Abstract
We focused on OGG1 Ser326Cys, MUTYH Gln324His, APEX1 Asp148Glu, XRCC1 Arg399Gln, and XRCC3 Thr241Met and examined the relationship between the different genotypes and survival of Japanese lung cancer patients. A total of 99 Japanese lung cancer patients were recruited into our study. Clinical data were collected, and genotypes of the target genes were identified by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). Survival analysis to verify the impact of these gene polymorphisms on the clinical outcome of lung cancer showed that lung squamous cell carcinoma patients with the Thr/Met genotype at XRCC3 had a significantly shorter survival time than those with the Thr/Thr genotype (13 months versus 48 months; log-rank test, p < 0.0001). Cox regression analysis showed that the carriers of XRCC3 genotypes were at a significantly higher risk [adjusted hazard ratio (HR) = 9.35, 95% confidence interval (CI) = 2.52–34.68, p = 0.001; adjusted HR = 9.05, 95% CI = 1.89–44.39, p = 0.006]. Our results suggest that XRCC3 Thr241Met may act as a favorable prognostic indicator for lung squamous cell carcinoma patients.
Keywords: gene polymorphisms, lung cancer, survival, DNA repair, XRCC3
1. Introduction
Lung cancer is a major cause of cancer mortality worldwide. The 5-year survival rate for lung cancer, particularly non-small-cell lung cancer (NSCLC), remains at less than 20% [1,2]. Genetic factors are considered to influence the outcome of lung cancer. Among genetic factors, DNA repair capacity is an important factor. The reason appears to be that DNA repair pathways, including nucleotide excision repair (NER), base excision repair (BER), and double-strand break repair (DSBR), play an important role in maintaining genetic stability through different pathways [3,4]. It is also possible that DNA repair capacity can affect the survival of lung cancer patients.
Four key proteins in the BER pathway are 8-oxoguanine DNA glycosylase (OGG1), Mut Y homolog (MUTYH/MYH), apurinic/apyrimidinic endonuclease-1(APEX1/APE1), and X-ray repair cross-complementing group 1 (XRCC1). Among the various DNA repair pathways, BER is considered to play a key role in removing DNA damage resulting from exposure to various endogenous and exogenous carcinogens. OGG1 and MUTYH recognize and remove the misincorporated oxidized nucleotide 8-OHdG and the adenine paired with 8-OHdG, respectively, and also prevent the occurrence of these events. OGG1 Ser326Cys is associated with the risk of lung cancer [5]. We have reported that MUTYH Gln324His was associated with increased risk of lung and colorectal cancers [6,7]. APEX1, the most stable product of oxidative DNA damage, exhibits 3′-phosphodiesterase activity that removes the abasic sites from cleaved DNA through OGG1 and MUTYH proteins [8]. Recently, we reported that genetic polymorphisms of APEX1 in DNA repair pathways contributed to lung cancer susceptibility, which was dependent on smoking status [6,9]. XRCC1 encodes a protein that complexes with DNA ligase to repair DNA gaps resulting from BER, and a polymorphism at codon 399 Arg to Gln of XRCC1 is associated with the risk of lung cancer.
In the DSBR pathway, the X-ray repair cross-complementing group 3 (XRCC3) is integral to DNA double-strand break recombination repair, and a polymorphism in codon 241 (Thr and Met) of XRCC3 has been associated with the level of bulky DNA adducts in leukocytes of healthy subjects [10].
Recently, there has been increasing evidence that reduced DNA repair capacity resulting from genetic polymorphisms of various DNA repair genes is associated with improved survival with platinum-based chemotherapy [11,12].
To our knowledge, few previous studies have examined the effect of these polymorphisms on the association between outcome and lung cancer in Japanese patients without chemotherapy. To determine the significance of these polymorphisms, we focused on OGG1 Ser326Cys (rs1052133), MUTYH Gln324His (rs3219489), APEX1 Asp148Glu (rs1130409), XRCC1 Arg399Gln (rs25487), and XRCC3 Thr241Met (rs861539) and examined the relationship between the different genotypes and the survival of Japanese lung cancer patients.
2. Results
The distribution of characteristics and clinical features of 99 lung cancer patients are shown in Table 1. The average age [± standard deviation (SD)] of the patients was 66.3 ± 9.3 years, the average tumor size (± SD) was 36.4 ± 9.3 mm, and the average pack-years (±SD) was 34.7 ± 31.9 years. Histological analysis of samples from these patients showed that 65.4% had adenocarcinoma, 29.8% had squamous cell carcinoma, and 4.8% had other types of carcinoma. A total of 52 patients had died. The overall median survival time (MST) was 63 months. Region of metastasis mainly involved lung, bone, or mediastinum lymph node. As shown in Table 1, males; patients ≥65 years of age; whose histological subtype was squamous cell carcinoma; those who smoked; and those who had advanced cancer (stage III and IV), and who had T stage (T2-4), lymph node metastasis, or recurrence had significantly shorter MSTs (log-rank test, p < 0.05). The median survival was 52 months for males and 81 months for females (log-rank test, p = 0.001).
Table 1.
Demographic and Clinical Characteristics of Lung Cancer Patients.
| Variable | Patients n | Median Survival (Months) | Log-rank p value | Adjusted HR (95% CI) | p value |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 65 | 52 | - | 1.00 | - |
| Female | 34 | 81 | 0.001 | 0.32 (0.16–0.65) | 0.001 |
| Age (years) | |||||
| <65 | 36 | 78 | 1.00 | ||
| ≥65 | 63 | 52 | 0.002 | 2.75 (1.41–5.36) | 0.003 |
| Histological subtype | |||||
| adenocarcinoma | 65 | 67 | - | 1.00 | - |
| squamous cell carcinoma | 29 | 49 | 0.020 | 1.93 (1.10–3.40) | 0.023 |
| others | 5 | 41 | - | - | - |
| Smoking status | |||||
| Non-smokers (Pack-years = 0) | 31 | 80 | - | 1.00 | - |
| Smokers (Pack-years > 0) | 67 | 53 | 0.002 | 3.02 (1.47–6.22) | 0.003 |
| No information | 1 | - | - | - | - |
| Stage | |||||
| I & II | 74 | 69 | - | 1.00 | - |
| III & IV | 20 | 41 | 0.001 | 2.60 (1.42–4.75) | 0.002 |
| No information | 5 | 15 | - | - | - |
| T stage | |||||
| T1 | 39 | 82 | - | 1.00 | - |
| T2&T3&T4 | 55 | 49 | <0.0001 | 3.68 (1.88–7.23) | <0.0001 |
| No information | 5 | 15 | - | - | - |
| Lymph node metastasis | |||||
| N0 | 66 | 68 | - | 1.00 | - |
| N1&N2 | 28 | 49 | 0.030 | 1.87 (1.05–3.33) | 0.034 |
| No information | 5 | 15 | - | - | - |
| Distant metastasis | |||||
| M0 | 89 | 66 | - | 1.00 | - |
| M1 | 5 | 50 | 0.783 | 1.18 (0.37–3.79) | 0.784 |
| No information | 5 | 14 | - | - | - |
| Recurrence | |||||
| No | 50 | 83 | 1.00 | ||
| Yes | 44 | 43 | <0.0001 | 6.15 (3.09–12.24) | <0.0001 |
| No information | 5 | 4 | - | - | - |
Multiple Cox regression analysis suggested that the risks of death from lung cancer were increased in patients with stages III and IV than in those with stages I and II (HR = 2.60, 95% CI = 1.42–4.75, p = 0.002), especially who had T stage (T2-4) and lymph node metastasis (HR = 3.68, 95% CI = 1.88–7.23, p < 0.0001 for T stage (T2&T3&T4); HR = 1.87, 95% CI = 1.05–3.33, p = 0.034 for lymph node metastasis; HR = 1.18, 95% CI = 0.37–3.79, p = 0.784 for distant metastasis). This analysis also showed that patients who were older (≥65 years of age), who smoked, whose histological subtype was squamous cell carcinoma, and who had recurrence had increased risks of death (HR = 2.75, 95% CI = 1.41–5.36, p = 0.003 for ≥65 years of age; HR = 1.93, 95% CI = 1.10–3.40, p = 0.023 for squamous cell carcinoma; HR = 3.02, 95% CI = 1.47–6.22, p = 0.003 for smoking status and HR = 6.15, 95% CI = 3.09–12.24, p < 0.0001 for recurrence). Females had significantly longer survival than males (HR = 0.32, 95% CI = 0.16–0.65, p = 0.001).
The associations between genotypes of 5 SNPs and survival of lung cancer patients are shown in Table 2. No associations were found between polymorphisms of these 5 genes and the overall survival of these patients. In the Cox regression mode, after adjusting for age, gender, tumor stage, metastasis, and recurrence, no associations were found between polymorphisms of these 5 genes and the overall risks of death of these patients.
Table 2.
DNA Repair Gene Polymorphisms and Patient Survival.
| Genotype | Patients n | MST (mon) | Log-rank p value | HR (95% CI) | Adjusted HR | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| p value | (95% CI) a | p value | |||||
| OGG1 | |||||||
| Ser/Ser | 25 | 58 | 1.00 | - | 1.00 | - | |
| Ser/Cys | 50 | 70 | 0.910 | 0.88 (0.45–1.72) | 0.712 | 0.96 (0.44–2.10) | 0.927 |
| Cys/Cys | 24 | 63 | 0.99 (0.46–2.10) | 0.972 | 0.90 (0.40–2.06) | 0.808 | |
| Ser/Cys, Cys/Cys | 74 | 66 | 0.784 | 0.92 (0.49–1.72) | 0.786 | 0.94 (0.47–1.89) | 0.854 |
| MUTYH | |||||||
| Gln/Gln | 20 | 58 | 1.00 | - | 1.00 | - | |
| Gln/His | 53 | 63 | 0.914 | 1.13 (0.55–2.33) | 0.731 | 0.96 (0.42–2.17) | 0.919 |
| His/His | 26 | 70 | 1.02 (0.45–2.32) | 0.972 | 0.85 (0.33–2.18) | 0.851 | |
| Gln/His, His/His | 79 | 63 | 0.797 | 1.09 (0.55–2.18) | 0.798 | 0.93 (0.42–2.03) | 0.845 |
| APEX | |||||||
| Asp/Asp | 40 | 58 | 1.00 | - | 1.00 | - | |
| Asp/Glu | 48 | 62 | 0.649 | 0.88 (0.50–1.55) | 0.652 | 1.02 (0.55–1.88) | 0.963 |
| Glu/Glu | 11 | 71 | 0.61 (0.21–1.78) | 0.37 | 0.51 (0.15–1.76) | 0.289 | |
| Asp/Glu, Glu/Glu | 59 | 64 | 0.505 | 0.83 (0.48–1.44) | 0.508 | 0.91 (0.50–1.66) | 0.766 |
| XRCC1 | |||||||
| Arg/Arg | 44 | 62 | - | 1.00 | - | 1.00 | - |
| Arg/Gln | 49 | 56 | 0.162 | 1.20 (0.69–2.08) | 0.524 | 0.85 (0.46–1.55) | 0.588 |
| Gln/Gln | 6 | 91 | 0.22 (0.03–1.62) | 0.136 | 0.37 (0.05–2.87) | 0.342 | |
| Arg/Gln, Gln/Gln | 55 | 60 | 0.897 | 1.04 (0.60–1.79) | 0.898 | 0.80 (0.44–1.46) | 0.470 |
| XRCC3 | |||||||
| Thr/Thr | 88 | 66 | - | 1.00 | - | 1.00 | |
| Thr/Met | 11 | 28 | 0.202 | 1.67 (0.75–3.72) | 0.209 | 1.94 (0.83–4.53) | 0.128 |
| Met/Met | 0 | - | - | - | - | - | - |
| Thr/Met, Met/Met | 11 | 28 | 0.202 | 1.67 (0.75–3.72) | 0.209 | 1.94 (0.83–4.53) | 0.128 |
HR adjusted for gender, age, smoking history, disease stage, metastasis, and reccurence.
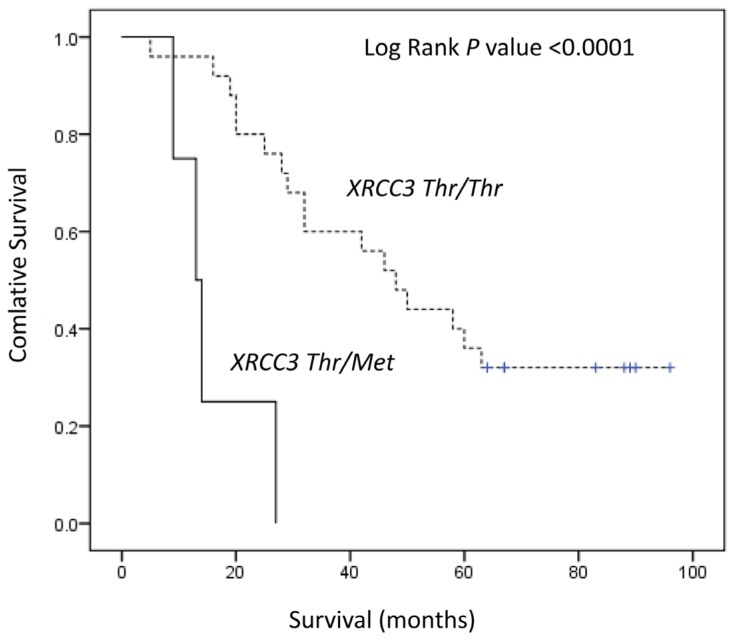
Table 3 summarizes the genotype distribution for lung adenocarcinomas and squamous cell carcinomas. No associations were found between polymorphisms of these 5 genes and the risks of death for adenocarcinoma patients. For squamous cell carcinoma, patients with Thr/Met genotype at XRCC3 showed a significantly shorter survival time than those with the Thr/Thr genotype (13 months versus 48 months; log-rank test, p < 0.0001) (Figure 1). Cox regression analysis showed that carriers of XRCC3 had a significantly a higher risk (crude HR = 9.35, 95% CI = 2.52–34.68, p = 0.001; adjusted HR = 9.05, 95% CI = 1.89–44.39, p = 0.006).
Table 3.
DNA Repair Gene Polymorphisms and Patient Survival in relation to Subtypes.
| Genotype | Patients n | MST (mon) | Log-rank p value | HR (95% CI) | Adjusted HR | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| p value | (95% CI) a | p value | |||||
| Adenocarcinoma | |||||||
| OGG1 | |||||||
| Ser/Ser | 16 | 62 | - | 1.00 | - | 1.00 | - |
| Ser/Cys, Cys/Cys | 49 | 68 | 0.784 | 0.89 (0.38–2.08) | 0.785 | 0.80 (0.29–2.26) | 0.803 |
| MUTYH | |||||||
| Gln/Gln | 13 | 70 | 1.00 | 1.00 | |||
| Gln/His, His/His | 52 | 66 | 0.65 | 1.25 (0.48–3.27) | 0.653 | 1.54 (0.44–5.36) | 0.498 |
| APEX | |||||||
| Asp/Asp | 28 | 63 | - | 1.00 | - | 1.00 | - |
| Asp/Glu, Glu/Glu | 37 | 70 | 0.549 | 0.80 (0.39–1.67) | 0.553 | 1.14 (0.48–2.73) | 0.763 |
| XRCC1 | |||||||
| Arg/Arg | 26 | 71 | 1.00 | - | 1.00 | - | |
| Arg/Gln, Gln/Gln | 39 | 64 | 0.522 | 1.28 (0.60–2.76) | 0.526 | 0.87 (0.35–2.20) | 0.775 |
| XRCC3 | |||||||
| Thr/Thr | 58 | 67 | - | 1.00 | - | 1.00 | - |
| Thr/Met, Met/Met | 7 | 64 | 0.995 | 1.00 (0.30–3.32) | 0.995 | 1.32 (0.38–4.64) | 0.661 |
| Squamous Cell Carcinoma | |||||||
| OGG1 | |||||||
| Ser/Ser | 8 | 52 | - | 1.00 | - | 1.00 | - |
| Ser/Cys, Cys/Cys | 21 | 48 | 0.824 | 1.11 (0.43–2.88) | 0.825 | 1.25 (0.44–3.55) | 0.670 |
| MUTYH | |||||||
| Gln/Gln | 5 | 64 | - | 1.00 | - | 1.00 | - |
| Gln/His, His/His | 24 | 45 | 0.377 | 1.72 (0.51–5.85) | 0.385 | 4.73 (0.51–5.85) | 0.153 |
| APEX | |||||||
| Asp/Asp | 11 | 40 | 1.00 | - | 1.00 | - | |
| Asp/Glu, Glu/Glu | 18 | 54 | 0.328 | 0.65 (0.27–1.56) | 0.334 | 0.43 (0.14–1.32) | 0.139 |
| XRCC1 | |||||||
| Arg/Arg | 16 | 50 | 1.00 | - | 1.00 | - | |
| Arg/Gln, Gln/Gln | 13 | 46 | 0.991 | 1.00 (0.42–2.37) | 0.991 | 1.17 (0.46–2.94) | 0.743 |
| XRCC3 | |||||||
| Thr/Thr | 25 | 48 | - | 1.00 | - | 1.00 | - |
| Thr/Met, Met/Met | 4 | 13 | <0.0001 | 9.35 (2.52–34.68) | 0.001 | 9.05 (1.89–44.39) | 0.006 |
HR adjusted for gender, age, smoking history, disease stage, metastasis, and reccurence.
Figure 1.
Kaplan–Meier survival curve of lung squamous cell carcinoma patients with the XRCC3 Thr241Met genotype.
3. Discussion
In this study, we assessed the OGG1 Ser326Cys, MUTYH Gln324His, APEX1 Asp148Glu, XRCC1 Arg399Gln, and XRCC3 Thr241Met gene polymorphisms that may influence DNA repair capacity and their association with the overall survival of lung cancer patients. The polymorphisms chosen for this study have also been shown to have functional significance and may be responsible for a low DNA repair capacity phenotype that is characteristic of cancer patients [13,14]. To our knowledge, this is the first report on these DNA repair gene polymorphisms in relation to survival without chemotherapy in Japanese lung cancer patients. In a previous study of Japanese patients, Takenaka et al. reported that the ERCC1 C8092A polymorphism may influence NSCLC prognosis regardless of ERCC1 protein expression and platinum sensitivity [15]. We explored the genotypes as well as pathological features of lung cancer patients in terms of their overall survival. In this study, XRCC3 Thr241Met might be an independent prognostic factor in squamous cell carcinoma. The adjusted HR for XRCC3 was 9.05 (p = 0.006), with the XRCC3 group being significant. This XRCC3 variant genotype was associated with significantly decreased survival in squamous cell carcinoma. In contrast, we observed that the patients carrying none of the adverse genotypes (OGG1 Ser326Cys, MUTYH Gln324His, APEX1 Asp148Glu, and XRCC1 Arg399Gln) had much better survival than those carrying variant alleles. For BER genes, several studies reported that variant alleles of XRCC1 399 and XRCC1 variant genotypes are significantly associated with poor survival [11,12,16–19]. In China, ERCC1 and XRCC1 were associated with the survival of non-smoker female lung adenocarcinoma patients [20]. Consistent with our study, Penas et al. showed that XRCC 3 is strongly associated with the survival of NSCLC patients treated with cisplatin/gemcitabine [21]. They suggested that the reduced efficacy of the XRCC3 protein, a consequence of the polymorphic variant, may have resulted in an impaired ability to repair cisplatin DNA damage [22]. Another report showed that the XRCC3 Met/Met genotype was significantly associated with increased risk of death among all patients, particularly males by univariate and multivariate analyses [23]. An explanation for these discordant results remains to be provided. However, there have been no previous reports on OGG1, MUTYH, and APEX1 with regard to survival in lung cancer. Thus, our report is the first on the detailed effects of DNA repair gene polymorphisms on the survival of Japanese lung cancer patients. In addition, numerous clinical features may play important roles in the survival of lung cancer patients. We found a significant association between survival and tumor histology. The disease stage at the time of diagnosis has a direct impact on the survival rate. Multivariate analysis showed that a higher stage (stages III and IV), male gender, older age, squamous cell carcinoma, smoking history, lymph node metastasis, and recurrence were independent prognostic factors associated with an increased risk of death. This higher tumor stage had a significant effect on survival, which is in accordance with other studies [18,24]. It is possible that individuals with these factors and with higher-stage disease already have too many genetic alterations during their tumor growth, which would reduce their survival. Our study has several limitations, especially the fact that the conclusion was based on several patients. Our data may be biased by the relatively small number of patients as a hospital-based case–control study. Therefore, further verification of these predictive biomarkers is required with a larger study population. The gene–environment interaction between smoking and these genotypes also needs to be clarified.
4. Experimental Section
4.1. Study Subjects
Study subjects included 99 lung cancer patients (65 with lung adenocarcinoma, 29 with lung squamous cell carcinoma, and 5 with other carcinomas) who were after surgical treatment but not receiving radiotherapy and chemotherapy and were included in previous studies that investigated the genetic polymorphisms of DNA repair proteins and metabolic enzymes [7,9,25]. These patients were recruited between April 2001 and July 2002 at the Hyogo Cancer Center. Informed consent was obtained from each patient. Detailed data on smoking were obtained by personal interviews. The study design was approved by the Ethics Review Committee on Genetic and Genomic Research, Kobe University Graduate School of Medicine. All samples were coded after the collection of blood and smoking frequency data. All patients were followed up for survival by November 2009 (the time of data analysis). Metastases were based on the status of pathological metastasis. The amount of smoke exposure was calculated as pack-years; the product of the number of years an individual smoked and the average number of cigarettes smoked per day (converted into a standard pack of 20 cigarettes).
4.2. Genotyping
Genomic DNA used for this study was isolated in previous studies [7,9,25]. The genotypes of OGG1 Ser326Cys, MUTYH Gln324His, APEX1 Asp148Glu, XRCC1 Arg399Gln, and XRCC3 Thr241Met were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis, as described previously [7,9].
4.3. Statistical Analysis
The effect of genetic polymorphisms on survival was also estimated using the Kaplan–Meier method and assessed using the log-rank test. The influence of clinical parameters on the outcomes of lung cancer patients was assessed by the log-rank test. The overall survival duration of lung cancer patients was calculated from the 1st day of treatment until either death or the last follow-up. A multiple Cox regression model was used to obtain the adjusted hazard ratio (HR) and 95% confidence interval (95% CI) for potential prognostic factors in lung cancer patients. All p values were calculated from 2-tailed statistical tests. Statistical analysis was performed with the PASW software package (version 17.0 for Windows; SPSS Japan, Inc., Tokyo, Japan). A p value of <0.05 was considered significant for an association between a genotype and lung cancer.
5. Conclusions
We analyzed the association between polymorphisms of five DNA repair genes and the outcome of Japanese lung cancer patients. Our results suggest that the XRCC3 Thr241Met gene polymorphism plays an important role in the overall survival of Japanese lung squamous cell carcinoma patients without chemotherapy. The XRCC3 Thr241Met gene polymorphism may be a prognostic factor in lung squamous cell carcinoma patients.
Abbreviations
- NER
nucleotide excision repair
- BER
base excision repair
- DSBR
double-strand break repair
- OGG1
8-oxoguanine DNA glycosylase
- MUTYH/MYH
Mut Y homolog
- APEX1/APE1
apurinic/apyrimidinic endonuclease-1
- XRCC1
X-ray repair cross-complementing group 1
- XRCC3
X-ray repair cross-complementing group 3
- CI
confidence interval
- NSCLC
non-small-cell lung cancer
- SD
standard deviation
- HR
hazard ratio
- MST
median survival time
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Pisani P., Parkin D.M., Ferlay J. Estimates of the worldwide mortality from eighteen major cancers in 1985. Implications for prevention and projections of future burden. Int. J. Cancer. 1993;55:891–903. doi: 10.1002/ijc.2910550604. [DOI] [PubMed] [Google Scholar]
- 2.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Yu Z., Chen J., Ford B.N., Brackley M.E., Glickman B.W. Human DNA repair systems: An overview. Environ. Mol. Mutagen. 1999;33:3–20. doi: 10.1002/(sici)1098-2280(1999)33:1<3::aid-em2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 4.Wood R.D., Mitchell M., Sgouros J., Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 5.Le Marchand L., Donlon T., Lum-Jones A., Seifried A., Wilkens L.R. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol. Biomark. Prev. 2002;11:409–412. [PubMed] [Google Scholar]
- 6.Kasahara M., Osawa K., Yoshida K., Miyaishi A., Osawa Y., Inoue N., Tsutou A., Tabuchi Y., Tanaka K., Yamamoto M., et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J. Exp. Clin. Cancer Res. 2008;27:49. doi: 10.1186/1756-9966-27-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyaishi A., Osawa K., Osawa Y., Inoue N., Yoshida K., Kasahara M., Tsutou A., Tabuchi Y., Sakamoto K., Tsubota N., et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J. Exp. Clin. Cancer Res. 2009;28:10. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett R.A., Wilson D.M., III, Wong D., Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl. Acad. Sci. USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osawa K., Miyaishi A., Uchino K., Osawa Y., Inoue N., Nakarai C., Tsutou A., Kido Y., Yoshimura M., Tsubota N., et al. APEX1 Asp148Glu gene polymorphism is a risk factor for lung cancer in relation to smoking in Japanese. Asian Pac. J. Cancer Prev. 2010;11:1181–1186. [PubMed] [Google Scholar]
- 10.Matullo G., Palli D., Peluso M., Guarrera S., Carturan S., Celentano E., Krogh V., Munnia A., Tumino R., Polidoro S., et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 11.Gurubhagavatula S., Liu G., Park S., Zhou W., Su L., Wain J.C., Lynch T.J., Neuberg D.S., Christiani D.C. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J. Clin. Oncol. 2004;22:2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 12.Ryu J.S., Hong Y.C., Han H.S., Lee J.E., Kim S., Park Y.M., Kim Y.C., Hwang T.S. Association between polymorphisms of ERCC1 and XPD and survival in non-small-celllung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44:311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Wei Q., Spitz M.R. The role of DNA repair capacity in susceptibility to lung cancer: A review. Cancer Metastasis Rev. 1997;16:295–307. doi: 10.1023/a:1005852211430. [DOI] [PubMed] [Google Scholar]
- 14.Osawa K. SNPs in ERCC1 and drug response to cisplatin in non-small-cell lung cancer patients. Pharmacogenomics. 2011;12:445–447. doi: 10.2217/pgs.11.15. [DOI] [PubMed] [Google Scholar]
- 15.Osawa K. Gene Polymorphisms and Chemotherapy in Non-small Cell Lung Cancer. Zhongguo Fei Ai Za Zhi. 2009;12:837–840. doi: 10.3779/j.issn.1009-3419.2009.08.01. [DOI] [PubMed] [Google Scholar]
- 16.Takenaka T., Yano T., Kiyohara C., Miura N., Kouso H., Ohba T., Kometani T., Shoji F., Yoshino I., Maehara Y. Effects of excision repair cross-complementation group 1 (ERCC1) single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients. Lung Cancer. 2010;67:101–107. doi: 10.1016/j.lungcan.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Wu X., Shell S.M., Yang Z., Zou Y. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006;66:2997–3005. doi: 10.1158/0008-5472.CAN-05-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreeja L., Syamala V.S., Syamala V., Hariharan S., Raveendran P.B., Vijayalekshmi R.V., Madhavan J., Ankathil R. Prognostic importance of DNA repair gene polymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln in lung cancer patients from India. J. Cancer Res. Clin. Oncol. 2008;134:645–652. doi: 10.1007/s00432-007-0328-4. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh W.C., Cheng Y.W., Lin C.J., Chou M.C., Chen C.Y., Lee H. Prognostic significance of X-ray cross-complementing group 1 T-77C polymorphism in resected non-small cell lung cancer. Jpn. J. Clin. Oncol. 2009;39:81–85. doi: 10.1093/jjco/hyn130. [DOI] [PubMed] [Google Scholar]
- 20.Liao W.Y., Shih J.Y., Chang G.C., Cheng Y.K., Yang J.C., Chen Y.M., Yu C.J. Genetic polymorphism of XRCC1 Arg399Gln is associated with survival in non-small-cell lung cancer patients treated with gemcitabine/platinum. J. Thorac. Oncol. 2012;7:973–981. doi: 10.1097/JTO.0b013e31824fe98c. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z., Zhou B., He Q., Li M., Guan P., Li X., Cui Z., Xue X., Su M., Ma R., et al. Association between polymorphisms in DNA repair genes and survival of non-smoking female patients with lung adenocarcinoma. BMC Cancer. 2009;9:439. doi: 10.1186/1471-2407-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De las Peñas R., Sanchez-Ronco M., Alberola V., Taron M., Camps C., Garcia-Carbonero R., Massuti B., Queralt C., Botia M., Garcia-Gomez R., et al. Spanish Lung Cancer Group. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann. Oncol. 2006;17:668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 23.Butkiewicz D., Rusin M., Sikora B., Lach A., Chorąży M. An association between DNA repair gene polymorphisms and survival in patients with resected non-small cell lung cancer. Mol. Biol. Rep. 2011;38:5231–5241. doi: 10.1007/s11033-010-0674-1. [DOI] [PubMed] [Google Scholar]
- 24.Ahrendt S.A., Hu Y., Buta M., McDermott M.P., Benoit N., Yang S.C., Wu L., Sidransky D. p53 mutations and survival in stage I non-small-cell lung cancer: Results of a prospective study. J. Natl. Cancer Inst. 2003;95:961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- 25.Osawa Y., Osawa K., Miyaishi A., Higuchi M., Tsutou A., Matsumura S., Tabuchi Y., Tsubota N., Takahashi J. NAT2 and CYP1A2 polymorphisms and lung cancer risk in relation to smoking status. Asian Pac. J. Cancer Prev. 2007;8:103–108. [PubMed] [Google Scholar]