SUMMARY
Human metapneumovirus (HMPV) and respiratory syncytial virus (RSV) cause lower respiratory infections. The virus fusion (F) glycoprotein promotes membrane fusion by refolding from a metastable pre-fusion to a stable post-fusion conformation. F is also a major target of the neutralizing antibody response. Here we show that a potently neutralizing anti-HMPV antibody (DS7) binds a structurally invariant domain of F, identifying a new epitope that could be targeted in vaccine development.
Human metapneumovirus (HMPV) and respiratory syncytial virus (RSV)1–3 define the Pneumovirus subfamily of the Paramyxoviridae and are major respiratory pathogens, causing significant morbidity in infants and the elderly4,5. Vaccine development for HMPV and RSV has been challenging, with no licensed vaccine for either. The paramyxovirus F protein is a class I viral fusion protein and a major target of the neutralizing antibody response6. F initially folds to a metastable, pre-fusion conformation6,7 that, upon activation, undergoes large-scale refolding6–8 coupled to membrane fusion. While major antigenic sites in HMPV and RSV F have been identified, our understanding of F neutralizing epitopes remains incomplete11,16,17. The anti-HMPV F DS7 Fab was identified in a human antibody phage display library9,10, exhibits subnanomolar affinity and is highly protective in vivo11. We set out to understand how this neutralizing antibody interacts with HPMV F.
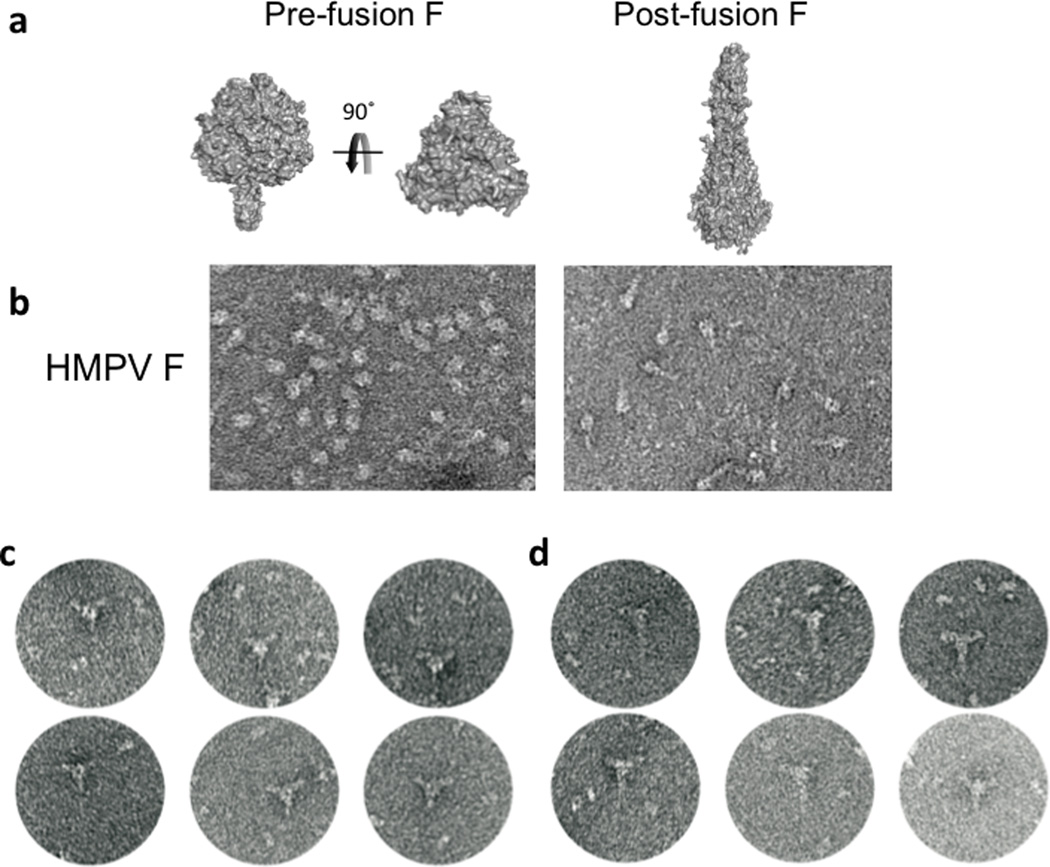
Previous EM studies have documented two conformations of the Parainfluenzavirus 5 (PIV5), human Parainfluenzavirus 3 (hPIV3) and Newcastle disease virus (NDV) F glycoproteins that match crystal structures of pre-fusion and post-fusion F conformations7,8,12,13. The pre-fusion HMPV F conformation was stabilized with a GCN4-derived trimerization tag, purified and visualized by negative stain EM (Figure 1). Two trimeric forms of the intact F consistent with pre- and post-fusion states were observed12,13.
Figure 1. Electron microscopy (EM) of pre- and post-fusion pneumovirus F proteins.
(a) The PIV5 F (pre-fusion) structure is shown in two views, rotated 90° from each other. One orientation shows the pre-fusion HRB stalk, and the second is oriented along the trimer axis and shows only the head region. The RSV F (post-fusion) trimer is shown, oriented perpendicular to the trimer axis. (b) EM images of HMPV F that appear consistent with the pre-fusion (left panel) and post-fusion (right panel) conformational states. (c, d) EM images of DS7 Fab bound to HMPV F in putative pre-fusion (c) and post-fusion (d) conformations.
Purified DS7 Fab formed stable complexes with the HMPV F (Supplementary Figure 1) and EM analysis revealed binding to the F head region (Figures 1c,d). Three Fabs bound to the F trimer, particularly visible for molecules oriented along the F trimer axis that appear to represent pre-fusion HMPV F (Figure 1c). Fab binding to the putative post-fusion conformation of HMPV F was also observed (Figure 1d), where the F orientation creates an asymmetric “T” shape.
DS7–F complexes prepared for crystallization exhibited spontaneous proteolysis, yielding an N-terminal ~45 kD fragment that co-crystallized with DS7 (Supplementary Figures 1, 2). The molecular weight of the complex (~110 kD, Supplementary Figure 1d) indicated that proteolysis led to dissociation of the trimer to a complex of 1 Fab bound to 1 F fragment. The structure was solved with heavy atom data and a Fab molecular replacement solution (Supplementary Tables 1, 2, 3). The final model consists of HMPV F DI, DII and DIII domains bound to DS7 (refinement statistics in Supplementary Table 1). DS7 and HMPV F DI-DII domains were completely modeled, while the F DIII lacked electron density for residues 95–113 and 165–177, corresponding to the fusion peptide and a connecting loop, respectively.
Consistent with the EM data, DS7 binds F residues located in the DI and DII head domains (Figure 2). The interface buries ~1600 Å2 of surface, involving 22 DS7 and 27 F residues. The DS7 heavy chain interacts exclusively with DI, contributing 2/3 of the total buried surface area. The heavy chain CDR3 inserts into a DI hydrophobic pocket formed by residues Leu24, Ile31, Pro282 and Trp284 (Figure 2c). The DS7 light chain spans the DI-DII interface, with CDR1 engaging residues that form the first two strands of DI and three residues in DII.
Figure 2. Structure of the DS7–HMPV F complex.
(a, b) The DS7 Fab engages a single F fragment consisting of DI (yellow), DII (red) and DIII (magenta) domains. The DS7 heavy chain (light blue) binds a hydrophobic pocket in DI. The DS7 light chain (pale green) buries residues in both DI and a loop in DII. (b) The complex is rotated 90° from the orientation in (a). (c) Residues in the DS7 binding site interactions are shown in stick format, with carbon atoms colored by domain as in (a). CDR3H inserts into a hydrophobic pocket on the broad side of DI, formed by Leu24, Ile31, Pro282, and Trp284, flanked by charged residues Lys20, Glu33, Lys312, Arg348 and Glu349. The light chain buries residues at the DI N-terminus and three residues in DII.
DS7 neutralizes representative strains of all HMPV genotypes (A1, A2, B1 and B2). Analysis of HMPV F sequence variability spanning a 20-year period reveals a mean of 96% amino acid identity across the four major subgroups14. F residues at the DS7 complex interface are highly conserved, with 25 of 27 identical amino acids in F proteins of A and B subgroups. The two F residues that vary (312 Gln or Lys and 348 Lys or Arg) flank the CDR3H binding pocket and appear compatible with binding (Figure 2c).
The DS7 epitope does not involve residues within known HMPV or RSV F major antigenic sites (Figure 3a). HMPV F sites fall into 6 groups, 5 of which have been mapped by selecting mAb-resistant virus mutants15–17, whereas six major antigenic sites (sites I – VI) were identified for RSV F16–18. These major antigenic sites are located predominantly in DII and DIII, and surround the DS7 epitope (Figure 3a). The central region of DI that defines the majority of the DS7 epitope is strikingly free of escape mutants, although it is a potent neutralizing site that would be expected be exposed and conserved in structure in the pre- and post-fusion states.
Figure 3. The DS7 epitope defines a novel antigenic site with conserved features in HMPV and RSV.
(a) Major antigenic sites in HMPV and RSV F are mapped onto the HMPV F structure, with the DS7 buried surface area shown as a transparent yellow surface. The view is from the perspective of the DS7 Fab, rotated ~90° from Figure 2a. Escape mutant sites are shown as side chains with dark blue (HMPV F) or magenta (RSV F) spheres. The individual HMPV residues are labeled, along with the HMPV antigenic sites 2–6 and the RSV antigenic sites I-VI. The RSV antigenic sites correspond to the following residues: Ag Site I: 357 (RSV - 389); Ag Site II: 232, 238, 242, 245 (RSV - 262, 268, 272, 275), Ag Sites IV, V, VI: 397, 400, 401, 404, 415 (RSV – 429, 432, 433, 436, 447). (b) The DS7 epitope surface is shown colored by atom type (yellow:carbon; blue:nitrogen; red:oxygen), with underlying residues in stick format. Asterisks mark residues conserved in HMPV and RSV F. A dotted line circumscribes a predominantly hydrophobic pocket that binds DS7 CDR3H. (c) The RSV F surface corresponding to the DS7 epitope is shown as in panel (b). The dotted line highlights the partially conserved hydrophobic pocket with Trp314 at its base.
Comparison of HMPV and RSV F surfaces shows that key features of the DS7 epitope are also present in RSV, suggesting that this site could be a target for RSV vaccine development. Seven out of 28 amino acids are conserved in the two proteins, including residues Pro282, Cys283, Trp284, and Asn351 (Figure 3b,c and Supplementary Table 4). These residues, along with Ile31 in HMPV and Val40 in RSV, line a hydrophobic pocket, which in HMPV F engages the DS7 CDR3H and could correspond to a “hot spot” for mAb binding to both proteins (Figures 3b,c). In RSV, the pocket is partially filled in by the substitution of Leu24 with RSV Tyr33. Surrounding residue differences would contribute to mAb specificity for RSV or HMPV F.
The refolding of paramyxovirus F proteins during membrane fusion and virus entry6,7 potentially has a major impact on neutralizing epitopes and thus antibody recognition. Neutralizing antibodies, like DS7, palivizumab/motavizumab and 101F, recognize structural features that are largely, if not completely, conserved in the pre- and post fusion F conformations. The DS7 complex structure highlights a neutralizing site that was not identified previously, which may represent a low frequency, sub-dominant antigenic site in the immune response to F. The position of the DS7 epitope, at the center of the structurally invariant DI domain and relatively distant from predicted subunit interfaces, suggests that DI domain- and scaffold-based vaccines might stimulate potent neutralizing responses to HMPV and RSV. Finally, we note that the HMPV F DIII retains a compact fold, similar to that of the pre-fusion PIV5 F DIII, while the overall conformation of the three domains (DI-DII-DIII) better matches post-fusion RSV F (Supplementary Discussion).
METHODS
HMPV F expression and purification
The MPV F gene from a B2 strain was engineered in the pcDNA3.1 expression vector with a full length GCN4 trimerization (GCNT) domain in-frame with the heptad repeat of the C-terminal HRB7,18. The GCNT domain contains 5 additional N-terminal residues (ARMKQ) compared to our previous PIV5 F construct7. The mature protein sequence includes HMPV F residues 19–541, along with the following C-terminal residues: ARMKQIEDKIEEILSKIYHIENEIARIKKLIGEAGSGENLYFQGGSGGHHHHHHHH. The additional residues include the GCNT domain (italics), a TEV cleavage site (underlined) and a His8-tag (bold). For protein production, the pcDNA3.1-F plasmid was transfected into 293F cells (Freestyle 293 expression system; Invitrogen) at a density of 1.5 million cells/mL using polyethylenimine. Supernatants were harvested 96 h post-transfection by centrifugation (15 min at 8000 × g at room temperature), filtered through 0.2 µm filters and dialyzed against 200 mM NaCl, 50 mM Na2HPO4 pH 7.4. The HMPV F protein was purified using Co2+ affinity chromatography (TALON Resin, BD Biosciences) and size exclusion chromatography using a Superdex-200 column equilibrated in 50 mM sodium phosphate, pH 7.4, and 200 mM NaCl.
DS7 Fab protein expression and characterization
DS7 lambda and heavy chain cDNA was obtained from GeneArt and cloned into pEE12.4 or pEE6.4 vectors (Lonza), respectively. The heavy chain vector was modified to contain an IgG1 constant chain with a stop codon immediately after the cysteine of the hinge disulfide. A double-gene construct was obtained by cloning the expression cassette of the heavy chain vector into pEE12.4. The double-gene plasmid was transformed into DH5α E. coli cells (Invitrogen). Plasmid DNA was prepared using a Giga Kit (Qiagen) and transfected into 293F cells (Invitrogen) using PolyFect reagent (Qiagen) in a 10 liter WAVE bioreactor bag (GE). After one week, supernatant was purified over a gravity column with CaptureSelect λ resin (BAC B.V., The Netherlands) and eluted with citrate buffer. DS7 Fab was concentrated with Amicon Ultra centrifugal filters with a 30 kD molecular weight cut-off (Millipore). The mammalian cell-expressed DS7 Fab was tested for in vitro plaque reduction neutralization in comparison to the original bacterial-expressed DS7 IgG, and for binding to native HMPV F by immunofluorescence, immunoblot, and flow cytometry (not shown).
Electron microscopy
Solutions of HMPV F were absorbed onto copper grids covered with a carbon film that had been freshly glow discharged. Grids were stained with a 1% aqueous solution of uranyl formate, freshly prepared and filtered immediately prior to use. Grids were observed in a JEOL 1230 electron microscope operated at 100kV and images were acquired with a Gatan 831 CCD camera. The pre-fusion HMPV F head region had a mean diameter of 7.48 nm (s.d. 1.1), with a range from 5.05–9.91 nm (n=116).
Complex formation and crystallization
Complexes of HMPV F and DS7 were observed using size exclusion chromatography with a Superdex-200 column (Supplementary Figure 1). Mixing ratios of HMPV F and DS7 were varied for crystallization to minimize the amount of free HMPV F and analyzed by size exclusion chromatography, native PAGE and SDS–PAGE. Complexes were dialyzed into 25 mM Tris-HCl and 125 mM NaCl pH 7.0 and concentrated to 6–8 mg/ml. Crystals were grown from hanging drops with a well solution containing 16% PEG 5000 MME, 15% glycerol, and 100 mM citrate pH 5.6. Crystals appeared after 7–15 days.
Data collection, structure determination and refinement
Native and heavy-atom-soaked crystals were transferred to a cryoprotectant solution of 19% PEG 5000 MME, 125 mM NaCl and 15% glycerol, 25 mM Tris-HCl pH 7.0, 100 mM citrate pH 5.6, followed by flash cooling in liquid nitrogen. Data were collected at the bl831 and bl822 beamlines at the Advanced Light Source, Lawrence Berkeley National Laboratory.
Crystals belong to space group P6322 and exhibited significant diffraction anisotropy. The native data were initially processed to 3.2 Å with HKL200019 and then submitted to the Diffraction Anisotropy Server20, which truncated the data to 3.4 Å along the c* axis and 3.8 Å along the a*/b* axes (Supplementary Table 1). Data from crystals soaked with 20 mM OsCl3 (osmium(III) chloride), 5 mM PiP (di-µ-iodobis(ethylenediamine)diplatinum(II) nitrate), 20 mM KAu(CN)2 (gold(I) potassium cyanide), 10 mM (C2H5HgO)2HPO2 (ethyl mercuric phosphate), 10 mM HgBr2 (mercury (II) bromide) and 10 mM C2H5HgCl (ethylmercury chloride)] were collected to resolution limits between 4.5 – 6.5 Å (Supplementary Table 2).
Molecular replacement searches were conducted with the program PHASER21 using the CCP4 suite of programs22. The B20-4 Fab model (2FJH)23 provided a single Fab solution with a Z-score of 17.5. Initial Refmac24 refinement, provided an Rfree of 54.97% and R factor of 55.18%. Heavy-atom sites for the derivative datasets were identified with the Fab model phases using the program SHARP25, followed by heavy atom refinement, phase calculation and density modification with DM and Solomon22. Using three platinum sites, one gold site, two osmium sites and six mercury datasets, we obtained an initial 4.5 Å density-modified map and associated phases. These were used to position individual domains (DI–DIII) and helices from the PIV5 F structure (2B9B)7 by using the six-dimensional phased translation search implemented in BRUTEPTF (http://zonker.bioc.aecom.yu.edu/server/NYSGRC.html). The search resulted in potential solutions for parts of each of DI–DIII. The original experimental phases after solvent flattening were used in automated building of the complex model with the CCP4 program BUCCANEER26, followed by manual building of a partial DIII model with the program COOT27. The preliminary DS7–F complex model phases were included in SHARP refinement of the heavy atoms to obtain an improved 4.5 Å density-modified map. The building and refinement was extended to 3.4 Å and additional heavy-atom sites were identified with SHARP (Supplementary Table 3), enabling completion of the DI-DIII and DS7 models.
The complex structure was refined using Phenix Refine28, followed by manual rebuilding with the program COOT27. The F model ends with residue 432 (of 541 in the ectodomain construct), consistent with the observed proteolytic F fragments. The final refinement statistics, native and heavy-atom data, and phasing statistics are summarized in Supplementary Tables 1, 2 and 3. Figures were generated with the program Pymol (http://www.pymol.org/).
Supplementary Material
ACKNOWLEDGMENTS
We thank past and present members of the Jardetzky, Crowe, Williams and Lamb Laboratories. This research was supported in part by NIH research grants to J.V.W. (AI-085062, AI-073697, AI-072414), R.A.L. (AI-23173), T.S.J. (GM-61050). R.A.L. is an Investigator of the Howard Hughes Medical Institute. J.E.C. is a Burroughs Wellcome Fund Clinical Scientist in Translational Research.
Footnotes
ACCESSION CODES
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession code XXXX.
AUTHOR CONTRIBUTIONS
X.W., J.C.K, G.P.L., R.G.C., R.A.L, J.W., J.E.C. and T.S.J designed experiments and reagents. X.W., J.C.K, G.P.L., R.G.C. and T.S.J performed experiments and contributed reagents. X.W., G.P.L AND T.S.J wrote the manuscript and supplementary information, and prepared the figures. X.W., J.C.K, G.P.L., R.G.C., R.A.L, J.W., J.E.C. and T.S.J analyzed data and edited the manuscript.
REFERENCES
- 1.van den Hoogen BG, et al. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 1. Wolters Kluwer: Lippincott WIlliams & Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 3.Collins PL, Crowe JE., Jr . Respiratory Syncytial Virus and Metapneumovirus. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Wolters Kluwer: Lippincott WIlliams & Wilkins; 2007. pp. 1601–1646. [Google Scholar]
- 4.Papenburg J, Boivin G. Rev Med Virol. 2011;20:245–260. doi: 10.1002/rmv.651. [DOI] [PubMed] [Google Scholar]
- 5.Williams JV, et al. N Engl J Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb RA, Jardetzky TS. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbas CF, 3rd, et al. Proc Natl Acad Sci U S A. 1992;89:10164–10168. doi: 10.1073/pnas.89.21.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson RA, et al. Proc Natl Acad Sci U S A. 1993;90:4141–4145. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JV, et al. J Virol. 2007;81:8315–8324. doi: 10.1128/JVI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Proc Natl Acad Sci U S A. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson K, et al. Virology. 2010;402:372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang CF, et al. Virol J. 2009;6:138. doi: 10.1186/1743-422X-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamelin ME, et al. Antiviral Res. 2010;88:31–37. doi: 10.1016/j.antiviral.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Ulbrandt ND, et al. J Gen Virol. 2008;89:3113–3118. doi: 10.1099/vir.0.2008/005199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulbrandt ND, et al. J Virol. 2006;80:7799–7806. doi: 10.1128/JVI.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbury PB, Kim PS, Alber T. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods in Enzymology : Macromolecular Crystallography, part A. Vol. 276. Academic Press; 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 20.Strong M, et al. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 22.CCP4. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Fuh G, et al. J Biol Chem. 2006;281:6625–6631. doi: 10.1074/jbc.M507783200. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 26.Cowtan K. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.