocol and parameters were first validated by docking GMP in the PDE5 catalytic site, which excellently reproduced the PDE5-GMP crystal complex conformation. The same protocol and parameters were then applied to the docking studies of SBA.
Experimental design and data analysis
For growth assays, the IC50 values were determined by testing a range of 8 concentrations with a minimum of 4 replicates per dose. COX and PDE inhibition assays used a minimum of 3 replicates. Statistical analysis was done using the unpaired 2-tailed Student t test. Significance was assumed for P < 0.05. Error bars represent SD.
Results
SBA inhibits tumor cell growth without COX inhibition
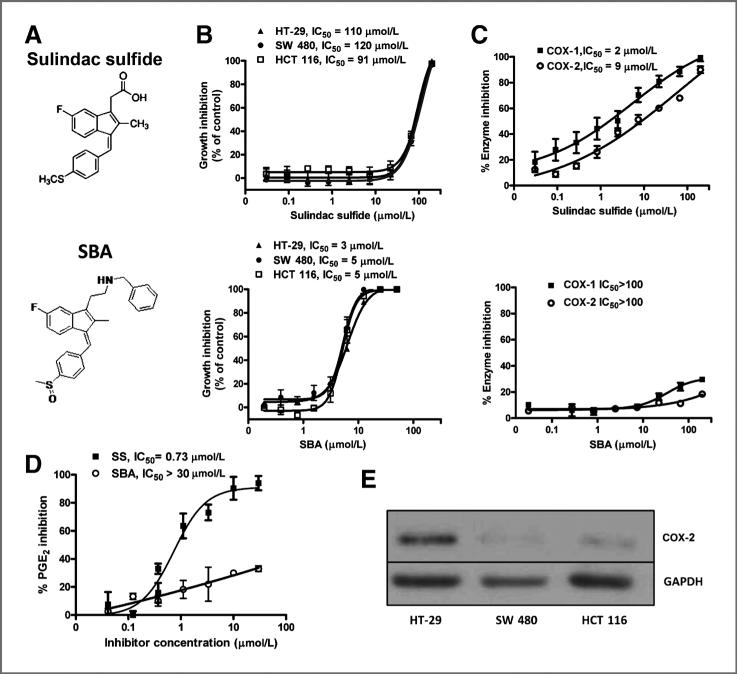
We previously reported that the carboxylic acid moiety of sulindac sulfide is essential for inhibiting COX-1 and COX-2, which could be chemically modified by substituting with a positively charged amide group to effectively block COX binding (31). Such derivatives were unexpectedly found to display enhanced potency to inhibit colon tumor cell growth compared with sulindac sulfide. To chemically optimize for COX-independent tumor cell growth inhibitory activity, we synthesized a large series of sulindac derivatives with various chemical modifications and screened for tumor cell growth and COX inhibitory activity. A benzylamine derivative was identified as shown in Fig. 1A that displayed high potency for inhibiting tumor cell growth, but did not inhibit COX-1 or COX-2.
Figure 1.
A, chemical structures of sulindac sulfide (SS) and SBA. B, tumor cell growth inhibitory activity of SS and SBA as measured by luciferase-based ATP assay after 72 hours of treatment. C, COX-1 and COX-2 inhibitory activity of SS, but not SBA. D, inhibition of PGE2 production by SS, but not SBA in LPS-stimulated U937 promonocytic cells. E, COX-2 protein expression in human HT-29, HCT 116, and SW480 colon cancer cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
As shown in Fig. 1B, SBA treatment for 72 hours inhibited the growth of human HT-29, SW480, and HCT 116 colon tumor cell lines with IC50 values ranging from 3 to 5 μmol/L, whereas sulindac sulfide was appreciably less potent with IC50 values of 90 to 120 μmol/L. SBA completely lacked inhibitory activity for COX-1 and COX-2 at concentrations up to 200 μmol/L (Fig. 1C). By comparison, sulindac sulfide inhibited COX-1 and COX-2 with IC50 values of 2 and 9 μmol/L, respectively. To determine whether SBA inhibited other enzymes involved in arachidonic acid metabolism, we measured treatments effects on the production of PGE2, which is the primary product in most tissues. Although we used a highly sensitive fluorescence-based ELISA assay for measuring PGE2 production, the levels produced by colon tumor cells (e.g., HT-29) did not result in a sufficient signal above noise to allow for inhibition studies. As an alternative method, we used human U937 promonocytic cells treated with LPS, which are commonly used for such assays. As shown in Fig. 1D, sulindac sulfide potently suppressed PGE2 production by these cells with an IC50 value of 0.7 μmol/L, whereas SBA was ineffective, which is consistent with its lack of COX-1 or COX-2 inhibitory activity. We also determined whether the sensitivity of the colon tumor cell lines to sulindac sulfide and SBA was related to the expression of COX-2. As shown in Fig. 1E, HT-29 cells expressed high levels of COX-2 as determined by Western blotting, but COX-2 was expressed to a lesser extent in SW480 or HCT 116 cells, despite no difference in their sensitivity to sulindac sulfide and SBA.
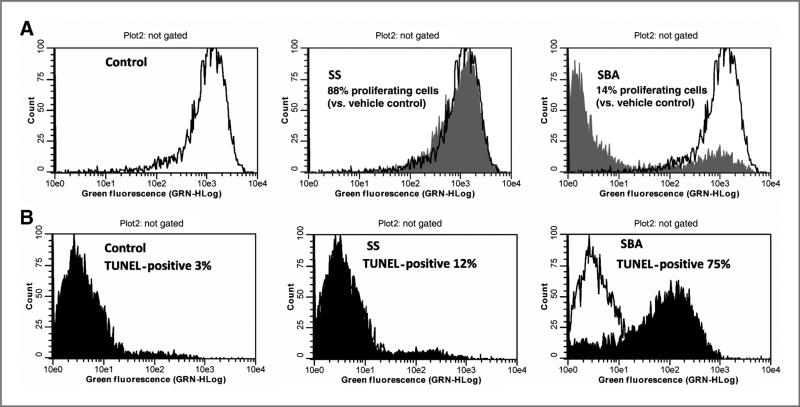
SBA inhibits colon tumor cell growth by inhibiting proliferating and inducing apoptosis
To determine the cellular basis for the tumor cell growth inhibitory activity of SBA, treatment effects on proliferation and apoptosis were measured and compared with sulindac sulfide. As shown in Fig. 2A, treatment of HCT 116 colon tumor cells with sulindac sulfide for 24 hours reduced the number of proliferating cells by approximately 12% relative to vehicle-treated cells. By comparison, treatment with SBA inhibited cell proliferation by 86% after the same treatment period. In addition, treatment with sulindac sulfide increased the number of apoptotic cells from 3% in the vehicle group to 12%, whereas SBA increased apoptotic cells to 75%, respectively (Fig. 2B). These results show that the enhanced potency of SBA compared with sulindac sulfide is associated with increased effectiveness to inhibit proliferation and induce apoptosis.
Figure 2.
A, inhibition of HCT 116 colon tumor cell proliferation after 24-hour treatment with 100 μmol/L SS (middle) or 50 μmol/L SBA (right) as determined by EdU incorporation and flow cytometry. B, apoptosis induction of HCT 116 colon tumor cells after 24-hour treatment with 100 μmol/L SS (middle) or 50 μmol/L SBA (right) as determined by TUNEL and flow cytometry.
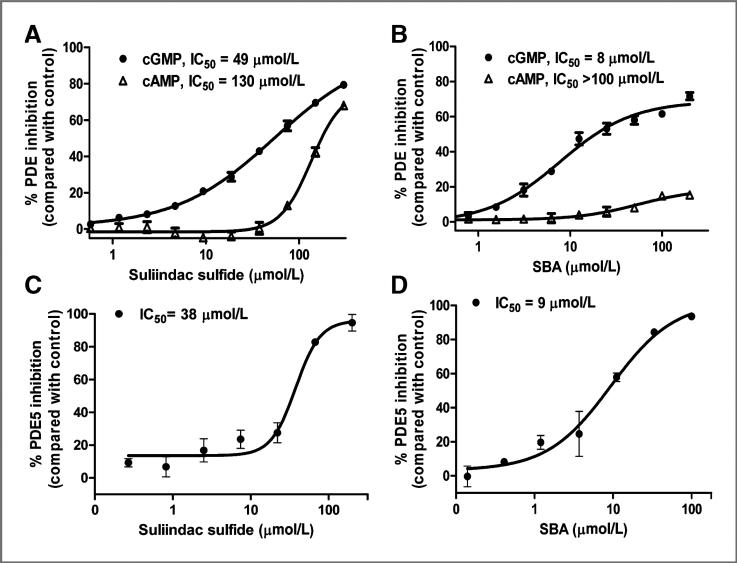
SBA selectively inhibits cGMP PDE
On the basis of previous studies that showed a close association between tumor cell growth inhibitory activity of sulindac sulfide and its ability to inhibit PDE5 (26–28, 34), we determined whether SBA has PDE inhibitory activity. For the initial studies we measured total cGMP and cAMP hydrolysis in whole-cell lysates from HT29 cells using a dual substrate assay of PDE activity as previously described (26). As shown in Fig. 3A, sulindac sulfide inhibited total cGMP hydrolysis with an IC50 value of 49 μmol/L, but appreciably higher concentrations were required to inhibit cAMP hydrolysis with an IC50 value of 133 μmol/L. Corresponding with its improved potency to inhibit tumor cell growth, SBA more potently inhibited cGMP hydrolysis with an IC50 value of 8 μmol/L as shown in Fig. 3B. In contrast with sulindac sulfide, SBA did not inhibit cAMP hydrolysis at concentrations as high as 200 μmol/L, which suggests the potential for improved isozyme selectivity compared with sulindac sulfide. The PDE5 inhibitory activity of SBA was next determined by conducting similar assays except using purified recombinant PDE5. As shown in Fig. 3C, sulindac sulfide inhibited PDE5 with an IC50 value of 38 μmol/L. SBA was appreciably more potent with an IC50 value of 9 μmol/L as shown in Fig. 3D. The potency values as determined using purified PDE5 were comparable with values as measured using whole tumor cell lysates, although less inhibition was observed in the latter, which is likely attributed to the presence of insensitive cGMP PDE degrading isozymes as described later.
Figure 3.
A, inhibition of cAMP and cGMP PDE activity by SS in HT-29 tumor cell lysate. B, inhibition of cAMP and cGMP PDE activity by SBA in HT-29 cell lysate. C, PDE5 inhibition by SS. D, PDE5 inhibition by SBA.
The PDE isozyme selectivity of sulindac sulfide and SBA was next measured using a panel of recombinant PDE isozymes. As summarized in Table 1, SBA was highly selective for PDE5. All other PDE isozymes were either insensitive or resulted in IC50 values that appreciably exceeded the concentration range required to inhibit colon tumor cell growth. PDE5 was also the most sensitive isozyme to sulindac sulfide, although sulindac sulfide also inhibited PDE2, PDE3, and PDE10 within the same concentration range as required for tumor cell growth inhibition, which suggests that the increased potency of SBA to inhibit tumor cell growth is associated with increased potency and selectivity to inhibit PDE5. However, because these studies are limited to a group of PDE isoforms that were commercially available, we cannot rule out the potential involvement of additional isozymes that may be expressed in colon tumor cells and sensitive to SBA.
Table 1.
Potency of SS and SBA to inhibit cAMP and cGMP hydrolysis by PDE isozymes
| SBA sensitivity |
SS sensitivity |
||||
|---|---|---|---|---|---|
| PDE family | Substrate | cGMP IC50, μmol/L | cAMP IC50, μmol/L | cGMP IC50, μmol/L | cAMP IC50, μmol/L |
| 1A | cAMP/cGMP | >100 | >100 | >100 | >100 |
| 1B | cAMP/cGMP | >100 | >100 | >100 | >100 |
| 1C | cAMP/cGMP | >100 | >100 | >100 | >100 |
| 2A | cAMP/cGMP | >100 | >100 | 97 | >100 |
| 3A | cAMP/cGMP | >100 | >100 | 84 | >100 |
| 3B | cAMP/cGMP | >100 | >100 | >100 | >100 |
| 4B | cAMP | ND | 75 | ND | >100 |
| 5A | cGMP | 9 | ND | 38 | ND |
| 6C | cGMP | >100 | ND | >100 | ND |
| 7A | cAMP | ND | >100 | ND | >100 |
| 8A | cAMP | ND | >100 | ND | >100 |
| 9A | cGMP | >100 | ND | >100 | ND |
| 10A | cAMP/cGMP | 62 | 77 | 70 | >100 |
| 11A | cAMP/cGMP | >100 | >100 | >100 | >100 |
Abbreviation: ND, not determined.
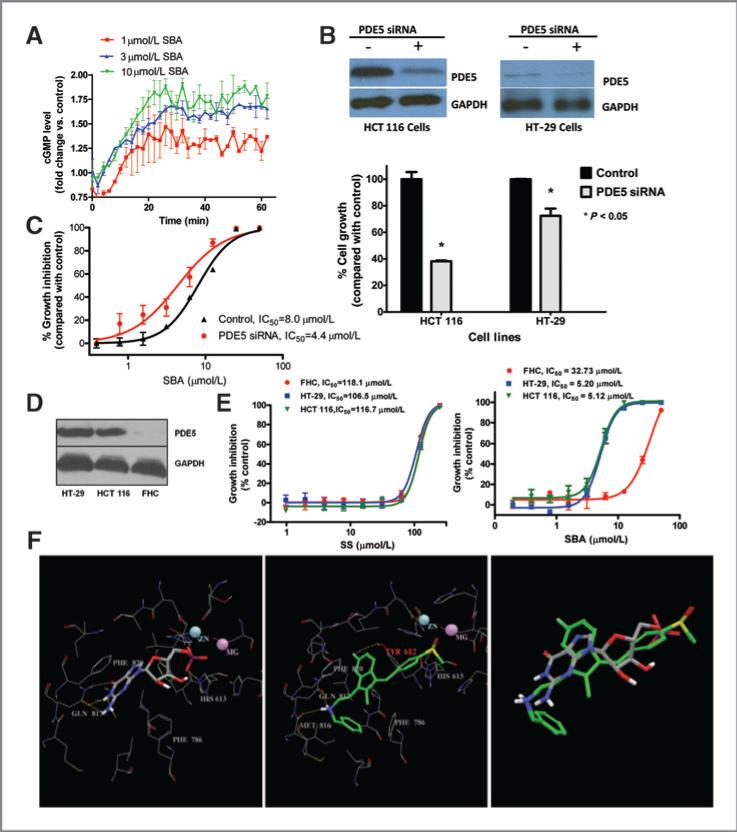
Elevation of intracellular cGMP by SBA
We previously reported that sulindac sulfide can increase intracellular cGMP levels as measured by a standard immunoassay method (26). To determine whether SBA can increase intracellular cGMP levels, we used HEK293 cells transfected with a construct of cGMP binding (GAF-A) protein fused to firefly luciferase. This luminescence assay in live cells allowed for kinetic measurements of intracellular cGMP levels in response to treatment. As shown in Fig. 4A, SBA treatment caused a rapid and sustained increase in luminescence that occurred within the same concentration range as required for cGMP PDE inhibition in cell lysates. Others studies confirmed the expression of PDE5 in HEK293 cells (data not shown), which suggest that cGMP elevation by SBA in this cell model results from PDE5 inhibition.
Figure 4.
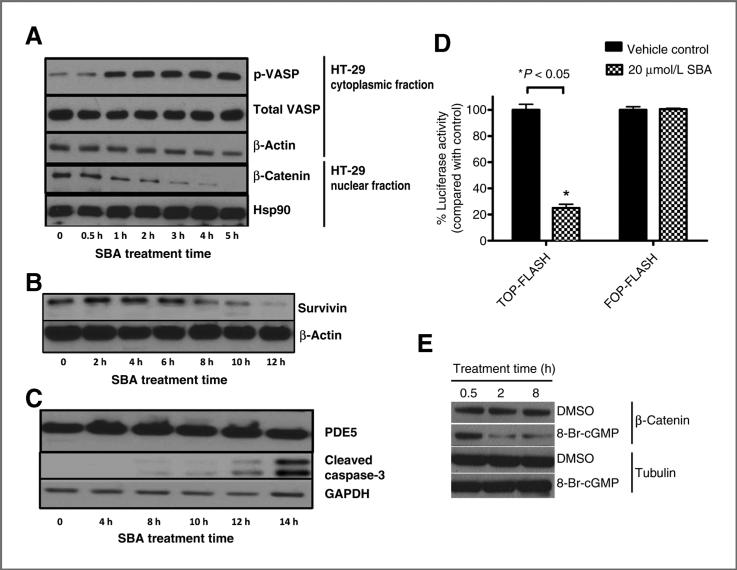
A, luciferase-based cGMP biosensor assay in HEK293 cells treated with 50 μmol/L SNP in the presence of 1, 3, or 10 μmol/L SBA. B, siRNA suppression of PDE5 and growth in human HCT 116 and HT-29 colon tumor cells following 72-hour transfection. C, growth inhibition by SBA of PDE5 siRNA knockdown HCT 116 cells compared with controls. D, PDE5 expression in human HCT 116 and HT-29 colon tumor cells and FHC. E, growth inhibitory activity of SBA and SS in human HCT 116 and HT-29 colon tumor cells compared with FHC. F, structural representation of GMP and SBA binding to PDE5. The panels show GMP-bound PDE5 (left), docked PDE5-SBA complex structure (middle), and the aligned GMP and SBA (right). Residues within 4 Å of the bound/docked molecules are shown by lines. GMP and SBA molecules are represented in solid sticks and their carbon atoms are colored in gray and green, respectively. Hydrogen-bonds are marked in yellow dashed lines.
PDE5 suppression by siRNA inhibits colon tumor cell growth and increases sensitivity to SBA
To assess the possibility that PDE5 is necessary for colon tumor cell growth, HCT 116 and HT-29 colon tumor cells were transfected with PDE5 siRNA to selectively suppress the expression of the enzyme. Western blot analysis as shown in Fig. 4B confirmed that PDE5 siRNA reduced PDE5 protein levels. PDE5 knockdown by siRNA inhibited the growth of HCT 116 and HT-29 cells by 60% and 30%, respectively, compared with cells transfected with nontargeted siRNA. In addition, PDE5 siRNA knockdown HCT 116 cells displayed increased sensitivity to SBA treatment as evident by a 2-fold decrease in its IC50 value compared with control cells as shown in Fig. 4C.
Tumor cell growth inhibition by SBA is associated with PDE5 expression
To further study the possibility that PDE5 expression can influence the sensitivity of colon tumor cells to SBA, PDE5 levels were measured in normal human colonocytes (FHC) and HT-29 and HCT 116 colon tumor cells. As shown in Fig. 4D by Western blotting, PDE5 was not be detected in FHC, whereas both colon tumor cell lines expressed relatively high levels of the enzyme. Consistent with these observations, SBA inhibited the growth of both tumor cell lines with an IC50 value of 5 μmol/L, whereas FHC were appreciably less sensitive with greater than a 6-fold higher IC50 value of 33 μmol/L (Fig. 4D). In contrast, sulindac sulfide did not show evidence of tumor selectivity, which may be attributed to its nonselective cGMP PDE inhibitory activity.
Mechanism of PDE5 inhibition
Molecular modeling studies were conducted to determine the mechanism by which SBA inhibits PDE5. For these studies we used a crystal structure of PDE5 complexed with the reaction product, GMP. Docking results as shown in Fig. 4F indicate that the heterocyclic ring scaffold of SBA effectively overlays with the guanine fragment of GMP that occupies the same central hydrophobic area within the catalytic site of PDE5 located between Phe829 and Phe786. Multiple hydrogen bonds were formed in the PDE5-GMP structure between the guanine of GMP and Gln817, which is an important amino acid residue that is necessary for substrate and inhibitor binding (33). Hydrogen bond interactions were also observed between Gln817 and the amine group of SBA as evident by the docked SBA-PDE5 complex structure. In addition, the sulfoxide group of SBA occupied the polar area near the 2 metal ions, similar to the phosphate group of GMP. Likely important for enzyme inhibition, SBA occupied a hydrophobic region through its phenyl group, which is close to the surface of PDE5 that is not occupied by the GMP molecule in the GMP-PDE5 complex structure. These results indicate that SBA directly binds the catalytic domain of PDE5.
PKG activation by SBA
To study the downstream events that may occur in response to cGMP PDE inhibition and cGMP elevation, we initially determined whether SBA could activate PKG in colon tumor cells. For these experiments, HT-29 cells were treated with SBA and the phosphorylation of the known PKG substrate, VASP, was measured using a phospho-specific VASP antibody that is selective for cGMP-stimulated phosphorylation at the Ser239 residue. As shown in Fig. 5A, the levels of phospho-VASP increased within 1 hour of SBA treatment at a concentration of 5 μmol/L and remained elevated for the duration of the experiment (5 hours). These experiments provide evidence that PKG activation occurs in response to SBA treatment at concentrations and times that paralleled those required for cGMP PDE inhibition and increased intracellular cGMP levels, respectively.
Figure 5.
A, increased VASP phosphorylation and time-dependent decrease in nuclear β-catenin levels in HT-29 cells treated with SBA. B, decreased β-catenin–regulated protein, survivin after 10 μmol/L treatment with SBA. C, caspase-3 cleavage and PDE5 expression in HT-29 cells treated with 10 μmol/L SBA. D, Wnt/β-catenin signaling reporter activity in HCT 116 cells treated with SBA for 24 hours. E, suppression of β-catenin by 8-BrcGMP (100 μmol/L) in HCT 116 cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Suppression of nuclear β-catenin levels and transcriptional activity by SBA
Previous studies have shown that PKG can phosphorylate β-catenin to reduce cytoplasmic and nuclear levels of β-catenin by a mechanism that appears to involve the activation of ubiquitin-mediated proteosomal degradation (23, 35). To determine whether SBA can reduce β-catenin levels within the same time period as PKG activation, nuclear β-catenin levels were measured in the same HT-29 cell lysates as used for experiments to measure PKG activation. As shown in Fig. 5A, SBA reduced nuclear levels of β-catenin within 1 hour of treatment, which paralleled the time required for activating PKG. The expression of the apoptosis regulatory protein, survivin, in which its synthesis is under the control of the β-catenin/Tcf-Lef transcription factor (36), was decreased after a slightly longer duration of treatment (Fig. 5B). Levels of cleaved caspase-3 were also measured as a biochemical marker of apoptosis and found to be increased by SBA treatment at time points that matched those where survivin levels were suppressed and occurred after the levels of nuclear β-catenin were decreased (Fig. 5C). There were no changes in PDE5 levels during the treatment period indicating that this enzyme remained stable during apoptosis.
To determine whether SBA can inhibit β-catenin–mediated Tcf transcriptional activity, we used the TOP/FOP-flash Tcf luciferase assay. As shown in Fig. 5D, SBA caused a significant decrease in β-catenin–mediated Tcf transcriptional activity. Finally, we confirmed that the PKG activator, 8-bromo-cGMP can also suppress β-catenin levels, which provides additional evidence that the cGMP and β-catenin pathways are interconnected (Fig. 5E).
Discussion
These results characterize the activity of a novel benzylamine derivative of sulindac that lack COX inhibitory activity but displays high potency to suppress the growth of colon tumor cells in vitro. The improved growth inhibitory potency of SBA was associated with increased effectiveness to inhibit proliferation and induce apoptosis of colon tumor cells. Similar to observations we previously reported for sulindac and other NSAIDs, the underlying biochemical mechanism responsible for the tumor cell growth inhibitory activity of SBA appears to involve cGMP PDE inhibition, although these results are novel as they show that derivatives can be synthesized that lack COX inhibitory activity, which display higher potency to inhibit colon tumor cell growth. The cGMP PDE inhibitory activity of SBA was evident by its ability to selectively inhibit cGMP hydrolysis by lysates from colon tumor cells as well as by recombinant PDE5 within the same concentration range as required for inhibition of colon tumor cell growth. Moreover, SBA increased intracellular cGMP levels and activated PKG within the same concentration range as required for cGMP PDE and tumor cell growth inhibition. Consistent with studies showing the ability of PKG to phosphorylate β-catenin to induce degradation, SBA reduced nuclear levels of β-catenin, its transcriptional activity, and survivin levels within a treatment period that preceded the induction of apoptosis. PDE5 siRNA knockdown experiments show that PDE5 is essential for growth of colon tumor cells that can impact the sensitivity to SBA. These observations provide strong evidence that PDE5 inhibition represents an important COX-independent mechanism for the anticancer properties of NSAIDs that can be targeted to develop potentially safer and more efficacious derivatives for CRC chemoprevention.
Our observations are consistent with studies by others who have also concluded that a COX-independent mechanism is fully responsible for or can contribute to the cancer chemopreventive activity of NSAIDs (10, 11, 37). For example, NSAIDs have been shown to suppress the growth of malignant cell lines that do not express COX-2 (37) and supplementation with prostaglandins do not reverse the inhibitory activity on cellular growth (38). In addition, the rank order potency among NSAIDs to inhibit prostaglandin synthesis and growth of tumor cells does not correlate (39) as higher doses of NSAIDs are generally required to inhibit tumor cell growth (40). Although the mechanism by which NSAIDs suppress tumorigenesis in vivo may involve both COX-dependent and independent effects, these results suggest that their intrinsic tumor cell growth inhibitory activity only involves a COX-independent mechanism.
Here, we show that SBA potently and selectively inhibits cGMP hydrolysis by colon tumor cell lysates without affecting cAMP hydrolysis. This was confirmed by studies using recombinant PDE isozymes where SBA selectively inhibited the cGMP-specific PDE5 isozyme. On the other hand, sulindac sulfide inhibited multiple cGMP PDE isozymes, including PDE2, 3, 5, and 10. Consistent with the importance of PDE5 as a target, we showed that this isozyme is essential for colon tumor cell growth by siRNA knockdown studies. PDE5 knockdown by siRNA also increased the sensitivity of colon tumor cells to SBA treatment. In addition, PDE5 was found to be elevated in human colon tumor cells compared with normal colonocytes and that the expression level was associated with the tumor selectivity of SBA. These observations are consistent with previous immunohistochemistry studies showing that PDE5 is overexpressed in human colon adenomas and adenocarcinomas as well as in bladder, lung, and breast tumors (25, 28, 41, 42). However, we cannot rule out the potential involvement of additional cGMP PDE isozymes that might be expressed in tumor cells and sensitive to this class of compounds. In support of this possibility, FDA-approved PDE5 inhibitors such as tadalafil used for the treatment of erectile dysfunction require high concentrations to inhibit tumor cell growth (micromolar levels) compared with concentrations required to inhibit PDE5 in cell-free assays (nanomolar levels; ref. 28), whereas others such as sildenafil are completely inactive.
Of relevance to our findings that cGMP elevation can inhibit growth and induce apoptosis of colon tumor cells, other investigators have shown an association between cGMP elevation and inhibition of colorectal tumorigenesis. Most notably, Shailubhai and colleagues showed that oral administration of the enteric peptide hormone, uroguanylin, which binds a membrane-associated guanylyl cyclase–coupled receptor to increase intracellular cGMP levels, inhibited tumor formation in the Apcmin mouse model, and increased rates of apoptosis within the tumors (43). These experiments also reported that uroguanylin levels were reduced in both colon adenomas and adenocarcinomas, suggesting that cGMP levels may be aberrantly low in colon tumors than in normal mucosa. Other studies have shown that human colon tumor cell lines transfected with constitutively activated mutants of PKG can undergo apoptosis and are unable to form colonies (44). PKG is also downregulated in many cancer types, including CRC (45), which suggests that suppression of the cGMP pathway may provide a growth or survival advantage to tumor cells.
Given that β-catenin is an important oncogenic protein involved in CRC, the observations that SBA can suppress β-catenin and its transcriptional activity support the possibility that this non-COX inhibitory derivative of sulindac (or related analogues) will be effective for CRC chemoprevention as has been established for other NSAIDs that inhibit COX-1 and/or COX-2. We found that SBA treatment can induce β-catenin degradation, which is consistent with previous studies showing that PKG can phosphorylate β-catenin to induce degradation (23, 46). The time-dependent activation of PKG by SBA paralleled the time required for cGMP elevation that is followed by the suppression of β-catenin nuclear and survivin levels. These events preceded caspase activation and suggest that the cGMP/PKG and Wnt/β-catenin pathways are interconnected to trigger conditions that lead to apoptosis induction.
In conclusion, these results support further studies to evaluate the efficacy and toxicity of SBA or other non-COX inhibitory derivatives of sulindac for cancer chemoprevention in animal models. Additional studies are also warranted to better define the role of cGMP and cGMP-degrading PDE isozymes in colorectal tumorigenesis.
Acknowledgments
Grant Support
This research was supported by NIH grants, NCI 1R01CA131378 and 1R01CA148817-01A1, and 1R21CA160280-01A1.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Authors’ Contributions
Conception and design: N. Li, A.H. Abadi, W.E. Grizzle, G. Piazza
Development of methodology: H.N. Tinsley, A.B. Keeton, A.H. Abadi, W.E. Grizzle, G. Piazza
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): J.D. Whitt, N. Li, H.N. Tinsley, Y. Li, B. Gary, W.E. Grizzle, G. Piazza
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.D. Whitt, N. Li, H.N. Tinsley, X. Chen, W. Zhang, A.B. Keeton, Y. Xi, A.H. Abadi, G. Piazza
Writing, review, and/or revision of the manuscript: J.D. Whitt, N. Li, H. N. Tinsley, X. Chen, W. Zhang, B. Gary, A.B. Keeton, Y. Xi, A.H. Abadi, W.E. Grizzle, G. Piazza
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): H.N. Tinsley, X. Chen, G. Piazza
Study supervision: G. Piazza
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JC. Risk factors and diagnosis of flat adenomas of the colon. Expert Rev Gastroenterol Hepatol. 2011;5:25–32. doi: 10.1586/egh.10.86. [DOI] [PubMed] [Google Scholar]
- 3.Johnson MD, Mackey R, Brown N, Church J, Burke C, Walsh RM. Outcome based on management for duodenal adenomas: sporadic versus familial disease. J Gastrointest Surg. 2010;14:229–35. doi: 10.1007/s11605-009-1091-4. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Rodriguez LA, Huerta-Alvarez C. Reduced risk of colorectal cancer among long-term users of aspirin and nonaspirin nonsteroidal antiinflammatory drugs. Epidemiology. 2001;12:88–93. doi: 10.1097/00001648-200101000-00015. [DOI] [PubMed] [Google Scholar]
- 5.Giardiello FM, Hamilton SR, Krush AJ, Piantadosi S, Hylind LM, Celano P, et al. Treatment of colonic and rectal adenomas with sulindac in familial adenomatous polyposis. N Engl J Med. 1993;328:1313–6. doi: 10.1056/NEJM199305063281805. [DOI] [PubMed] [Google Scholar]
- 6.Rigas B, Goldman IS, Levine L. Altered eicosanoid levels in human colon cancer. J Lab Clin Med. 1993;122:518–23. [PubMed] [Google Scholar]
- 7.Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183–8. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 8.Hwang DH, Fung V, Dannenberg AJ. National Cancer Institute workshop on chemopreventive properties of nonsteroidal anti-inflamma-tory drugs: role of COX-dependent and -independent mechanisms. Neoplasia. 2002;4:91–7. doi: 10.1038/sj.neo.7900226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigas B, Kashfi K. Cancer prevention: a new era beyond cyclooxygenase-2. J Pharmacol Exp Ther. 2005;314:1–8. doi: 10.1124/jpet.104.080564. [DOI] [PubMed] [Google Scholar]
- 10.Alberts DS, Hixson L, Ahnen D, Bogert C, Einspahr J, Paranka N, et al. Do NSAIDs exert their colon cancer chemoprevention activities through the inhibition of mucosal prostaglandin synthetase? J Cell Biochem Suppl. 1995;22:18–23. doi: 10.1002/jcb.240590804. [DOI] [PubMed] [Google Scholar]
- 11.Piazza GA, Rahm AL, Krutzsch M, Sperl G, Paranka NS, Gross PH, et al. Antineoplastic drugs sulindac sulfide and sulfone inhibit cell growth by inducing apoptosis. Cancer Res. 1995;55:3110–6. [PubMed] [Google Scholar]
- 12.Piazza GA, Rahm AK, Finn TS, Fryer BH, Li H, Stoumen AL, et al. Apoptosis primarily accounts for the growth-inhibitory properties of sulindac metabolites and involves a mechanism that is independent of cyclooxygenase inhibition, cell cycle arrest, and p53 induction. Cancer Res. 1997;57:2452–9. [PubMed] [Google Scholar]
- 13.Piazza GA, Alberts DS, Hixson LJ, Paranka NS, Li H, Finn T, et al. Sulindac sulfone inhibits azoxymethane-induced colon carcinogenesis in rats without reducing prostaglandin levels. Cancer Res. 1997;57:2909–15. [PubMed] [Google Scholar]
- 14.Charalambous D, O'Brien PE. Inhibition of colon cancer precursors in the rat by sulindac sulphone is not dependent on inhibition of pros-taglandin synthesis. J Gastroenterol Hepatol. 1996;11:307–10. doi: 10.1111/j.1440-1746.1996.tb01376.x. [DOI] [PubMed] [Google Scholar]
- 15.Reddy BS, Kawamori T, Lubet RA, Steele VE, Kelloff GJ, Rao CV. Chemopreventive efficacy of sulindac sulfone against colon cancer depends on time of administration during carcinogenic process. Cancer Res. 1999;59:3387–91. [PubMed] [Google Scholar]
- 16.Stoner GD, Budd GT, Ganapathi R, DeYoung B, Kresty LA, Nitert M, et al. Sulindac sulfone induced regression of rectal polyps in patients with familial adenomatous polyposis. Adv Exp Med Biol. 1999;470:45–53. doi: 10.1007/978-1-4615-4149-3_5. [DOI] [PubMed] [Google Scholar]
- 17.Arber N, Kuwada S, Leshno M, Sjodahl R, Hultcrantz R, Rex D. Sporadic adenomatous polyp regression with exisulind is effective but toxic: a randomised, double blind, placebo controlled, dose-response study. Gut. 2006;55:367–73. doi: 10.1136/gut.2004.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang ES, Strate LL, Ho WW, Lee SS, Chan AT. Long-term use of aspirin and the risk of gastrointestinal bleeding. Am J Med. 2011;124:426–33. doi: 10.1016/j.amjmed.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee D, Nissen SE, Topol EJ. Risk of cardiovascular events associated with selective COX-2 inhibitors. Jama. 2001;286:954–9. doi: 10.1001/jama.286.8.954. [DOI] [PubMed] [Google Scholar]
- 20.Koornstra JJ, Rijcken FE, Oldenhuis CN, Zwart N, van der Sluis T, Hollema H, et al. Sulindac inhibits beta-catenin expression in normal-appearing colon of hereditary nonpolyposis colorectal cancer and familial adenomatous polyposis patients. Cancer Epidemiol Biomarkers Prev. 2005;14:1608–12. doi: 10.1158/1055-9965.EPI-05-0112. [DOI] [PubMed] [Google Scholar]
- 21.Boon EM, Keller JJ, Wormhoudt TA, Giardiello FM, Offerhaus GJ, van der Neut R, et al. Sulindac targets nuclear beta-catenin accumulation and Wnt signalling in adenomas of patients with familial adenomatous polyposis and in human colorectal cancer cell lines. Br J Cancer. 2004;90:224–9. doi: 10.1038/sj.bjc.6601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice PL, Kelloff J, Sullivan H, Driggers LJ, Beard KS, Kuwada S, et al. Sulindac metabolites induce caspase- and proteasome-dependent degradation of beta-catenin protein in human colon cancer cells. Mol Cancer Ther. 2003;2:885–92. [PubMed] [Google Scholar]
- 23.Thompson WJ, Piazza GA, Li H, Liu L, Fetter J, Zhu B, et al. Exisulind induction of apoptosis involves guanosine 3′,5′-cyclic monophosphate phosphodiesterase inhibition, protein kinase G activation, and attenuated beta-catenin. Cancer Res. 2000;60:3338–42. [PubMed] [Google Scholar]
- 24.Clapper ML, Coudry J, Chang WC. beta-catenin-mediated signaling: a molecular target for early chemopreventive intervention. Mutat Res. 2004;555:97–105. doi: 10.1016/j.mrfmmm.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Piazza GA, Thompson WJ, Pamukcu R, Alila HW, Whitehead CM, Liu L, et al. Exisulind, a novel proapoptotic drug, inhibits rat urinary bladder tumorigenesis. Cancer Res. 2001;61:3961–8. [PubMed] [Google Scholar]
- 26.Tinsley HN, Gary BD, Keeton AB, Zhang W, Abadi AH, Reynolds RC, et al. Sulindac sulfide selectively inhibits growth and induces apoptosis of human breast tumor cells by phosphodiesterase 5 inhibition, elevation of cyclic GMP, and activation of protein kinase G. Mol Cancer Ther. 2009;8:3331–40. doi: 10.1158/1535-7163.MCT-09-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tinsley HN, Gary BD, Thaiparambil J, Li N, Lu W, Li Y, et al. Colon tumor cell growth-inhibitory activity of sulindac sulfide and other nonsteroidal anti-inflammatory drugs is associated with phosphodiesterase 5 inhibition. Cancer Prev Res. 2010;3:1303–13. doi: 10.1158/1940-6207.CAPR-10-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tinsley HN, Gary BD, Keeton AB, Lu W, Li Y, Piazza GA. Inhibition of PDE5 by sulindac sulfide selectively induces apoptosis and attenuates oncogenic Wnt/{beta}-catenin-mediated transcription in human breast tumor cells. Cancer Prev Res. 2011;4:1275–84. doi: 10.1158/1940-6207.CAPR-11-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beavo JA. Cyclic nucleotide phosphodiesterases: functional implications of multiple isoforms. Physiol Rev. 1995;75:725–48. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 30.Lincoln TM, Cornwell TL. Intracellular cyclic GMP receptor proteins. Faseb J. 1993;7:328–38. doi: 10.1096/fasebj.7.2.7680013. [DOI] [PubMed] [Google Scholar]
- 31.Piazza GA, Keeton AB, Tinsley HN, Gary BD, Whitt JD, Mathew B, et al. A novel sulindac derivative that does not inhibit cyclooxygenases but potently inhibits colon tumor cell growth and induces apoptosis with antitumor activity. Cancer Prev Res. 2009;2:572–80. doi: 10.1158/1940-6207.CAPR-09-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer. 1976;17:565–77. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 33.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 34.Zoraghi R, Corbin JD, Francis SH. Phosphodiesterase-5 Gln817 is critical for cGMP, vardenafil, or sildenafil affinity: its orientation impacts cGMP but not cAMP affinity. J Biol Chem. 2006;281:5553–8. doi: 10.1074/jbc.M510372200. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Li H, Underwood T, Lloyd M, David M, Sperl G, et al. Cyclic GMP-dependent protein kinase activation and induction by exisu-lind and CP461 in colon tumor cells. J Pharmacol Exp Ther. 2001;299:583–92. [PubMed] [Google Scholar]
- 36.Kim PJ, Plescia J, Clevers H, Fearon ER, Altieri DC. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–9. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- 37.Hanif R, Pittas A, Feng Y, Koutsos MI, Qiao L, Staiano-Coico L, et al. Effects of nonsteroidal anti-inflammatory drugs on proliferation and on induction of apoptosis in colon cancer cells by a prostaglandin-independent pathway. Biochem Pharmacol. 1996;52:237–45. doi: 10.1016/0006-2952(96)00181-5. [DOI] [PubMed] [Google Scholar]
- 38.Kusuhara H, Matsuyuki H, Matsuura M, Imayoshi T, Okumoto T, Matsui H. Induction of apoptotic DNA fragmentation by nonsteroidal anti-inflammatory drugs in cultured rat gastric mucosal cells. Eur J Pharmacol. 1998;360:273–80. doi: 10.1016/s0014-2999(98)00679-7. [DOI] [PubMed] [Google Scholar]
- 39.de Mello MC, Bayer BM, Beaven MA. Evidence that prostaglandins do not have a role in the cytostatic action of anti-inflammatory drugs. Biochem Pharmacol. 1980;29:311–8. doi: 10.1016/0006-2952(80)90506-7. [DOI] [PubMed] [Google Scholar]
- 40.Williams CS, Watson AJ, Sheng H, Helou R, Shao J, DuBois RN. Celecoxib prevents tumor growth in vivo without toxicity to normal gut: lack of correlation between in vitro and in vivo models. Cancer Res. 2000;60:6045–51. [PubMed] [Google Scholar]
- 41.Pusztai L, Zhen JH, Arun B, Rivera E, Whitehead C, Thompson WJ, et al. Phase I and II study of exisulind in combination with capecitabine in patients with metastatic breast cancer. J Clin Oncol. 2003;21:3454–61. doi: 10.1200/JCO.2003.02.114. [DOI] [PubMed] [Google Scholar]
- 42.Whitehead CM, Earle KA, Fetter J, Xu S, Hartman T, Chan DC, et al. Exisulind-induced apoptosis in a non-small cell lung cancer orthotopic lung tumor model augments docetaxel treatment and contributes to increased survival. Mol Cancer Ther. 2003;2:479–88. [PubMed] [Google Scholar]
- 43.Shailubhai K, Yu HH, Karunanandaa K, Wang JY, Eber SL, Wang Y, et al. Uroguanylin treatment suppresses polyp formation in the Apc (Min/+) mouse and induces apoptosis in human colon adenocarcinoma cells via cyclic GMP. Cancer Res. 2000;60:5151–7. [PubMed] [Google Scholar]
- 44.Deguchi A, Thompson WJ, Weinstein IB. Activation of protein kinase G is sufficient to induce apoptosis and inhibit cell migration in colon cancer cells. Cancer Res. 2004;64:3966–73. doi: 10.1158/0008-5472.CAN-03-3740. [DOI] [PubMed] [Google Scholar]
- 45.Kwon IK, Schoenlein PV, Delk J, Liu K, Thangaraju M, Dulin NO, et al. Expression of cyclic guanosine monophosphate-dependent protein kinase in metastatic colon carcinoma cells blocks tumor angiogenesis. Cancer. 2008;112:1462–70. doi: 10.1002/cncr.23334. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Liu L, David ML, Whitehead CM, Chen M, Fetter JR, et al. Proapoptotic actions of exisulind and CP461 in SW480 colon tumor cells involve beta-catenin and cyclin D1 down-regulation. Biochem Pharmacol. 2002;64:1325–36. doi: 10.1016/s0006-2952(02)01345-x. [DOI] [PubMed] [Google Scholar]