Abstract
Autophagy is a homeostatic process common to all eukaryotic cells that serves to degrade intracellular components. Among three classes of autophagy, macroautophagy is best understood, and is the subject of this Review. The function of autophagy is multifaceted, and includes removal of long-lived proteins and damaged or unneeded organelles, recycling of intracellular components for nutrients, and defense against pathogens. This process has been extensively studied in yeast, and understanding of its functional significance in human disease is also increasing. This Review explores the basic machinery and regulation of autophagy in mammalian systems, methods employed to measure autophagic activity, and then focuses on recent discoveries about the functional significance of autophagy in respiratory diseases, including chronic obstructive pulmonary disease, cystic fibrosis, tuberculosis, idiopathic pulmonary fibrosis, pulmonary arterial hypertension, acute lung injury, and lymphangioleiomyomatosis.
Keywords: autophagy, chronic obstructive pulmonary disease, idiopathic pulmonary fibrosis, epithelial cells, fibroblasts
Autophagy is an evolutionarily conserved process first described more than 50 years ago that is vital to cellular homeostasis. Many of the molecular events and pathways involved in autophagy were discovered in yeast cells, but knowledge of its function and regulation in mammalian systems has increased greatly in the last decade. Best studied in cancer and neurodegenerative diseases, its role in respiratory diseases is becoming increasingly important with each passing year. To date, it has been studied in chronic obstructive pulmonary disease (COPD), pulmonary fibrosis, cystic fibrosis (CF), pulmonary arterial hypertension (PAH), tuberculosis, acute lung injury, and lymphangioleiomyomatosis. This Review first focuses on the basic mechanisms and regulation of autophagy, as well as the various techniques employed in its measurement in cells and tissues. We then turn to the functional significance of autophagy in the aforementioned respiratory diseases (Table 1). Given the cell- and environment-dependent nature of autophagy, the discussion of these diseases here are within the context of the major respiratory cell types. Continued elucidation of the functional role of autophagy in these lung diseases and others will allow better understanding of their pathophysiologic underpinnings and offer potential new diagnostic and/or therapeutic targets.
TABLE 1.
MAJOR STUDIES OF AUTOPHAGY IN RESPIRATORY DISEASE
| Chronic Obstructive Pulmonary Disease | |
| Chen and colleagues, 2008 (48) | Egr-1 regulates autophagy in cigarette smoke–induced chronic obstructive pulmonary disease. |
| Chen and colleagues, 2010 (49) | Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke–induced emphysema. |
| Monick and colleagues, 2010 (53) | Identification of an autophagy defect in smokers’ alveolar macrophages. |
| Pulmonary Arterial Hypertension | |
| Lee and colleagues, 2010 (64) | Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. |
| Cystic Fibrosis | |
| Luciani and colleagues, 2010 (50) | Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. |
| Abdulrahman and colleagues, 2011 (54) | Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. |
| Lymphangioleiomyomatosis | |
| Parkhitko and colleagues, 2011 (46) | Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. |
| Lung Immunity | |
| Gutierrez and colleagues, 2004 (60) | Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. |
| Deretic and colleagues, 2006 (58) | Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defense mechanism. |
| Singh and colleagues, 2006 (59) | Human IRGM induces autophagy to eliminate intracellular mycobacteria. |
| Ponpuak and colleagues, 2010 (61) | Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. |
| Nakahira and colleagues, 2011 (62) | Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. |
| Idiopathic Pulmonary Fibrosis | |
| Mi and colleagues, 2011 (51) | Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. |
| Patel and colleagues, 2012 (63) | Autophagy in idiopathic pulmonary fibrosis. |
| Acute Lung Injury | |
| Tanaka and colleagues, 2012 (52) | Hyperoxia-induced LC3B interacts with the Fas apoptotic pathway in epithelial cell death. |
Numbers in parentheses are reference numbers.
Classes of Autophagy
The term autophagy, derived from the Greek and meaning “self eating,” comprises three related but distinct cellular processes that entail delivery of cytoplasmic components to lysosomes for degradation and recycling. Microautophagy involves nonselective envelopment of cytoplasmic components directly by lysosomes. Chaperone-mediated autophagy consists of selectively transporting cargo tagged by a pentapeptide motif to the lysosome, binding to lysosomal receptors, and translocation across the membrane. The third category of autophagy is macroautophagy. This process begins with the formation of a double-membraned autophagosome enveloping proteins or organelles to be recycled followed by fusion with a lysosome to form an autolysosome. Macroautophagy can involve bulk degradation of cytoplasm or degradation of specific components. This latter process has introduced new subclasses of macroautophagy into the nomenclature (i.e., mitophagy [mitochondria] [1], ERphagy or reticulophagy [endoplasmic reticulum (ER)] [2], pexophagy [peroxisomes] [3], ribophagy [ribosomes] [4], and xenophagy [microbes] [5]). Macroautophagy (henceforth, referred to as autophagy) is the most well studied mode of autophagy, and is the subject of this Review.
The Molecular Machinery of Autophagy
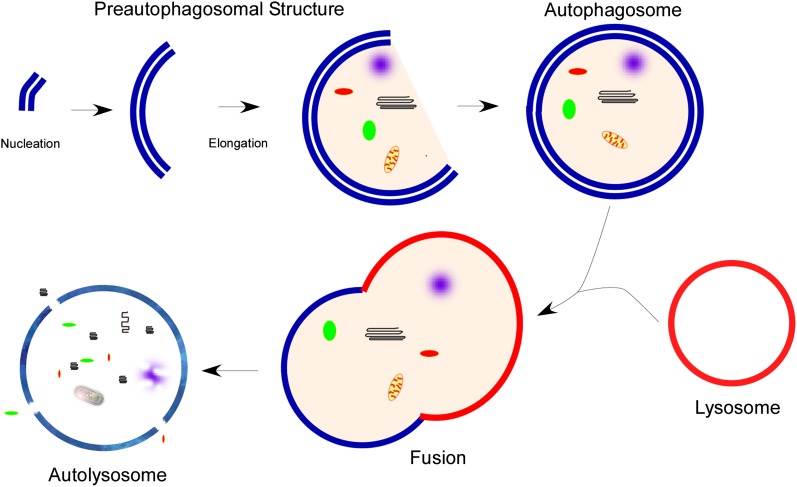
Autophagy is a complex process involving multiple proteins and steps, including: formation of an initiation complex and development of a double-membraned phagophore (nucleation); elongation of the membrane and completion of an autophagosome vesicle around cargo; lysosomal fusion; dissolution of the inner membrane allowing hydrolases to degrade the cargo; and recycling of the components (Figure 1). This process is best studied in yeast, but, over the last decade, mammalian orthologs of many autophagy proteins have been discovered.
Figure 1.
Overview of macroautophagy. Macroautophagy (autophagy) begins with the nucleation and elongation of a preautophagosomal structure. This structure begins to encompass the cargo intended for degradation, followed by completion of the double-membraned autophagosome. The autophagosome fuses with a lysosome, forming the autolysosome. Lysosomal hydrolases then degrade the contents, which are recycled back to the cytosol through membrane permeases.
The exact source of the initial autophagosome membrane formation remains under investigation. This membrane may arise de novo from building blocks in the cytoplasm or from other membranous structures. There is now evidence that the ER contributes to the formation of the phagophore (6–8); however, mitochondria (9) and Golgi (10), as well as the plasma membrane (11), may also be sources.
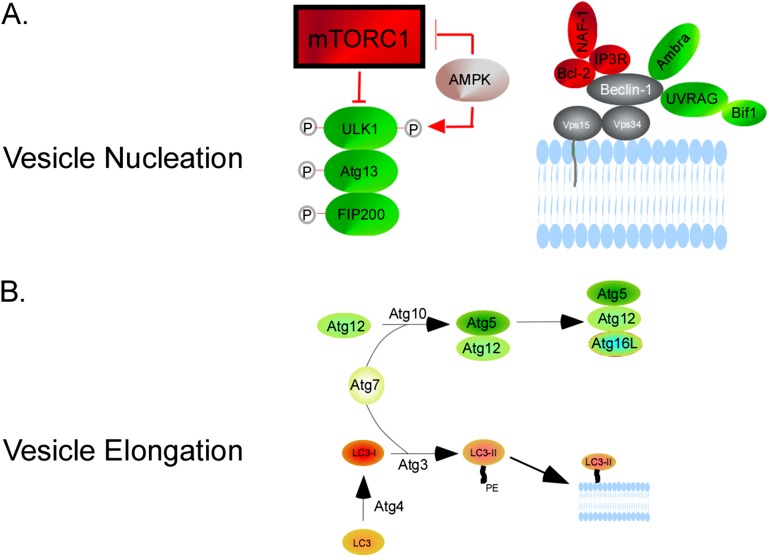
The current paradigm holds that the most upstream portion of the autophagy pathway involves UNC-51–like kinase (ULK) 1 (12). This kinase is part of a complex with Fak-family kinase–interacting protein and Atg13. Under nutrient-replete conditions, mammalian target of rapamycin complex (mTORC) 1 interaction with ULK1 maintains the latter in a phosphorylated and inactive state. When mTORC1 is down-regulated, ULK1 is dephosphorylated, allowing it to phosphorylate Fak-family kinase–interacting protein-200 and Atg13, which initiates vesicle nucleation (Figure 2A) (13). Autophagosome nucleation also requires Vps34, a class III phosphatidylinositol-3-kinase (PI3K) (14). Vps34 is a component of the PI3K complex, additional members of which include beclin-1, Atg14, and p150/Vps15. Through enhancing affinity to its binding partners, beclin-1 potentiates Vps34 activity (15). Beclin-1 activity can be modulated through several binding partners: binding of Ambra-1, ultraviolet (UV) radiation resistance–associated gene (UVRAG), and bif-1 induces autophagy, whereas binding of the antiapoptotic protein, Bcl-2, prevents autophagosome initiation (16). The latter interaction can be further modulated by inositol 1,4,5-triphosphate receptor and nutrient deprivation–autophagy factor, as well as starvation-induced activation of c-Jun NH2-terminal kinase (JNK1), which phosphorylates Bcl-2 and releases beclin-1 (17). Rubicon is another negative regulator through its interaction with the PI3K Complex (18). The product of this complex is phosphatidylinositol-3-phosphate, which recruits factors, such as WD-repeat protein interacting with phosphoinositides-1/2 and double FYVE-containing protein 1 to the preautophagosomal structure.
Figure 2.
Molecular machinery of vesicle nucleation and elongation. (A) Vesicle nucleation begins with mammalian target of rapamycin complex (mTORC) 1 down-regulation, which can be affected by AMP-activated protein kinase (AMPK) via TSC2. This results in UNC-51–like kinase (ULK) 1 dephosphorylation and subsequent phosphorylation of Fak-family kinase–interacting protein (FIP)-200 and Atg13. AMPK can also activate ULK1 directly by phosphorylation at a different site than mTORC1. Nucleation also involves the phosphatidylinositol-3-kinase (PI3K) complex, which includes Vps34, Beclin-1, Atg14, and p150/Vps15. Beclin-1 potentiates Vps34 activity. Beclin-1 activity can be modulated through several binding partners: Ambra-1, ultraviolet (UV) radiation resistance–associated gene (UVRAG), and bif-1 binding induces autophagy; Bcl-2 binding prevents autophagosome initiation. The latter interaction is enhanced by inositol 1,4,5-triphosphate receptor (IP3R), nutrient deprivation–autophagy factor-1 (NAF-1). (B) Vesicle elongation involves two ubiquitin-like reactions. Through the action of Atg7 and Atg10, Atg12 is covalently bound to Atg5, forming Atg12-Atg5. This complex conjugates with Atg16L1, forming Atg12-Atg5-Atg16L1. The second reaction revolves around microtubule-associated protein 1 light chain 3/Atg8 (LC3). Atg4B cleaves the LC3 precursor to form cytosolic LC3-I. Atg7 and Atg3 lipidate LC3-I with phosphatidylethanolamine (PE) to form LC3-II, which is targeted to the forming autophagosome.
The vesicle elongation step involves two ubiquitin-like reactions (Figure 2B). In the first, through the sequential action of Atg7 and Atg10, Atg12 is covalently bound to Atg5, forming Atg12-Atg5 (19). This complex then forms a conjugate with Atg16L1, forming Atg12-Atg5-Atg16L1, which is required for elongation. The second reaction (the better-characterized pathway) revolves around microtubule-associated protein 1 light chain 3 (Atg8), commonly referred to LC3. The LC3 precursor is cleaved by Atg4B to form cytosolic LC3-I. Atg7 and Atg3 then lipidate LC3-I with phosphatidylethanolamine to form LC3-II (20). LC3-II is targeted to the forming autophagosome, studding both the cytoplasmic and luminal membranes of the double-membraned structure. The outer leaflet LC3-II can be delipidated and recycled, but the inner membrane LC3-II is degraded after fusion with lysosomes. By its presence on the mature autophagosome, LC3-II can serve as a marker of this structure.
Completed autophagosomes move along microtubules in a dynein-dependent fashion, and then fuse with late endosomes and lysosomes (21, 22). Fusion involves several proteins, including Rab7, class Vps proteins, and ultraviolet radiation resistance–associated gene (23). The lysosomal hydrolases then degrade the cargo and permeases channel degraded molecules back into the cytoplasm.
Regulatory Pathways in Autophagy
The best-studied regulator of autophagy is mTORC1. The inhibitory, and thus regulatory, effect of mTORC1 relies upon phosphorylation of ULK1. Once mTORC1 is down-regulated and its kinase activity fails to keep ULK1 phosphorylated, the autophagic cascade can begin. A number of other signaling pathways and proteins also regulate autophagy, some of which work through mTORC1 (24). The cellular energy sensor, AMP-activated protein kinase (AMPK), is of particular importance. Activation of AMPK under low ATP:AMP ratios induces phosphorylation of TSC2, which then inhibits mTORC1 and induces autophagy (25). Recently, AMPK has also been shown to regulate autophagy by a second mechanism: it can interact directly with and phosphorylate ULK1 at sites different than mTORC1, resulting in its activation (26). Egan and colleagues (27) also demonstrated that AMPK is required for autophagy, and that, in its absence, p62 accumulates and mitophagy is deficient.
Autophagy can also be induced by ER stress through mechanisms that are still being understood. In several studies, autophagy has been shown to help cells adapt to ER stress in a prosurvival capacity (28). One possible signaling mechanism responsible for this induction involves JNK1. ER stress activates a transmembrane protein, IRE1, which leads to a mitogen-activated protein kinase signaling cascade activating JNK1. Subsequently, JNK1 phosphorylation of beclin-1 induces autophagy, as described previously here (17). Death-associated protein kinase activation by ER stress has also been shown to phosphorylate beclin-1 (29). Other molecules that may be involved include PKR-like ER kinase (PERK1) and eIF2α, the latter of which is phosphorylated under ER stress conditions (30).
The well studied tumor suppressor gene, p53, also has the ability to modulate autophagy, although in a unique way: it can activate and inhibit autophagy (31). The nature of the interaction between p53 and autophagy is likely cell- and context-dependent, and can involve AMPK, PTEN, and TSC1 in an mTORC1-dependent pathway or damage-regulated autophagy modulator in an mTORC1-independent pathway. Through these signaling molecules, genotoxic stress–mediated induction of p53 can stimulate autophagy. However, in the absence of such a stress, basal levels of p53 appear to suppress autophagy (31).
A more recently discovered pathway that can also regulate autophagy occurs through the tp53-induced glycolysis and apoptosis regulator (TIGAR). TIGAR is an isoform of phospho-fructokinase (pFK2) that can shunt glucose from glycolysis to the pentose phosphate pathway. Recent work by Bensaad and colleagues (32) shows that TIGAR inhibits autophagy through quenching of reactive oxygen species (ROS). Although p53 regulates TIGAR, they demonstrated that TIGAR can modulate autophagy independent of p53.
How to Measure Autophagy: A Moving Target
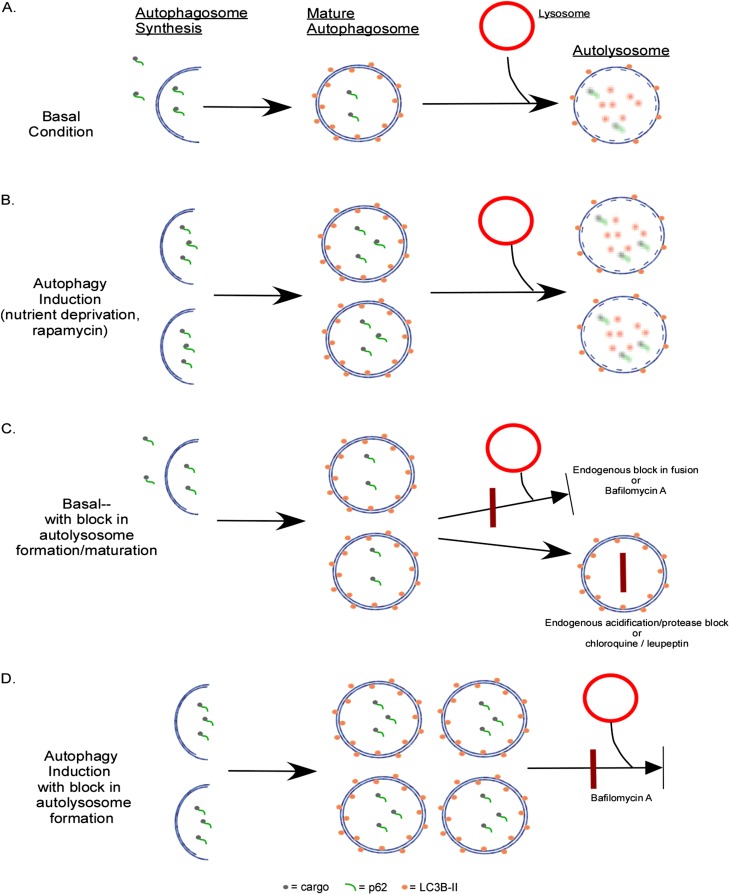
As the field of autophagy has blossomed, the methods for measuring autophagy have also evolved. Historically, the gold standard for analyzing autophagy has been to observe and quantify autophagosomes by electron microscopy (EM) (33, 34). Although this remains a valid method, it can be cumbersome, labor intensive, and expensive to perform EM for every experiment. Therefore, investigators have turned to practical alternate approaches to monitor this process. Most commonly, this involves measuring protein levels of various Atg proteins. Atg8 or microtubule-associated protein 1 light chain 3 (LC3B) is frequently used in this manner. LC3B is present throughout the cytosol as the unprocessed LC3B-I form. As it becomes incorporated into the autophagosomal membrane, LC3B-I becomes lipidated with phosphatidylethanolamine to form LC3B-II. These two isoforms exhibit differential migration by SDS-PAGE, and therefore the presence and density of LC3B-II can be used as a marker of the number of autophagosomes (35). However, this remains a static measure of autophagosome number, and does not measure the activity of the pathway. Increased LC3B-II or increased autophagosomes by EM can occur in the presence of either high autophagic activity (Figure 3B) or a downstream block in the system (Figure 3C). The latter results in an accumulation of early autophagosomes and LC3B-II protein, even though autophagic degradation is diminished. Therefore, most investigators are currently employing activity or flux measurements to supplement their analyses. One of these methods involves p62/sequestosome I, a cytosolic chaperone protein that has an LC3B binding domain (36). It binds to polyubiquitinated protein aggregates and carries them to the autophagosome (as well as the proteasome), where it binds to LC3B. Then the cargo and p62 itself are degraded by autophagy (Figure 3). Thus, accumulation of p62 in the cell suggests decreased autophagic activity (37).
Figure 3.
Measurement of autophagic flux. (A) Under basal conditions, there is a low level of autophagosome formation and maturation into an autolysosome. p62 and its cargo, as well as LC3-II along the inner membrane, is degraded in the autolysosome. (B) When autophagy is induced, there is increased autophagosome formation and completion of the pathway, resulting in increased autolyosomes. (C) Under basal conditions, there is a low level of autophagosome formation. However, in the presence of downstream blocks to autolysosome formation or function, autophagosome numbers are increased. p62 does not undergo degradation, and accumulates in this condition. The block can occur endogenously or can be introduced with chemical inhibitors. Bafilomycin A prevents autophagosome and lysosome fusion, chloroquine prevents autolysosomal acidification, and leupeptin inhibits lysosomal protease function. (D) To elucidate whether increased numbers of autophagosomes are due to autophagy induction (B) or downstream block (C), a chemical inhibitor is employed. In this case, introducing bafilomycin to a system where autophagy is induced results in increased numbers of autophagosomes (compared with [B]).
One can also measure flux by exogenously inhibiting autophagosome–lysosome fusion or autolysosome function and measuring accumulation of LC3B-II. Several agents can be used for this approach, including leupeptin (lysosomal proteases), chloroquine (lysosomal acidification), and bafilomycin A (autophagosome–lysosome fusion). The greater the difference in LC3B-II between samples with and without the inhibiting agent, the more is being formed, suggesting higher activity of that autophagic pathway (Figure 3D). The flux method can be easily employed for in vitro experiments, and techniques have recently been developed and validated in vivo as well (38). However, measuring flux in human lung tissue is not yet possible, as the techniques rely on live cells and tissue.
The use of green fluorescent protein (GFP)–labeled autophagy proteins, such as LC3B, has allowed another set of measurement techniques (39, 40). Under basal conditions, the cytosolic distribution of LC3B results in a diffuse green fluorescent signal. When autophagy is induced and LC3B becomes incorporated into autophagosomes, the GFP signal coalesces into larger puncta. The formation of these puncta correlates with autophagosome formation. This technique has been employed in vitro by transfecting cells with a vector expressing the GFP-LC3 fusion protein, and a transgenic mouse expressing the fusion protein constitutively on a CAG-promoter has also been developed (41). Furthermore, the fusion protein undergoes cleavage under the acidic conditions of the lysosome, and this can be used to measure flux. By immunoblotting against GFP, one can detect the larger fusion protein that exists before the lysosome and the smaller GFP fragment that is created after lysosomal activity (42). The formation of the latter can be used to monitor completion of autophagic degradation.
Functions of Autophagy in Disease
Autophagy has several key functions that serve to maintain homeostasis in all cells. One of these functions is as a response to metabolic stresses, such as hypoxia, nutrient deprivation, or growth factor reduction. In these conditions, the bulk degradation and recycling of cytoplasmic contents produces amino acids and fatty acids, which can then be used in de novo synthesis of proteins that may be critical in the stress response or to generate ATP through the TCA cycle.
Another basic function of autophagy is the clearance of cytoplasmic components that are not necessary, not functioning, or harmful to the cell, such as protein aggregates, organelles, or pathogens. It has been shown in human disease, particularly in certain neurodegenerative diseases, such as Huntington’s disease, Alzheimer’s disease, Parkinson’s disease, and spinocerebellar ataxia, that deficient autophagy contributes to accumulation of protein aggregates, resulting in cell dysfunction and death. For example, Huntington’s disease involves a mutant protein, huntingtin, which contains polyglutamine expansion tracts that render the protein aggregate prone; these aggregates contribute to neuronal cell toxicity. In cell and animal models, it has been demonstrated that blocking autophagy aggravates aggregation and toxicity; conversely, inducing autophagy is beneficial (43, 44).
Finally, though autophagy has a clear role in proadaptive cell survival, it may also be involved in nonapoptotic programmed cell death. The specific nature and importance of this function is complex and controversial (45). This feature of autophagy was postulated based on the presence of large numbers of autophagosomes in some dying cells. However, the question is whether these autophagosomes are attempting salvage or promoting death. In addition, pathways involved in apoptosis and autophagy have significant commonality. The complexity of the relationship between cell death and autophagy is highlighted in cancer. A synthesis of current available evidence highlights the importance of context and cell-specificity. It is believed that autophagy has tumor suppressor properties in normal cells that are based on facilitation of nonapoptotic cell death of premalignant cells and prevention of DNA damage by limiting ROS. Despite this tumor suppressor potential, there is also evidence that autophagy is necessary for established tumor cells to survive by assisting in energy production in low-nutrient settings. Recently, in tuberous sclerosis complex, which includes lymphangioleiomyomatosis, it has been shown that inhibiting autophagy decreased survival of TSC2-null cells and the growth of TSC2-null tumors (46).
The functional significance of autophagy in respiratory disorders is complex, and our understanding continues to evolve. Investigations to date have tended to focus on specific cell types involved in the disease of interest. This is a sound approach, because autophagy may have different roles in the different cell types in the same disease process. This Review, therefore, analyzes the significance of autophagy in various cell types that are dysfunctional in respiratory diseases. For each cell type, we explore what is known about autophagy in that cell in the context of well known pulmonary disorders.
Epithelial Cells
For the purpose of this Review, we discuss epithelial cells as one category, recognizing, of course, that epithelial cells in the respiratory system are heterogeneous. The form and function of epithelia change as one moves down the tracheo-bronchio-alveolar tree to serve various roles, from mucous secretion and clearance to gas exchange. These cells are involved in sensing environmental cues from one side and internal cues from the other. In doing so, they interact with numerous other cell types, including macrophages, fibroblasts, and endothelial cells. Epithelial cells are of clear importance in emphysema, CF, and acute lung injury. In emphysema, epithelial cell death likely contributes to the loss of alveolar units and development of the disease. CF involves a mutated membrane channel primarily found on epithelial cells. In addition, the current paradigm holds epithelial cells responsible for the onset of idiopathic pulmonary fibrosis (IPF), perhaps due to increased fragility or abnormal wound healing (47).
COPD is one of the most extensively studied lung diseases in the field of autophagy. In the early studies by Chen and colleagues (48), epithelial cell autophagy was investigated after it was discovered that COPD human lung samples had evidence of activated autophagy with increased numbers of autophagosomes by EM and increased LC3B-II protein levels. Furthermore, there was a correlation with disease stage (by Global Initiative for Chronic Obstructive Lung Disease criteria) and autophagy markers. Follow-up in vitro work demonstrated that, in response to cigarette smoke extract (CSE), various epithelial cell lines had increased expression of LC3B-II. EM of Beas-2B cells treated with CSE also demonstrated increased autophagosomes compared with room air–exposed cells. In the murine emphysema model, lung homogenate from wild-type (WT) mice exposed to whole cigarette smoke for 6 months also revealed an increased LC3B-II:I ratio. Delving into the mechanism by which cigarette smoke induces autophagy in epithelial cells, the authors found that the transcription factor, Egr-1, along with cofactor, E2F-4, had increased activity and induced expression of LC3B. This was occurring through reduction in HDAC activity, leading to acetylation of Egr-1. A chronic smoke exposure experiment in Egr-1 knockout mice showed basal airspace enlargement, but no change in the smoke-exposed cohort, demonstrating that, in the absence of Egr-1, cigarette smoke cannot induce autophagy and subsequent apoptosis. Further investigation explored the link between LC3B and apoptosis (49). In LC3B−/− mice treated with 3 months of cigarette smoke, apoptosis markers were significantly decreased compared with LC3B+/+ mice. Immunoprecipitation and immunofluorescence showed an interaction between LC3B and Fas that became disrupted by CSE and that appears to be mediated by lipid raft protein, caveolin-1. Thus, autophagy induction in epithelial cells plays a significant role in COPD pathogenesis. In addition, we subsequently discuss another important aspect of autophagy in COPD in our exploration of alveolar macrophages.
More recently, Luciani and colleagues (50) explored autophagy in epithelial cells in CF. Analysis of airway epithelial cells expressing mutant CF transmembrane conductance regulator (CFTR) demonstrated fewer autophagosomes, less LC3-II, and increased p62 compared with cells expressing WT CFTR, all consistent with impaired autophagy. It stands to reason that autophagy would play a significant role in CF, as many of the mutations in the gene lead to a misfolded protein, and autophagy functions to clear these kinds of proteins. However, it had not been shown that autophagy is impaired in CF. Further experiments in the airway epithelial cells showed that the key autophagy protein, beclin-1, was localized in aggresomes in the CF mutant cells. The authors postulated that this entrapment of beclin-1 in aggresomes prevented autophagy initiation. They tested this hypothesis by overexpressing beclin-1 in these cells and found evidence of restored autophagy measured by LC3-II, p62 protein, and GFP-LC3 puncta. The investigators further discovered that beclin-1 becomes aggresome bound secondary to crosslinking by transglutaminase-2, an effect mediated by increased ROS in CF epithelial cells. Antioxidant treatment in these cells reversed the phenotype. This was a key study that provides the basis by which epithelial cells expressing mutant CFTR may be rescued by inducing autophagy.
The first study to explore the relationship between autophagy and epithelial cells in pulmonary fibrosis was performed by Mi and colleagues (51). They discovered that IL-17A is an inhibitor of autophagy in mouse lung epithelial-12 cells. Although the exact mechanism for this effect remains to be explored, in the presence of IL-17A, there was a reduction in some autophagy proteins, including beclin-1. Interestingly, antagonism of IL-17A was able to restore autophagy and consequently decrease transforming growth factor (TGF)-β1–induced collagen production by epithelial cells. In the murine bleomycin model of pulmonary fibrosis, the authors demonstrated that blocking IL-17A induced autophagy in the lung and protected the mice against fibrosis. This beneficial effect was abrogated if the animals were simultaneously treated with a chemical inhibitor of autophagy, 3-methyladenine. This very interesting study provides evidence for a protective effect of autophagy in epithelial cells in pulmonary fibrosis, which is similar to what we explore subsequently here in fibroblasts.
Finally, the recent work of Tanaka and colleagues (52) demonstrates the role of LC3B in epithelial cell death under hyperoxia. In epithelial cells subject to hyperoxic stress and in mice under hyperoxia, LC3B was found to be activated and transcriptionally regulated via the JNK pathway. Furthermore, LC3B was found to interact with and cross-regulate the Fas pathway to prevent apoptotic cell death, in contrast to the findings in COPD. Overexpression of LC3B demonstrated a protective effect on epithelial cells during hyperoxia. Thus, autophagy may be cytoprotective in hyperoxia-induced lung injury.
In the realm of epithelial cells, we can see that there is a disease-specific role for autophagy. In COPD, it appears to be responsible for disease pathogenesis. In contrast, autophagy may be beneficial in CF, pulmonary fibrosis, and acute lung injury.
Alveolar Macrophages
Although there is an emerging recognition of the importance of autophagy in various functions of innate and adaptive immunity, this Review focuses solely on macrophages. Like epithelial cells, “pulmonary” macrophages are not a uniform population, but we discuss the alveolar macrophage, which is the predominant macrophage in the lung. This cell serves as the first line of phagocytic defense against microbial pathogens and inhaled substances, such as cigarette smoke, but it normally resides in a relatively quiescent state until activated by invaders. In the latter situation, macrophages are key in maintaining the normal sterility of the alveolar microenvironment.
Although the study by Chen and colleagues described above demonstrated increased autophagy in epithelial cells in COPD (48), Monick and colleagues (53, 54) explored the modulation of autophagy in alveolar macrophages during smoking. Macrophages isolated from active smokers have been shown in the past to have impaired immune function (55). The study by Monick demonstrates that such alveolar macrophages also have evidence of impaired autophagy, including increased p62. As a result, these cells accumulate protein aggregates, dysfunctional mitochondria, and have decreased ability to transport bacteria to lysosomes. This study showed increased numbers of autophagosomes and LC3-II in smokers’ macrophages, as well as increased LC3-II in nonsmoker macrophages exposed to 2% CSE. Although this finding could represent activation of autophagy, the investigators proceeded to measure autophagic flux by treating the macrophages with inhibitors of autolysosome function, bafilomycin A or leupeptin, and measuring LC3-II turnover. They found that macrophages from smokers had less flux, even though baseline LC3-II levels were higher than nonsmokers’ macrophages. This finding is supported by the fact that high molecular weight p62 aggregates were also increased in smokers’ macrophages. In a functional study, the authors demonstrate impaired xenophagy in smokers’ macrophages, with decreased delivery of nanoparticles and bacteria to the lysosome. This work provides substantial evidence for impaired autophagy in alveolar macrophages from smokers that may render these macrophages less capable of fighting microbes and thus predisposing smokers to pneumonias. Hence, although increased autophagic activity in epithelial cells contributes to apoptosis and development of emphysema, dysfunctional autophagy in macrophages may explain a different aspect of the disease—the abnormal immunological and inflammatory phenotypes present in COPD.
Abdulrahman and colleagues (54) investigated macrophage dysfunction in clearance of Burkholderia cepacia in CF. B. cepacia is a common pathogen in patients with CF (56), and previous studies have shown that intracellular persistence of the organism in CF macrophages is responsible for the infectious burden (57). In this study, the authors found that B. cepacia infection resulted in down-regulation of several autophagy genes in WT and ΔF508 macrophages, including Atg5, Atg12, and Atg8 (LC3B), with a more pronounced effect in the latter. Treating ΔF508 macrophages infected with B. cepacia with rapamycin induced autophagy and decreased bacterial number. In vivo studies with ΔF508 mice infected with B. cepacia found dramatically reduced bacterial burden in lungs of mice treated with rapamycin. Immunofluorescence showed increased colocalization with LC3 and Lysotracker Green in rapamycin-treated cells, supporting increased autophagic clearance of the bacteria. This finding is particularly surprising given rapamycin’s known immunosuppressive properties, and demonstrates the importance of autophagy in microbial clearance. Thus, autophagy induction may be of therapeutic benefit in patients with CF by two mechanisms in two different cell types: (1) promoting clearance of CF mutant protein aggregates in epithelial cells; and (2) enhancing bacterial clearance by macrophages.
Macrophages also provide the initial defense in the lungs against Mycobacterium tuberculosis invasion. The mycobacteria infect macrophages and then are either degraded or persist in arrested phagosomes. The arrest results from the ability of M. tuberculosis to inhibit formation of PI3-P, which is required for phagolysosome biogenesis and mycobacterial killing (58). Macrophages stimulated with IFN-γ are able to overcome this block, and IFN-γ, through a GTPase called IRGM, also induces autophagy (59). Induction of autophagy by IFN-γ or rapamycin has also been shown to enhance mycobacterial killing (60). One of the mechanisms for this action involves delivery of cytosolic proteins by p62 to lysosomes where they become potent tuberculosis antibiotics (61).
A recent study by Nakahira and colleagues (62) provides intriguing insight into the link between autophagy and the inflammasome. They demonstrated that, under autophagy-deficient conditions, the lack of selective mitochondrial autophagy (mitophagy) creates an accumulation of dysfunctional mitochondria in macrophages. This creates increased ROS and release of mtDNA into the cytosol, which then activates the NALP3 inflammasome, inducing IL-1β and IL-18 production. In the murine LPS model, LC3B-deficient mice had higher expression of these cytokines and increased mortality. Thus, autophagy, and specifically mitophagy, in macrophages plays a key role in limiting the inflammasome response.
In summary, the role of autophagy in alveolar macrophages demonstrates that autophagy is not simply a housekeeping process, but also has important functions in immunity. This function provides another therapeutic target in infectious respiratory diseases or chronic lung diseases characterized by impaired immunity.
Fibroblasts
Lung fibroblasts are a relative late comer to the field of autophagy in lung diseases. As important components of the interstitium, producers of extracellular matrix, and participants in wound healing, fibroblasts are important to all lung diseases. However, they gain increased importance in the study of fibrotic lung diseases, such as IPF, in which we have already seen the role of autophagy in epithelial cells.
Our group recently completed work on the role of autophagy in IPF (63). We initially expected that autophagy would be induced in IPF based on several pathways that are both up-regulated in IPF and known to activate autophagy: ER stress, HIF-1α, oxidative stress, and AMPK. However, upon examination for autophagosomes of lung tissues from patients with IPF by EM and LC3B and p62 protein levels, we found these autophagy markers were not increased at all. In fact, they are not significantly different from control samples. To understand the mechanism behind this disconnect, we investigated the effect of TGF-β1 on autophagy in fibroblasts, and found an inhibitory effect. As a major profibrotic cytokine, TGF-β1 may be responsible for the lack of induction of autophagy in human lung fibroblasts. In addition, we explored the potential mechanism by which autophagy may be beneficial in fibrogenesis, and discovered that silencing key autophagy proteins, LC3 or beclin-1, potentiated the TGF-β1–induced expression of fibronectin and myofibroblast marker α–smooth muscle actin in fibroblasts. Thus, a lack of autophagy may be potentiating the effects of this cytokine with respect to extracellular matrix production and transformation to a myofibroblast phenotype. Furthermore, in the murine bleomycin model, treatment of mice with rapamycin partially protected these animals against lung fibrosis, an effect that may be mediated by rapamycin’s ability to induce autophagy. Based on these findings, inducing autophagy may have beneficial antifibrotic effects.
Endothelial Cells
Vascular endothelial cells play a critical role in the development of PAH. Lee and colleagues (64) investigated the effects of hypoxia on autophagy in these cells. Using pulmonary artery endothelial cells exposed to hypoxia for 24 hours, they found increased expression of LC3B-II and increased GFP-LC3 puncta formation. Analysis of lungs from patients with idiopathic PAH demonstrated elevated LC3B-II protein levels. Immunohistochemistry and immunofluorescence also showed higher LC3B expression in both vascular endothelial and vascular smooth muscle cells. These findings raised the question of whether this induction of autophagy is detrimental or protective. To test these two possibilities, LC3B−/− mice were used in a chronic hypoxia–induced pulmonary hypertension model. The genetically deficient mice demonstrated evidence of increased pulmonary hypertension compared with WT mice. In addition, silencing of LC3B in pulmonary artery endothelial cells enhanced hypoxic cell proliferation. Together, these findings support a protective role for autophagy in this disease process.
Conclusions
Our understanding of the science of autophagy in human disease is evolving rapidly, and knowledge about the role of autophagy in respiratory disease is still scant. The field has matured to the level that we are now able to appreciate different roles for autophagy in different cell types, in certain cases within the same disease. This is an important paradigm, as one would expect such a basic and ubiquitous cellular process to be extremely cell- and environment-dependent. Exogenous modulation of autophagy in an effort to modify disease will need to be similarly selective, and, although the proper tools may not yet all be at our disposal, this area holds great promise for the discovery of new diagnostic targets and/or treatment modalities.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health (NIH) grants 5T32HL007633-25 (A.S.P.), R01HL087122-01 (D.M.), and P01 HL108801, R01 and HL060234, and a FAMRI Clinical Innovator Award (A.M.K.C.).
Originally Published in Press as DOI: 10.1165/rcmb.2012-0282TR on September 13, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tolkovsky AM. Mitophagy. Biochim Biophys Acta 2009;1793:1508–1515 [DOI] [PubMed] [Google Scholar]
- 2.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 2007;3:285–287 [DOI] [PubMed] [Google Scholar]
- 3.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta 2006;1763:1767–1775 [DOI] [PubMed] [Google Scholar]
- 4.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the UBP3P/BRE5P ubiquitin protease. Nat Cell Biol 2008;10:602–610 [DOI] [PubMed] [Google Scholar]
- 5.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005;120:159–162 [DOI] [PubMed] [Google Scholar]
- 6.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008;182:685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009;11:1433–1437 [DOI] [PubMed] [Google Scholar]
- 8.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009;5:1180–1185 [DOI] [PubMed] [Google Scholar]
- 9.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010;141:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006;119:3888–3900 [DOI] [PubMed] [Google Scholar]
- 11.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010;12:747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2010;22:132–139 [DOI] [PubMed] [Google Scholar]
- 13.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008;181:497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000;275:992–998 [DOI] [PubMed] [Google Scholar]
- 15.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005;1:46–52 [DOI] [PubMed] [Google Scholar]
- 16.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit beclin 1–dependent autophagy. Cell 2005;122:927–939 [DOI] [PubMed] [Google Scholar]
- 17.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008;30:678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two beclin 1-binding proteins, Atg14l and rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009;11:385–396 [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 1998;273:33889–33892 [DOI] [PubMed] [Google Scholar]
- 20.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004;36:2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 2008;33:109–122 [DOI] [PubMed] [Google Scholar]
- 22.Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006;7:129–145 [DOI] [PubMed] [Google Scholar]
- 23.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class c vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008;10:776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meijer AJ, Codogno P. AMP-activated protein kinase and autophagy. Autophagy 2007;3:238–240 [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of ULK1. Nat Cell Biol 2011;13:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (HATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011;331:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006;26:9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase–mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-xl and induction of autophagy. EMBO Rep 2009;10:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/EIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 2007;14:230–239 [DOI] [PubMed] [Google Scholar]
- 31.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008;10:676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J 2009;28:3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 2005;1:23–36 [DOI] [PubMed] [Google Scholar]
- 34.Swanlund JM, Kregel KC, Oberley TD. Investigating autophagy: quantitative morphometric analysis using electron microscopy. Autophagy 2010;6:270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007;3:542–545 [DOI] [PubMed] [Google Scholar]
- 36.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. P62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131–24145 [DOI] [PubMed] [Google Scholar]
- 37.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009;452:181–197 [DOI] [PubMed] [Google Scholar]
- 38.Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA, Choi AM. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy 2011;7:629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast APG8P, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using APG5-deficient mouse embryonic stem cells. J Cell Biol 2001;152:657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004;15:1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett 2006;580:2623–2629 [DOI] [PubMed] [Google Scholar]
- 43.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet 2006;15:433–442 [DOI] [PubMed] [Google Scholar]
- 44.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 2004;36:585–595 [DOI] [PubMed] [Google Scholar]
- 45.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 2008;9:1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkhitko A, Myachina F, Morrison TA, Hindi KM, Auricchio N, Karbowniczek M, Wu JJ, Finkel T, Kwiatkowski DJ, Yu JJ, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)–dependent. Proc Natl Acad Sci USA 2011;108:12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc 2006;3:364–372 [DOI] [PubMed] [Google Scholar]
- 48.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, et al. Egr-1 regulates autophagy in cigarette smoke–induced chronic obstructive pulmonary disease. PLoS ONE 2008;3:e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen ZH, Lam HC, Jin Y, Kim HP, Cao J, Lee SJ, Ifedigbo E, Parameswaran H, Ryter SW, Choi AM. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke–induced emphysema. Proc Natl Acad Sci USA 2010;107:18880–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, Gavina M, Pulze L, Giardino I, Pettoello-Mantovani M, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol 2010;12:863–875 [DOI] [PubMed] [Google Scholar]
- 51.Mi S, Li Z, Yang HZ, Liu H, Wang JP, Ma YG, Wang XX, Liu HZ, Sun W, Hu ZW. Blocking IL-17a promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1–dependent and –independent mechanisms. J Immunol 2011;187:3003–3014 [DOI] [PubMed] [Google Scholar]
- 52.Tanaka A, Jin Y, Lee SJ, Zhang M, Kim HP, Stolz DB, Ryter SW, Choi AM. Hyperoxia-induced LC3B interacts with the Fas apoptotic pathway in epithelial cell death. Am J Respir Cell Mol Biol 2012;46:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monick MM, Powers LS, Walters K, Lovan N, Zhang M, Gerke A, Hansdottir S, Hunninghake GW. Identification of an autophagy defect in smokers’ alveolar macrophages. J Immunol 2010;185:5425–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, Newland C, Rosales-Reyes R, Kopp B, McCoy K, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy 2011;7:1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green GM. Cigarette smoke: protection of alveolar macrophages by glutathione and cysteine. Science 1968;162:810–811 [DOI] [PubMed] [Google Scholar]
- 56.Ledson MJ, Gallagher MJ, Jackson M, Hart CA, Walshaw MJ. Outcome of Burkholderia cepacia colonisation in an adult cystic fibrosis centre. Thorax 2002;57:142–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamothe J, Huynh KK, Grinstein S, Valvano MA. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell Microbiol 2007;9:40–53 [DOI] [PubMed] [Google Scholar]
- 58.Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, Davis A, de Haro S, Naylor J, Lee HH, et al. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol 2006;8:719–727 [DOI] [PubMed] [Google Scholar]
- 59.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006;313:1438–1441 [DOI] [PubMed] [Google Scholar]
- 60.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004;119:753–766 [DOI] [PubMed] [Google Scholar]
- 61.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin IV HW, Kyei GB, Johansen T, Vergne I, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 2010;32:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011;12:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, Rosas IO, Morse D. Autophagy in idiopathic pulmonary fibrosis. PLoS ONE 2012;7:e41394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SJ, Smith A, Guo L, Alastalo TP, Li M, Sawada H, Liu X, Chen ZH, Ifedigbo E, Jin Y, et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am J Respir Crit Care Med 2010;183:649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.