Abstract
Background
Mycobacterium avium subspecies paratuberculosis (Map) is the aetiological agent of Johne’s disease or paratuberculosis and is included within the Mycobacterium avium complex (MAC). Map strains are of two major types often referred to as ‘Sheep’ or ‘S-type’ and ‘Cattle’ or ‘C-type’. With the advent of more discriminatory typing techniques it has been possible to further classify the S-type strains into two groups referred to as Type I and Type III. This study was undertaken to genotype a large panel of S-type small ruminant isolates from different hosts and geographical origins and to compare them with a large panel of well documented C-type isolates to assess the genetic diversity of these strain types. Methods used included Mycobacterial Interspersed Repetitive Units - Variable-Number Tandem Repeat analysis (MIRU-VNTR), analysis of Large Sequence Polymorphisms by PCR (LSP analysis), Single Nucleotide Polymorphism (SNP) analysis of gyr genes, Pulsed-Field Gel Electrophoresis (PFGE) and Restriction Fragment Length Polymorphism analysis coupled with hybridization to IS900 (IS900-RFLP) analysis.
Results
The presence of LSPA4 and absence of LSPA20 was confirmed in all 24 Map S-type strains analysed. SNPs within the gyr genes divided the S-type strains into types I and III. Twenty four PFGE multiplex profiles and eleven different IS900-RFLP profiles were identified among the S-type isolates, some of them not previously published. Both PFGE and IS900-RFLP segregated the S-type strains into types I and III and the results concurred with those of the gyr SNP analysis. Nine MIRU-VNTR genotypes were identified in these isolates. MIRU-VNTR analysis differentiated Map strains from other members of Mycobacterium avium Complex, and Map S-type from C-type but not type I from III. Pigmented Map isolates were found of type I or III.
Conclusion
This is the largest panel of S-type strains investigated to date. The S-type strains could be further divided into two subtypes, I and III by some of the typing techniques (IS900-RFLP, PFGE and SNP analysis of the gyr genes). MIRU-VNTR did not divide the strains into the subtypes I and III but did detect genetic differences between isolates within each of the subtypes. Pigmentation is not exclusively associated with type I strains.
Background
The aetiologic agent of Johne’s disease or paratuberculosis, M. avium subsp. paratuberculosis (Map), is one of the subspecies included in the Mycobacterium avium Complex (MAC). Based on the comparison of whole-genomes of Map, a biphasic evolution scheme has been proposed distinguishing two major lineages, a sheep lineage and a cattle lineage [1]. In addition to genotypic differences [2,3], strains belonging to these two lineages exhibit phenotypic differences including growth rate [2-4], utilization of different iron metabolic pathways [4], profile of cytokine responses induced in bovine macrophages [5] or transcriptional profiles in a human macrophage model [6]. The association of each lineage with either the sheep or cattle host is not exclusive since strains representative of either lineage can cause disease in all types of ruminants. Historically, strains belonging to the sheep lineage have been referred to as ‘Sheep or S-type’ and those of the cattle lineage ‘Cattle or C-type’ according to the species from which they were first isolated. As the technologies for molecular typing advanced and more genotyping studies were undertaken, greater genetic diversity was detected within both the S- and C-type strains. Pulsed-field gel electrophoresis (PFGE) revealed three strain types designated Types I, II and III [7,8]. Type II is synonymous with C-type and types I and III comprise the S-type. In this paper we will use the term S-type to describe collectively type I and III strains and have designated the types I and III as subtypes. S-type strains have not been characterized to the same extent as C-type strains due to the difficulty in culturing the strains in vitro resulting in a limited number of strains available for such studies. Here we undertook the first comprehensive genotyping study of a large representative panel of S-type strains using various typing methods that have been applied to Map strains, individually or in combinations, to draw a portrait of S-type strains. We studied both inter and intra-subtype genotypic strain differences using restriction fragment length polymorphism analysis coupled with hybridization to IS900 (IS900 RFLP), PFGE and various PCRs based on variable-number tandem repeat (VNTR) loci and mycobacterial interspersed repetitive units (MIRUs) [9,10] MIRU-VNTR typing [11], the presence or absence of large sequence polymorphisms (LSPs) [12] and the gyrA and B genes [13]. Our panel of S-type strains comprised strains from different geographic origins with different restriction enzyme profiles and includes pigmented strains. We also incorporated typing data obtained for additional Map C-type isolates to represent the all diversity of the genotypes described and Mycobacterium. avium subsp. avium (Maa) Mycobacterium. avium subsp. silvaticum (Mas) and Mycobacterium avium subsp. hominissuis (Mah) for comparison.
Methods
Panel of strains
A total of 24 Map S-type isolates were obtained from Scotland (n = 6), Spain (n = 11), Canada (n = 2), New Zealand (n = 2), Faroe Islands (n = 2) and Iceland (n = 1), isolated from sheep, goats and a pig (see Table 1 and Additional file 1: Table S1). Isolates were propagated on slopes of one of the following media depending on what was used routinely in the supply laboratories: modified Middlebrook 7H11 supplemented with 20% (vol/vol) heat-inactivated newborn calf serum, 2.5% (vol/vol) glycerol, 2 mM asparagine, 10% (vol/vol) Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment medium (Becton Dickinson, Oxford, Oxfordshire, United Kingdom), Selectatabs (code MS 24; MAST Laboratories Ltd., Merseyside, United Kingdom), and 2 μg ml-1 mycobactin J (Allied Monitor, Fayette, Mo.); Herrold’s egg yolk medium with 2 μg ml−1 mycobactin J or Lowenstein-Jensen medium with 2 μg ml−1 mycobactin J. In addition, genotyping information obtained previously [11,18,19] from 148 Map C-type strains, 31 Maa isolates, 4 Mas isolates and 82 Mah isolates were used for comparison and phylogenetic analyses and are described in Additional file 1: Table S1.
Table 1.
Synthesis of information and genotyping data of S strains of Map by subtype
| Strain | Subtype |
Origin |
Profile |
MIRU- VNTR Patterns5 | SNP Gyr A & B | LSP6A20 4 | Pigmentation | Strains References | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Host | Country1 regions | RFLP2 | PFGE3 | MIRU- VNTR4 | |||||||||
| 6756 |
|
Ovine |
NZ |
S1 |
nd |
INMV |
72 |
41331118 |
A/C C/C |
- |
+ |
|
[14] |
| 6759 |
|
Ovine |
NZ |
S1 |
nd |
INMV |
72 |
41331118 |
A/C C/C |
- |
+ |
|
[14] |
| P133/79 |
|
Ovine |
FO |
S2 |
nd |
INMV |
70 |
71331118 |
A/C C/C |
- |
+ |
|
[15] |
| 21P |
|
Ovine |
FO |
S2 |
[9] |
INMV |
70 |
71331118 |
A/C C/C |
- |
+ |
+ |
[8] |
| 235 G |
I |
Ovine |
UK, Shetland |
S2 |
[75–8] |
INMV |
70 |
71331118 |
A/C C/C |
- |
+ |
+ |
[8] |
| M189 |
|
Ovine |
UK, Scotland |
S2 |
[7] |
INMV |
21 |
51331118 |
A/C C/C |
- |
+ |
+ |
[8] |
| M15/04 |
|
Ovine |
UK, Scotland |
S2 |
[nd-70] |
INMV |
70 |
71331118 |
A/C C/C |
- |
+ |
+ |
[8] |
| M254/04 |
|
Ovine |
UK, Scotland |
S2 |
nd |
INMV |
32 |
61331118 |
A/C C/C |
- |
+ |
+ |
This study |
| M71/03 |
|
Ovine |
UK, Scotland |
S2 |
[80–71] |
INMV |
70 |
71331118 |
A/C C/C |
- |
+ |
+ |
This study |
| M72/03 |
|
Ovine |
UK, Scotland |
S2 |
[77–70] |
INMV |
70 |
71331118 |
A/C C/C |
- |
+ |
+ |
This study |
| 22 G |
|
Ovine |
ES, Basque |
A |
[69–50] |
INMV |
84 |
91331118 |
A/T C/T |
- |
+ |
|
[16] |
| OVICAP16 |
|
Caprine |
ES, Andalucia |
A |
[65–61] |
INMV |
85 |
(11)1331118 |
A/T C/T |
- |
+ |
|
[16] |
| OVICAP49 |
|
Ovine |
ES, Navarra |
A |
[57–57] |
INMV |
70 |
71331118 |
A/T C/T |
- |
+ |
+ |
This study |
| 21I |
|
Ovine |
ES, Basque |
B |
[61–47] |
INMV |
70 |
71331118 |
A/T C/T |
- |
+ |
|
[16] |
| PCR311 |
|
Caprine |
ES, Balearic |
B |
[16–47] |
INMV |
70 |
71331118 |
A/T C/T |
- |
+ |
|
[16] |
| 19I |
|
Ovine |
ES, Basque |
C |
[79–55] |
INMV |
70 |
71331118 |
A/T C/T |
- |
+ |
|
[16] |
| 85/14 |
III |
Ovine |
CA |
C |
nd |
INMV |
27 |
81331118 |
A/T C/T |
- |
+ |
|
[14] |
| OVICAP34 |
|
Ovine |
ES, Basque |
D |
[66–62] |
INMV |
21 |
51331118 |
A/T C/T |
- |
+ |
|
This study |
| 18I |
|
Ovine |
ES, Basque |
E |
[67–51] |
INMV |
84 |
91331118 |
A/T C/T |
- |
+ |
|
[16] |
| FO21 |
|
Ovine |
ES, Aragon |
F |
[56–56] |
INMV |
84 |
91331118 |
A/T C/T |
- |
+ |
+ |
This study |
| LN20 |
|
Porcine |
CA |
I1 |
nd |
INMV |
71 |
51131118 |
A/T C/T |
- |
+ |
|
[14] |
| 269OV |
|
Ovine |
ES, Basque |
I10 |
[69–54] |
INMV |
72 |
41331118 |
A/T C/T |
- |
+ |
|
[16] |
| M284/08 |
|
Ovine |
ES, Basque |
I10 |
[71–64] |
INMV |
72 |
41331118 |
A/T C/T |
- |
+ |
|
[16] |
| P465 | Ovine | IS | I2 | nd | INMV | 73 | 20331118 | A/T C/T | - | + | This study | ||
1 Country and regions: ES, Spain; CA, Canada; UK, United Kingdom; FO, Faroe Island; IS, Iceland; NZ, New Zealand.
2 nd, not determined; alphanumeric nomenclature as defined by Pavlik et al., 1999 [17], alphabetic nomenclature correspond to new profiles identified in this study.
3 Nomenclature as defined by Stevenson et al., 2002 [8].
4 Nomenclature as defined by Thibault et al., 2007 [11].
5 Number of repeats at locus 292-X3-25-47-3-7-10-32 defined by Thibault et al., 2007 [11].
6 +, presence; -, absence.
IS900-RFLP method
Map strains were typed by IS900-RFLP as described previously [11]. Profiles were designated according to nomenclature previously described [17,20-22]. Profiles were analysed using Bionumerics™ software version 6.5 (Applied Maths, Belgium).
PFGE analysis
PFGE analysis was carried out using SnaBI and SpeI according to the published standardized procedure of Stevenson et al. [8] with the following modifications. Plugs were prepared to yield a density of 1.2 × 1010 cells ml-1 and the incubation time in lysis buffer was increased to 48 hr. The concentration of lysozyme was increased to 4 mg ml-1. Incubation with proteinase K was carried out for a total of seven days and the enzyme was refreshed after four days. Restriction of plug DNA by SpeI was performed with 10U overnight after which the enzyme was refreshed and incubated for a further 6 hr. The parameters for electrophoresis of SpeI restriction fragments were changed to separate fragments of between 20 and 250Kb as determined by the CHEF MAPPER and electrophoresis was performed for 40 hr. Gel images were captured using an Alphaimager 2200 (Alpha Innotech). Profiles were analysed using Bionumerics™ software version 6.5 (Applied Maths, Belgium).
SNP analysis of gyrA and gyrB genes
Primers (Additional file 2: Table S2) were designed for both gyrA (GenBank accession no. 2720426 [Genome number: NC_002944.2]) and gyrB genes (GenBank accession no. 2717659 [Genome number: NC_002944.2]). The PCR mixture was composed as follows using the GoTaq Flexi DNA polymerase (Promega). Two microliters of DNA solution was added to a final volume of 50 μl containing 0.2 μl of GoTaq Flexi DNA polymerase (5 U/μl), 2 mM (each) dATP, dCTP, dGTP, and dTTP (Promega); 10 μl of 5x PCR buffer supplied by the manufacturer; 1 μM of each primers; and 1.5 mM of MgCl2. The reactions were carried out using a TC-512 thermal cycler (Techne). PCR conditions were as follows: 1 cycle of 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; and 1 cycle of 7 min at 72°C. PCR products were visualized by electrophoresis using 1.5% agarose gels (agarose electrophoresis grade; Invitrogen), purified using NucleoSpin® Extract II (Macherey-Nagel) and sequenced by GenomExpress (Grenoble, France). Sequence analysis and SNP detection were performed by using the Bionumerics™ software version 6.5 (Applied Maths, Belgium).
LSP analysis
Primers were used according to Semret et al. [12] and described in Additional file 2: Table S2. The PCR mixture comprised 2 μl of DNA solution added to a final volume of 50 μl containing 0.2 μl of GoTaq Flexi DNA polymerase (5 U/μl Promega), 2 mM (each) dATP, dCTP, dGTP, and dTTP (Promega); 10 μl of 5x PCR buffer supplied by the manufacturer; 1 μM of each primers; 1 μL of dimethyl sulfoxide (Sigma) and 1.5 mM of MgCl2. The reactions were carried out using a TC-512 thermal cycler (Techne). PCR conditions were as follows: 1 cycle of 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and 1 cycle of 7 min at 72°C. To detect presence or absence of each LSP, PCR products were analyzed by electrophoresis using 1.5% agarose gels.
MIRU-VNTR analysis
DNA in agarose plugs prepared for PFGE analysis was used for MIRU-VNTR analysis according to Stevenson et al. [23]. Small pieces of agarose plug, approximately 2 mm thick, were washed in TE buffer (pH 8) to remove residual EDTA in the storage buffer. One hundred microlitres of TE buffer were added to the agarose and the sample boiled for 10 min to melt the agarose. Five microlitres were used for PCR and MIRU-VNTR analysis interrogating eight polymorphic loci was performed as described by Thibault et al.[11]. The allelic diversity (h) at a locus was calculated by using Nei’s index (see Additional file 3: Table S4) h = 1 − ∑ xi2n/(n − 1)], where xi is the frequency of the ith allele at the locus, and n the number of isolates [24].
Calculation of the discriminatory power
The Simpson Discrimination Index (DI) described by Hunter and Gaston [25] was used as a numerical index for the discriminatory power of each typing method PFGE, IS900-RFLP and MIRU-VNTR and combinations of the typing methods (see Table 2 and Additional file 3: Table S4). The DI was calculated using the following formula:
Table 2.
Discriminatory Index of IS900 RFLP, MIRU-VNTR and SnaB1, Spe1 PFGE typing used alone and in combination
| Typing methods | S type | C type1 | All types | |||
|---|---|---|---|---|---|---|
| |
Subtypes |
I |
III |
I + III |
II |
|
| IS900 RFLP |
No.2 |
10 |
14 |
24 |
35 |
59 |
| DI |
0.356 |
0.934 |
0.873 |
0.644 |
0.856 |
|
| PFGE SnaB1 |
No. |
5 |
10 |
15 |
24 |
39 |
| DI |
1.000 |
0.956 |
0.990 |
0.895 |
0.960 |
|
| PFGE Spe1 |
No. |
5 |
10 |
15 |
24 |
39 |
| DI |
1.000 |
0.978 |
0.990 |
0.801 |
0.924 |
|
| PFGE (SnaB1-Spe1) |
No. |
5 |
10 |
15 |
24 |
39 |
| DI |
1.000 |
1.000 |
1.000 |
1.000 |
1.000 |
|
| MIRU-VNTR |
No. |
10 |
14 |
24 |
35 |
59 |
| DI |
0.644 |
0.89 |
0.801 |
0.876 |
0.925 |
|
| IS900 RFLP + MIRU-VNTR |
No. |
10 |
14 |
24 |
35 |
59 |
| DI | 0.644 | 0.736 | 0.935 | 0.965 | 0.977 | |
1: Panel of strains selected to represented the whole diversity of RFLP and MIRU-VNTR profiles of type C strains.
2: No. Number of strains analyzed.
| (1) |
Where N is the total number of isolates in the typing scheme, s is the total number of distinct patterns discriminated by each typing method and strategy, and nj is the number of isolates belonging to the jth pattern.
Results
LSP analysis
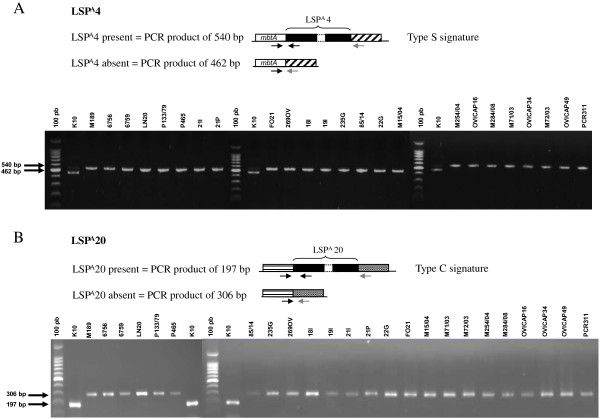
The existence of two major Map lineages was previously supported by the distribution of two LSPs specific for Map as described by Semret et al. [12,26,27]: the region corresponding to LSPA20 is specifically absent from strains of the sheep lineage whereas that corresponding to LSPA4 is specifically absent from strains of the cattle lineage. The distribution of these LSPs was thus investigated across our representative panel of Map S-type strains from various origins. As shown in Figure 1, analysis by PCR supports the association of the LSPA20 region with C-type strains whereas the LSPA4 region is present in all S-type strains. Presence of the LSPA4 region was not related to PFGE subtype I versus III, of the country of origin and pigmentation status (Table 1).
Figure 1.
Detection of types and subtypes of strains based on of the absence or presence of large sequences LSPA4 (A) and LSPA20 (B) investigated by PCR.
SNP analysis
Since SNPs found in gyrA and B genes have been reported to be subtype (I, II, III)-specific, the panel of Map S-type strains was subjected to SNP analysis and compared to C type K-10 strain. As shown in Table 3, consensus sequences obtained matched those previously published and distinguished types I, II and III of Map.
Table 3.
SNPs found in gyrA and gyrB genes for M. avium subsp. paratuberculosis strain K-10 and M. avium subsp. paratuberculosis types I and III
| Strains | Type | IS900 RFLP profiles |
gyrA |
gyrB |
||
|---|---|---|---|---|---|---|
| position 1822 | 1986 | 1353 | 1626 | |||
| K10* |
II |
R01 |
CCCGAGGAGCGGATCGCT- |
ACTCGTGGGCGCGGTGTTGT |
CCGGTCGACCGATCCGCGC- |
CCAGCACATCTCGACGCTGT |
| 6756 |
I |
S1 |
.....A....- |
.......... |
......C...- |
.......... |
| 6759 |
I |
S1 |
.....A....- |
.......... |
......C...- |
.......... |
| P133/79 |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| 21P |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| 235 G |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| M189 |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| M15/04 |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| M254/04 |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| M71/03 |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| M72/03 |
I |
S2 |
.....A....- |
.......... |
......C...- |
.......... |
| 22 G |
III |
A |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| OVICAP16 |
III |
A |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| OVICAP49 |
III |
A |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| 21I |
III |
B |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| PCR311 |
III |
B |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| 19I |
III |
C |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| 85/14 |
III |
C |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| OVICAP34 |
III |
D |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| 18I |
III |
E |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| FO21 |
III |
F |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| LN20 |
III |
I1 |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| 269OV |
III |
I10 |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| M284/08 |
III |
I10 |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| P465 |
III |
I2 |
.....A....- |
.....T..... |
......C...- |
.....T..... |
| Consensus |
I |
|
CCCGAGGAGAGGATCGCT- |
ACTCGTGGGCGCGGTGTTGT |
CCGGTCGACCGACCCGCGC- |
CCAGCACATCTCGACGCTGT |
| Consensus |
III |
|
CCCGAGGAGAGGATCGCT- |
ACTCGTGGGTGCGGTGTTGT |
CCGGTCGACCGACCCGCGC- |
CCAGCACATTTCGACGCTGT |
| K10 | II | CCCGAGGAGCGGATCGCT- | ACTCGTGGGCGCGGTGTTGT | CCGGTCGACCGATCCGCGC- | CCAGCACATCTCGACGCTGT | |
*K10 sequenced strain type II used as sequence reference.
PFGE typing
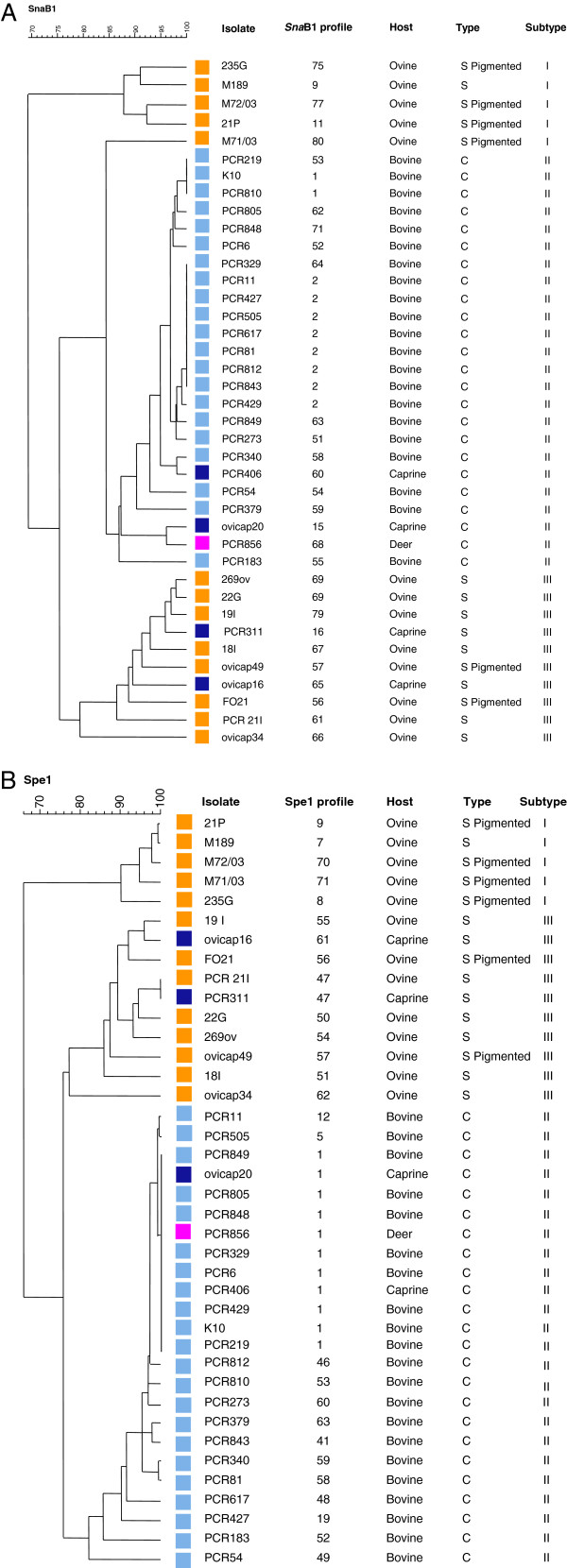
PFGE analysis results were obtained for 15 S-type and 24 C-type strains (Figure 2A and 2B). The sequenced K10 type II strain was also included. SnaB1 or SpeI analyses segregated strains according to the two sheep and cattle lineages and at the subtype level I, II and III. With SnaBI and SpeI individually, 5 different profiles were obtained for the 5 type I strains and 9 different profiles for the 10 type III strains. The type II strains exhibited 15 different SnaBI profiles, with profile [2] being the most frequent (8 strains) and 14 different SpeI profiles with profile [1] being the most frequent (11 strains). The DI of the subtype I and subtype III were respectively 1 and 0.956 for SnaB1 and 1 and 0.978 for SpeI and that of C-type (Type II) was 0.895 for SnaBI and 0.801 for SpeI (see Table 2 and Additional file 3: Table S4). DI of 0.96 and 0.924 for SnaBI and SpeI respectively was achieved for the 39 Map strains presented in Figure 2A and 2B. The combination of both enzymes gave 39 unique multiplex profiles (see Table 1 and Additional file 1: Table S1).
Figure 2.
UPGMA Dendrogram showing the profiles of Map strain obtained by PFGE using SnaB1 (A) or (B) Spe1. The numbering codes of the profiles obtained for each enzyme were assigned according to the nomenclature available at http://www.moredun.org.uk/PFGE-mycobacteria. The colored squares indicate the animal origin of strains: cattle (sky blue), sheep (orange), goat (dark blue) and deer (purple).
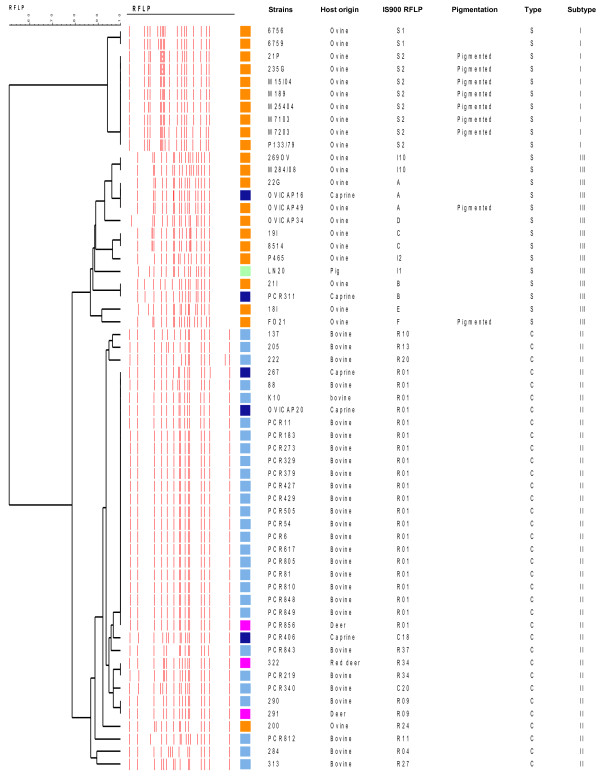
IS900-RFLP typing
IS900-RFLP typing clearly separated the strains into three groups that correlate with the PFGE subtypes I, II and III (Figure 3). Ten strains of S-type, subtype I cluster into two groups of profiles S1 (n = 2) and S2 (n = 8). The 14 strains of S-type, subtype III display more polymorphism with 9 profiles, including 6 new ones. Profiles previously described included I1 (n = 1), I2 (n = 1) and I10 (n = 2). The new profiles were called A (n = 3), B (n = 2), C (n = 2), D, E and F (n = 1 each) (indicated in the Additional file 4: Figure S1). The strains of C-type were well distinguished from S-type and were not highly polymorphic. In this panel of strains the most widely distributed profile R01 was found for 21 strains, then R09 (n = 2) and R34 (n = 2) and 10 profiles were identified in only one isolate, R04, R10, R11, R13, R20, R24, R27, R37, C18 and C20. With this Map panel of strains the discrimination index (DI) of RFLP was shown very variable depending on the type and the subtype of the strains. The DI of the subtype I was very low (0.356), for the subtype III high (0.934) and that of C-type (Type II) was low (0.644) (Table 2). A DI of 0.856 was achieved for the 59 Map strains presented in Figure 3.
Figure 3.
UPGMA dendrogram based on IS900 RFLP typing, using Bst EII on a panel of strains of S-type and C-types. 28 strains were isolated from cattle (sky blue), 22 from sheep (orange), 5 from goat (dark blue), 3 from deer (purple) and one from pig (light green).
MIRU-VNTR typing
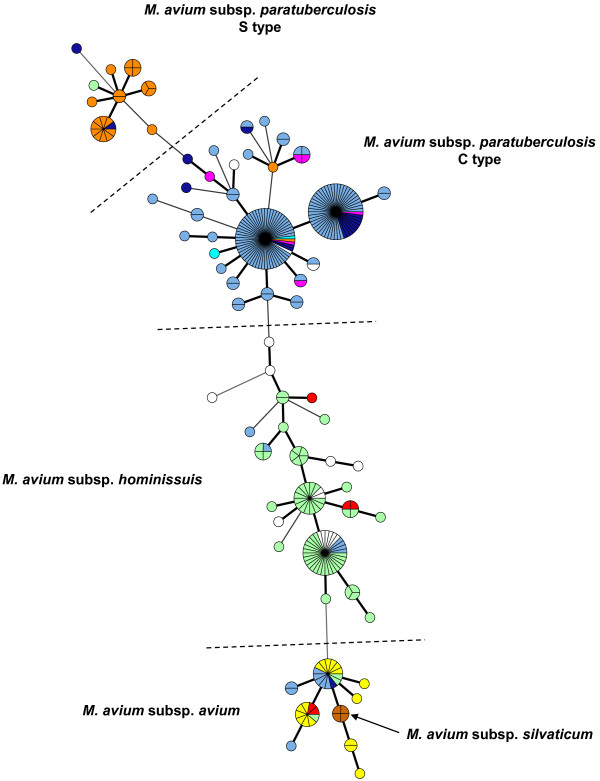
The result of MIRU-VNTR typing of the S-type strains is shown in Table 1. MIRU-VNTR data from 148 C-type (type II) strains previously described [11,18,19] were included in the analysis (see Additional file 1: Table S1). MIRU-VNTR using the eight markers described previously [11] could differentiate between S- and C-type strains but not between the subtypes I and III. On this panel of strains, type III strains were the most polymorphic with a DI of 0.89 compared to 0.644 for type I strains and 0.876 for type II strains selected to represent the diversity of INMV profiles described. INMV profiles 21, 70 and 72 were shared by both type I and III strains. As described previously [11] IS900 RFLP and MIRU-VNTR typing may be used in combination to gain higher resolution. This was verified also on this panel of strains including S-type. In total, the combination of the two methods distinguished 32 distinct patterns comprising 59 isolates. Therefore, using carefully on the same set of strains, a DI of 0.977 was achieved for this panel by using IS900 RFLP and MIRU-VNTR typing in combination compared to 0.856 for IS900 RFLP typing alone and 0.925 for MIRU-VNTR typing (Table 2 and Additional file 3: Table S4). Because MIRU-VNTR is applicable to all members of the MAC, we wanted to know how the INMV profiles segregated within the MAC. None of the INMV profiles identified in the S-type strains matched those of other MAC members. The results presented by the minimum spanning tree in Figure 4, show that Map S-type strains are clearly separated from Map C-type strains, including 113 strains previously typed, and also from any strains belonging to the other subspecies hominissuis, avium or silvaticum. The allelic diversities of the various loci are shown in Additional file 5: Table S3. Five markers were monomorphic in Map S subtype III and 7 in Map S subtype I. In terms of the discriminatory hierarchy, locus 292 displayed the highest allelic diversity for both S- and C-type strains. This study shows that genotyping with MIRU-VNTR can distinguish MAC isolates to the species level and also distinguish with MAP subspecies to the strain type level.
Figure 4.
Minimum spanning tree based on MIRU-VNTR genotypes among Mycobacterium avium subsp. paratuberculosis of types S and C, Mycobacterium avium subsp. avium, Mycobacterium avium subsp. hominissuis, and Mycobacterium avium subsp. silvaticum. 135 strains were isolated from cattle (sky blue), 23 strains from sheep (orange), 17 strains from goat (dark blue), 63 strains from pigs (light green), 17 strains from birds (yellow), 17 strains from humans (white), 6 strains from deer (purple), 5 strains from other sources (red), 4 strains from wood pigeons (brown), and 2 different vaccine strains (316 F from France and United Kingdom) (light blue). Each genotype is displayed as a pie chart, the size of which is proportional to the number of strains, with color-coded distribution of the strain origins. The number of loci differing between the genotypes is indicated by the style of the connecting lines: thick and short, 1 difference; intermediate, 2 differences; thin and long: 3 differences.
Discussion
In comparison to Map C-type strains, investigation of the epidemiology and genetics of S-type strains has been hampered due to difficulties in their isolation and their extremely slow growth-rate in laboratory culture [28,29]. Indeed, the isolation and maintenance of Map S-type strains continues to be a challenge for laboratories worldwide and relative to Map C-type strains a paltry number are available for study. Nowadays representative genome sequences are available for both C- and S-type subtype III Map strains [30,31]. This has facilitated the identification of specific genetic elements that can be used to identify isolates and discriminate between types and, in some cases subtypes of strains [14,16,22,32-34]. In this study we assembled a panel of S-type strains from different geographic origins and host species and undertook extensive molecular typing to improve our knowledge on the genetic diversity of these strains and their phylogenetic relationship with respect to Map C-type strains and other members of MAC. This is the largest panel of S-type strains investigated to date. Additionally, the study also permitted identification of the most efficient typing techniques for S-type strains. The results of the study coupled with previous results on genotypic and phenotypic characterization of Map strains concur with the division of this subspecies into two major lineages comprising S-type and C-type strains. However, the results of IS900-RFLP, PFGE and SNP analysis of the gyr genes clearly divide Map strains into three subtypes, Type II or C strains, Type I and Type III strains. But from the data available on these strains, the two subtypes do not seem to be associated with a particular phenotype and may just reflect regional genetic differences. Type I was first proposed to describe a group of ovine pigmented Map strains with distinctive PFGE profiles [8]. However, as more ovine strains were typed by PFGE, it became apparent that there was another cluster of non-pigmented ovine Map strains that were designated Type III strains [7]. The pigmented phenotype consequently became associated with the Type I strains. However, in this study we included two pigmented strains originating from different geographic locations, which were typed as type III by SNP analysis of the gyr genes, IS900 RFLP and PFGE. The pigmentation phenotype is not therefore restricted to type I and there is no other obvious phenotype currently known to differentiate between types I and III. MIRU-VNTR, despite being highly discriminatory between strains did not separate the S-type strains into the two types I and III. There is therefore an argument for simplifying the current nomenclature for Map strain grouping [14]. Due to the historical nomenclature, to the absence of other comprehensive studies including all strain types and typing methods, to the inability of several techniques to distinguish between Type I and III and to the genetic and phenotypic similarities found between them in previous studies, we propose that S- and C-type nomenclature could be used to denote the two major groups or lineages and the Type I and III used to distinguish subtypes within S-type strains as we have done in this paper. In agreement with previous studies both PFGE and IS900-RFLP revealed little heterogeneity between isolates of the S subtype I. By comparison, this study shows that strains of S subtype III are more polymorphic. Diverse genotypes clustered within S subtype III have been identified circulating in small regional areas in Spain or even in the same farm [34], making more evident the higher heterogeneity of these strains. Interestingly, as far as we know no evidence of S subtype I strains has been found in Spain, a country with a significant sample of S-type strains in our panel and in previous works [8,16].
For molecular epidemiology (i.e. strain tracking), of the typing techniques used MIRU-VNTR would be the preferred technique for studying S-type strains. This technique gave a high discriminatory index with the eight loci employed in this study and could segregate the different members of MAC and the Map S- and C-type strains, although it has limitations in that it cannot differentiate between the subtypes I and III. For detecting genetic variability between S-type strains the number of loci used could be reduced to 3 (292, X3 and 25). The greatest genetic variation occurred at locus 292 with S-type strains typically having a much higher number of repeats than C-type strains (up to 11 were detected in this study). No more than 4 repeats at locus 292 were detected in C-type strains. The locus 292 locus is flanked by loci MAP2920c and MAP2921c referenced as acetyltransferase and quinone oxidoreductase, respectively. There has been only one other report of MIRU-VNTR typing of S-type strains [22]. In the latter study MIRU-VNTR loci 3 and 7 were thought to be of special importance for identifying subtype III strains but only two subtype III strains were typed. In our study all 14 subtype III and 10 subtype I strains had the same, one-repeat unit alleles at each of these two loci, as found in the two strains typed previously [22]. Although uncommon, a few C-type strains in this study were also found with a single copy at these loci so this is even not unique to S-type strains. All Mah, Maa and Mas strains tested in this study also had one repeat unit at locus 3 and all Maa and 61% of Mah strains had a single copy at locus 7. The discriminatory power of MIRU-VNTR to differentiate between the subtypes I and III could be improved by identifying additional loci. Although MIRU-VNTR cannot distinguish between the subtypes I and III, currently it is the only PCR-based typing technique to reveal significant genetic diversity between S-type strains useful for epidemiological investigations. The technique requires only a small amount of DNA and can therefore be carried out on single colonies as well as cell pellets from liquid culture systems. LSP analysis rapidly differentiates the S-type from C-type strains by the absence of LSPA20 and presence of LSPA4 but provides no information regarding genetic diversity within S-type strains. SNP analysis of the gyr genes is more complex requiring sequencing of the PCR product to differentiate between S- and C-types and between subtypes I and III [13]. However, the S subtype information would be of limited value for epidemiological studies and tracing the source of infection. Furthermore, as we become better at isolating S-type strains and type more strains it is likely that further S subtypes will become apparent. PFGE and IS900-RFLP both give good discrimination between the Map strain types and subtypes but require larger amounts of high quality DNA, which necessitates in vitro growth of the strains and therefore is not ideal for S-type strains.
Conclusions
This is the largest panel of S-type strains investigated to date. The S-type strains can be further divided into two types, I and III, by some (IS900-RFLP, PFGE and SNP analysis of the gyr genes) but not all (not by MIRU-VNTR typing) of the typing techniques. Pigmentation is not exclusively associated with S subtype I strains. Therefore, a simplified nomenclature is proposed designating types I and III as subtypes of S-type strains. The epidemiological and phylogenetic significance of S type subdivision into I and III subtypes needs, however, to be further clarified. Molecular typing using IS900-RFLP, PFGE and MIRU-VNTR demonstrates that S-type strains are genetically diverse, subtype III being the most heterogeneous group. Due to the scarcity of S-type strains in culture, typing techniques have been largely optimized using C-type strains. Further genomic sequencing of S-type strains should reveal variable genetic loci unique to S-type strains that could be exploited to further improve discrimination of S-type strains. Genome sequence data of isolates belonging to subtypes I and III should ultimately clarify the phylogeny and provide a framework to classify different phenotypic, pathogenic and epidemiological characteristics of Map strains.
Competing interests
The authors have no competing interests.
Authors’ contributions
FB, IS and KS conceived of the study, participated in its design and coordination, collated and analysed the data and drafted the manuscript. TC, LL, JG, IH, JM and VT participated in the laboratory and field work. RJ, TC, LL, PS participated in analysing the data. All authors read, criticized and approved the final manuscript.
Supplementary Material
Table S1. Description of the strains used in this study indicating their origins and details of their genotypes and phenotypes data.
Table S2. The table shows the primers sequences used in this study.
Table S4. The table details the calculation of the Discriminatory Index for each typing methods including IS900 RFLP, MIRU-VNTR and PFGE (SnaB1, Spe1) used alone and in combination. The table details the calculation of the allelic diversity (h) at a locus MIRU-VNTR using Nei’s index.
Figure S1. The figure shows the new IS900 RFLP profiles obtained from analysis with strains S of subtype III.
Table S3. The table describes the MIRU-VNTR allelic distribution among the strains of Map of type S and C and other Mac members.
Contributor Information
Franck Biet, Email: Franck.Biet@tours.inra.fr.
Iker A Sevilla, Email: isevilla@neiker.net.
Thierry Cochard, Email: Thierry.Cochard@tours.inra.fr.
Louise H Lefrançois, Email: llefrancois@tours.inra.fr.
Joseba M Garrido, Email: jgarrido@neiker.net.
Ian Heron, Email: ianheron23@hotmail.com.
Ramón A Juste, Email: rjuste@neiker.net.
Joyce McLuckie, Email: joyce.mcluckie@moredun.ac.uk.
Virginie C Thibault, Email: virginie.thibault@univ-tours.fr.
Philip Supply, Email: philip.supply@ibl.fr.
Desmond M Collins, Email: des.collins@agresearch.co.nz.
Marcel A Behr, Email: marcel.behr@mcgill.ca.
Karen Stevenson, Email: karen.stevenson@moredun.ac.uk.
Acknowledgements
FB, TC, LL and VT were supported by the Institut National de la Recherche Agronomique. KS, IH and JM were funded by the Scottish Government Rural and Environment Science and Analytical Services Division. The work of IS, JG and RJ was supported by the Departamento de Medio Ambiente, Planificación Territorial, Agricultura y Pesca del Gobierno Vasco.
References
- Alexander DC, Turenne CY, Behr MA. Insertion and deletion events that define the pathogen mycobacterium avium subsp. Paratuberculosis. J Bacteriol. 2009;19(3):1018–1025. doi: 10.1128/JB.01340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DM, Gabric DM, de Lisle GW. Identification of two groups of mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J Clin Microbiol. 1990;28(7):1591–1596. doi: 10.1128/jcm.28.7.1591-1596.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lisle GW, Collins DM, Huchzermeyer HF. Characterization of ovine strains of mycobacterium paratuberculosis by restriction endonuclease analysis and DNA hybridization. Onderstepoort J Vet Res. 1992;59(2):163–165. [PubMed] [Google Scholar]
- Janagama HK, Kumar S, Bannantine JP, Kugadas A, Jagtap P, Higgins L, Witthuhn B, Sreevatsan S. Iron-sparing response of mycobacterium avium subsp. Paratuberculosis is strain dependent. BMC Microbiol. 2010;10(1):268. doi: 10.1186/1471-2180-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janagama HK, Jeong K, Kapur V, Coussens P, Sreevatsan S. Cytokine responses of bovine macrophages to diverse clinical mycobacterium avium subspecies paratuberculosis strains. BMC Microbiol. 2006;6:10. doi: 10.1186/1471-2180-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala AS, Janagama HK, Paustian ML, Zhu X, Bannantine JP, Kapur V, Sreevatsan S. Comparative transcriptional analysis of human macrophages exposed to animal and human isolates of mycobacterium avium subspecies paratuberculosis with diverse genotypes. Infect Immun. 2006;74(11):6046–6056. doi: 10.1128/IAI.00326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan L, Alvarez J, Aranaz A, Rodriguez A, Romero B, Bezos J, Mateos A, Dominguez L. Molecular epidemiology of types I/III strains of mycobacterium avium subspecies paratuberculosis isolated from goats and cattle. Vet Microbiol. 2006;115(1–3):102–110. doi: 10.1016/j.vetmic.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Stevenson K, Hughes VM, de Juan L, Inglis NF, Wright F, Sharp JM. Molecular characterization of pigmented and nonpigmented isolates of mycobacterium avium subsp. Paratuberculosis. J Clin Microbiol. 2002;40(5):1798–1804. doi: 10.1128/JCM.40.5.1798-1804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the mycobacterium tuberculosis genome. Mol Microbiol. 2000;36(3):762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- Thibault VC, Grayon M, Boschiroli ML, Hubbans C, Overduin P, Stevenson K, Gutierrez MC, Supply P, Biet F. New variable number tandem repeat markers for typing M. avium subsp. paratuberculosis and M. avium strains: comparison with IS900 RFLP and IS1245 RFLP typing. J Clin Microbiol. 2007;45(8):2404–2410. doi: 10.1128/JCM.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semret M, Turenne CY, de Haas P, Collins DM, Behr MA. Differentiating host-associated variants of mycobacterium avium by PCR for detection of large sequence polymorphisms. J Clin Microbiol. 2006;44(3):881–887. doi: 10.1128/JCM.44.3.881-887.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Aranaz A, Romero B, de Juan L, Alvarez J, Bezos J, Rodriguez S, Stevenson K, Mateos A, Dominguez L. Polymorphisms in gyrA and gyrB genes among mycobacterium avium subsp. Paratuberculosis type I, II, and III isolates. J Clin Microbiol. 2007;45(10):3439–3442. doi: 10.1128/JCM.01411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turenne CY, Collins DM, Alexander DC, Behr MA. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J Bacteriol. 2008;190(7):2479–2487. doi: 10.1128/JB.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lisle GW, Yates GF, Collins DM. Paratuberculosis in farmed deer: case reports and DNA characterization of isolates of Mycobacterium paratuberculosis. J Vet Diagn Invest. 1993;5(4):567–571. doi: 10.1177/104063879300500411. [DOI] [PubMed] [Google Scholar]
- Sevilla I, Garrido JM, Geijo M, Juste RA. Pulsed-field gel electrophoresis profile homogeneity of Mycobacterium avium subsp. paratuberculosis isolates from cattle and heterogeneity of those from sheep and goats. BMC Microbiol. 2007;7:18. doi: 10.1186/1471-2180-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlik I, Horvathova A, Dvorska L, Bartl J, Svastova P, du Maine R, Rychlik I. Standardisation of restriction fragment length polymorphism analysis for mycobacterium avium subspecies paratuberculosis. J Microbiol Methods. 1999;38(1–2):155–167. doi: 10.1016/s0167-7012(99)00091-3. [DOI] [PubMed] [Google Scholar]
- Radomski N, Thibault VC, Karoui C, de Cruz K, Cochard T, Gutierrez C, Supply P, Biet F, Boschiroli ML. Determination of genotypic diversity of mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol. 2010;48(4):1026–1034. doi: 10.1128/JCM.01869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault VC, Grayon M, Boschiroli ML, Willery E, Allix-Beguec C, Stevenson K, Biet F, Supply P. Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of mycobacterium avium subsp. Paratuberculosis isolates. J Clin Microbiol. 2008;46(12):4091–4094. doi: 10.1128/JCM.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DM, Cavaignac S, de Lisle GW. Use of four DNA insertion sequences to characterize strains of the mycobacterium avium complex isolated from animals. Mol Cell Probes. 1997;11(5):373–380. doi: 10.1006/mcpr.1997.0131. [DOI] [PubMed] [Google Scholar]
- Overduin P, Schouls L, Roholl P, van der Zanden A, Mahmmod N, Herrewegh A, van Soolingen D. Use of multilocus variable-number tandem-repeat analysis for typing mycobacterium avium subsp. Paratuberculosis. J Clin Microbiol. 2004;42(11):5022–5028. doi: 10.1128/JCM.42.11.5022-5028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobius P, Fritsch I, Luyven G, Hotzel H, Kohler H. Unique genotypes of Mycobacterium avium subsp. paratuberculosis strains of Type III. Vet Microbiol. 2009;139(3–4):398–404. doi: 10.1016/j.vetmic.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Stevenson K, Alvarez J, Bakker D, Biet F, de Juan L, Denham S, Dimareli Z, Dohmann K, Gerlach GF, Heron I. et al. Occurrence of mycobacterium avium subspecies paratuberculosis across host species and european countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol. 2009;9:212. doi: 10.1186/1471-2180-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89(3):583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semret M, Alexander DC, Turenne CY, de Haas P, Overduin P, van Soolingen D, Cousins D, Behr MA. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J Clin Microbiol. 2005;43(8):3704–3712. doi: 10.1128/JCM.43.8.3704-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semret M, Zhai G, Mostowy S, Cleto C, Alexander D, Cangelosi G, Cousins D, Collins DM, van Soolingen D, Behr MA. Extensive genomic polymorphism within mycobacterium avium. J Bacteriol. 2004;186(18):6332–6334. doi: 10.1128/JB.186.18.6332-6334.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins DV, Evans RJ, Francis BR. Use of BACTEC radiometric culture method and polymerase chain reaction for the rapid screening of faeces and tissues for mycobacterium paratuberculosis. Aust Vet J. 1995;72(12):458–462. doi: 10.1111/j.1751-0813.1995.tb03489.x. [DOI] [PubMed] [Google Scholar]
- Whittington RJ, Marsh I, Turner MJ, McAllister S, Choy E, Eamens GJ, Marshall DJ, Ottaway S. Rapid detection of Mycobacterium paratuberculosis in clinical samples from ruminants and in spiked environmental samples by modified BACTEC 12B radiometric culture and direct confirmation by IS900 PCR. J Clin Microbiol. 1998;36(3):701–707. doi: 10.1128/jcm.36.3.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Wu CW, Hsu C, Zhou S, Schwartz DC, Bayles DO, Paustian ML, Alt DP, Sreevatsan S, Kapur V. et al. Genome sequencing of ovine isolates of mycobacterium avium subspecies paratuberculosis offers insights into host association. BMC Genomics. 2012;13(1):89. doi: 10.1186/1471-2164-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA. 2005;102(35):12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Aranaz A, Gould KA, Linedale R, Stevenson K, Alvarez J, Dominguez L, de Juan L, Hinds J, Bull TJ. Discovery of stable and variable differences in the mycobacterium avium subsp. Paratuberculosis type I, II, and III genomes by pan-genome microarray analysis. Appl Environ Microbiol. 2009;75(3):676–686. doi: 10.1128/AEM.01683-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Romero B, Rodriguez S, de Juan L, Bezos J, Mateos A, Dominguez L, Aranaz A. Molecular characterization of mycobacterium avium subspecies paratuberculosis types II and III isolates by a combination of MIRU-VNTR loci. Vet Microbiol. 2010;144(1–2):118–126. doi: 10.1016/j.vetmic.2009.12.028. [DOI] [PubMed] [Google Scholar]
- Sevilla I, Li L, Amonsin A, Garrido JM, Geijo MV, Kapur V, Juste RA. Comparative analysis of Mycobacterium avium subsp. paratuberculosis isolates from cattle, sheep and goats by short sequence repeat and pulsed-field gel electrophoresis typing. BMC Microbiol. 2008;8:204. doi: 10.1186/1471-2180-8-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Description of the strains used in this study indicating their origins and details of their genotypes and phenotypes data.
Table S2. The table shows the primers sequences used in this study.
Table S4. The table details the calculation of the Discriminatory Index for each typing methods including IS900 RFLP, MIRU-VNTR and PFGE (SnaB1, Spe1) used alone and in combination. The table details the calculation of the allelic diversity (h) at a locus MIRU-VNTR using Nei’s index.
Figure S1. The figure shows the new IS900 RFLP profiles obtained from analysis with strains S of subtype III.
Table S3. The table describes the MIRU-VNTR allelic distribution among the strains of Map of type S and C and other Mac members.