Abstract
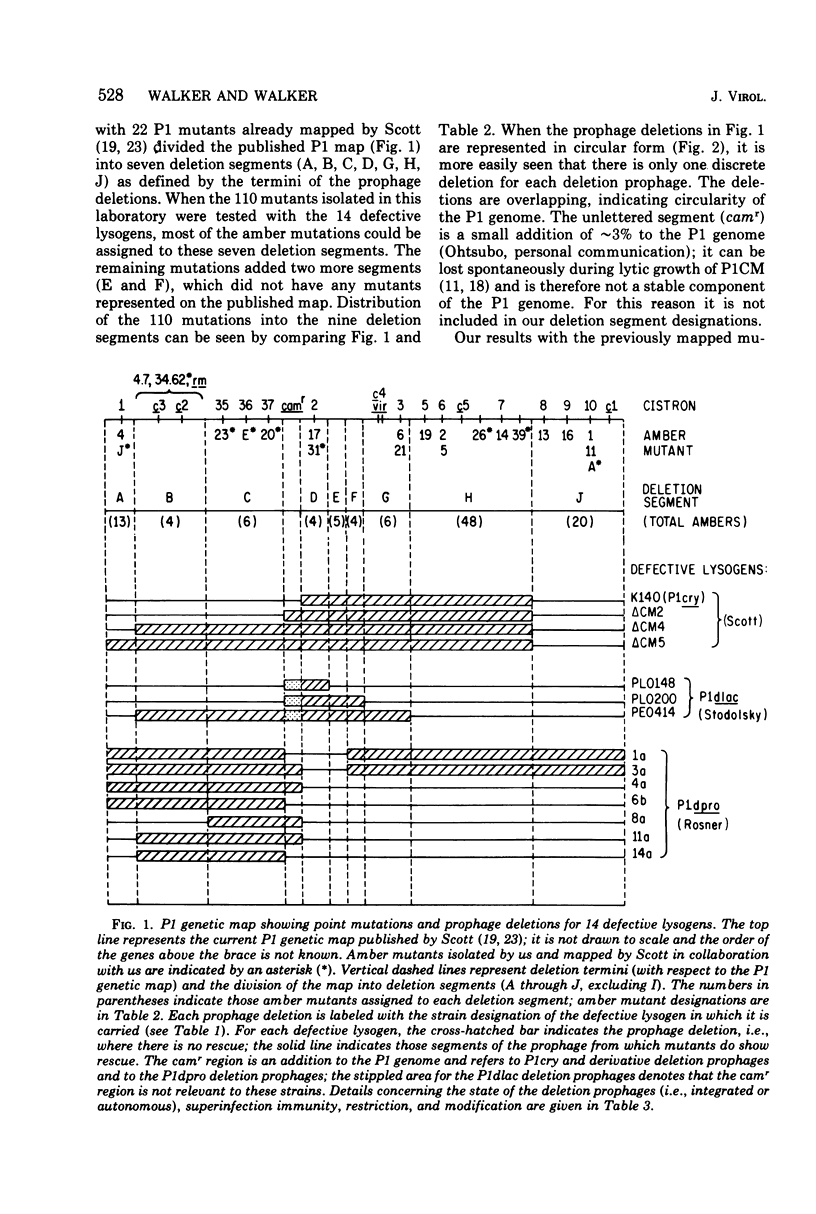
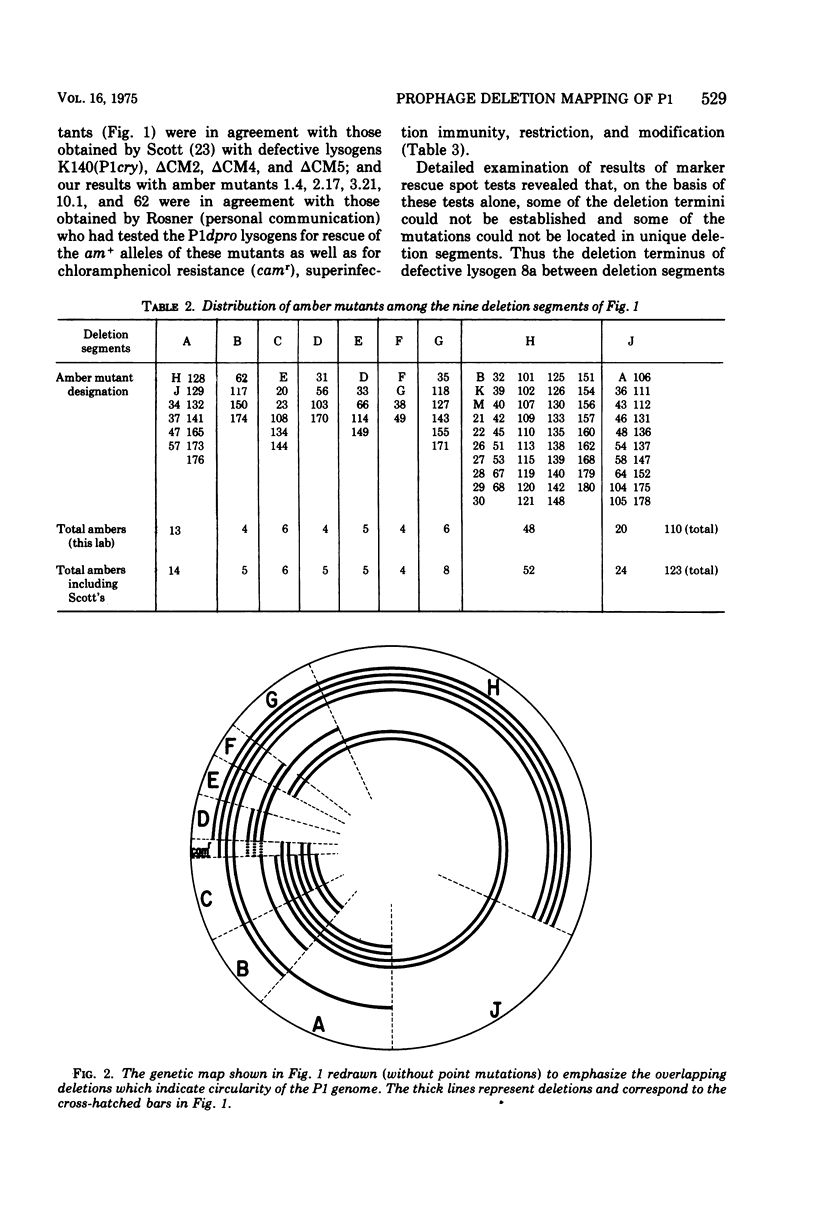
One hundred and ten amber mutants of coliphage P1 were isolated and localized into groups with respect to the existing genetic map by use of nonpermissive Escherichia coli K-12 strains lysogenic for P1 with deletions. These lysogens contain one of three types of deletion prophages: P1cry and its derivatives, P1dlacs, and P1dpros. Fourteen such lysogens were tested for their ability to rescue the amber mutants which were then assigned to one of nine deletion segments of the P1 genome defined by the termini of the various prophage deletions. The relationship of the nine deletion segments with the published P1 map is described, two new segments having been added. The deletions of the 14 prophages overlapped sufficiently to indicate that the P1 genetic prophage map should be represented in circular form, which is consistent with the fact that P1 is normally a circular plasmid in the prophage state. The distribution of mutants into deletion segments is nonrandom for at least one segment. In addition, the deletion termini of the 14 defective prophages coincided in five out of nine regions separating the nine deletion segments. Various possible explanations are discussed for the nonrandom recurrence of these deletion termini, including the evidence of hot spots of recombination.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J., Boram W., Bukhari A. I., Faelen M., Howe M., Metlay M., Taylor A. L., Toussaint A., Van de Putte P., Westmaas G. C. Summary of the genetic mapping of prophage Mu. Virology. 1973 Jul;54(1):90–92. doi: 10.1016/0042-6822(73)90117-7. [DOI] [PubMed] [Google Scholar]
- Adhya S., Cleary P., Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci U S A. 1968 Nov;61(3):956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D. Genetics of bacteriophage P22. I. Isolation of prophage deletions which affect immunity to superinfection. Virology. 1972 Jul;49(1):257–267. doi: 10.1016/s0042-6822(72)80027-8. [DOI] [PubMed] [Google Scholar]
- Hertman I., Scott J. R. Recombination of phage P1 in recombination deficient hosts. Virology. 1973 Jun;53(2):468–470. doi: 10.1016/0042-6822(73)90227-4. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Azure mutants: a type of host-dependent mutant of the bacteriophage f2. Science. 1967 Jun 23;156(3782):1618–1623. doi: 10.1126/science.156.3782.1618. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- KONDO E., MITSUHASHI S. DRUG RESISTANCE OF ENTERIC BACTERIA. IV. ACTIVE TRANSDUCING BACTERIOPHAGE P1 CM PRODUCED BY THE COMBINATION OF R FACTOR WITH BACTERIOPHAGE P1. J Bacteriol. 1964 Nov;88:1266–1276. doi: 10.1128/jb.88.5.1266-1276.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LURIA S. E., ADAMS J. N., TING R. C. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology. 1960 Nov;12:348–390. doi: 10.1016/0042-6822(60)90161-6. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Otsubo E., Deonier R. C., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. V. ilv+ Deletion mutants of F14. J Mol Biol. 1974 Nov 15;89(4):585–597. doi: 10.1016/0022-2836(74)90037-0. [DOI] [PubMed] [Google Scholar]
- Rae M. E., Stodolsky M. Chromosome breakage, fusion and reconstruction during P1dl transduction. Virology. 1974 Mar;58(1):32–54. doi: 10.1016/0042-6822(74)90139-1. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Modification-deficient mutants of bacteriophage P1. I. Restriction by P1 cryptic lysogens. Virology. 1973 Mar;52(1):213–222. doi: 10.1016/0042-6822(73)90410-8. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A defective P1 prophage with a chromosomal location. Virology. 1970 Jan;40(1):144–151. doi: 10.1016/0042-6822(70)90386-7. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A new gene controlling lysogeny in phage P1. Virology. 1972 Apr;48(1):282–283. doi: 10.1016/0042-6822(72)90139-0. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Clear plaque mutants of phage P1. Virology. 1970 May;41(1):66–71. doi: 10.1016/0042-6822(70)90054-1. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Genetic studies on bacteriophage P1. Virology. 1968 Dec;36(4):564–574. doi: 10.1016/0042-6822(68)90188-8. [DOI] [PubMed] [Google Scholar]
- Walker D. H., Jr, Anderson T. F. Morphological variants of coliphage P1. J Virol. 1970 Jun;5(6):765–782. doi: 10.1128/jvi.5.6.765-782.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]



