Abstract
Recent studies indicate that cyclic AMP (cAMP) induces cytotoxic T lymphocyte antigen (CTLA) 4. CTLA4 is expressed in T cells, and is a negative regulator of T cell activation. CTLA4 expression is regulated by T cell receptor plus CD28 (adaptive immune signaling) at both the transcriptional and post-transcriptional level. Here, we examine the pathways by which cAMP regulates CTLA4 expression, focusing on transcriptional activation. Elevating intracellular cAMP levels by cell-permeable cAMP analogs, the adenylyl cyclase activator, forskolin, or phosphodiesterase inhibitors increases CTLA4 mRNA expression in EL4 murine T cells and primary CD4+ T cells. Activation of protein kinase A (using the protein kinase A–selective agonist, N6-phenyladenosine-cAMP), but not exchange proteins activated by cAMP (using the exchange proteins activated by cAMP–selective 8-pCPT-2Me-cAMP), increases CTLA4 promoter activity. Mutation constructs of the CTLA4 promoter uncover an enhancer binding site located within the −150 to −130 bp region relative to the transcription start site. Promoter analysis and chromatin immunoprecipitation assays suggest that cAMP response element–binding is a putative transcription factor induced by cAMP. We have previously shown that CTLA4 mediates decreased pulmonary inflammation in an LPS-induced murine model of acute lung injury (ALI). We observed that LPS can induce CTLA4 transcription via the same cAMP-inducible promoter region. The immunosuppressant, rapamycin, decreases cAMP and LPS-induced CTLA4 transcription in vitro. In vivo, LPS induces cAMP accumulation in bronchoalveolar lavage fluid, bronchoalveolar lavage cells, and lung tissues in ALI. We demonstrate that rapamycin decreases cAMP accumulation and CTLA4 expression in ALI. Together, these data suggest that cAMP may negatively regulate pulmonary inflammatory responses in vivo and in vitro by altering CTLA4 expression.
Keywords: cytotoxic T lymphocyte antigen 4, cAMP, acute lung injury, protein kinase A, rapamycin
Clinical Relevance
These data suggest a potential mechanism by which cyclic AMP (cAMP) regulates immune suppression and a potential link between innate and adaptive immune signaling. cAMP may increase cytotoxic T lymphocyte antigen 4 as a means of negatively regulating pulmonary inflammatory responses in vivo and in vitro.
Activated T cells express negative regulators, including cytotoxic T lymphocyte antigen (CTLA) 4, a negative regulator of T cell activation (1–3). CTLA4 binds to CD80 or CD86 with greater affinity than CD28 to block costimulatory signals, leading to inhibition of cell cycle progression and IL-2 production (4). The blockade of CTLA4 by antibody prevents the accumulation of inducible cyclic AMP (cAMP) early repressor and cAMP response element modulator, and leads to the rescue of IL-2 expression (5, 6). cAMP has a similar inhibitory effect on proliferation and IL-2 production in T cells (7, 8).
cAMP is an important second messenger in many biological processes. cAMP is synthesized from ATP by adenylyl cyclase and degraded by phosphodiesterase (PDE). In physiological conditions, a wide range of signaling molecules, such as epinephrine, glucagon, and prostaglandin E2, increase cAMP accumulation via G protein–coupled receptor. cAMP can also be activated in response to microbial challenges or LPS (9). The main targets of cAMP are protein kinase (PK) A and exchange proteins activated by cAMP (Epac). Elevated cAMP levels are associated with suppression of innate immune functions, including the production of proinflammatory mediators (10, 11), phagocytosis (12, 13), and microbial killing (14). In particular, cAMP inhibits T cell activation and proliferation, likely mediated by regulatory T cells (Tregs) (7). In primary pulmonary alveolar macrophages, PKA modulates the inhibition of TNF-α and IL-12 (15, 16), whereas Epac mediates the inhibition of phagocytosis (17). Both PKA and Epac mediate the inhibition of bacterial killing by the generation of reactive oxygen species (17). Furthermore, in dendritic cells, inflammatory mediators are modulated by both PKA and Epac pathways, suggesting that cAMP plays a key role in modulating immune responses (10).
Our prior studies indicate that CTLA4 expression is regulated by adaptive immune signaling at the level of transcription (1, 18). The pathways by which other signaling modulates CTLA4 expression remain ill defined. Recent studies indicate that cAMP induces CTLA4 in human CD4+ T lymphocytes (19, 20), and here we focus on dissecting the pathways by which cAMP regulates CTLA4 in vitro and in vivo.
Materials and Methods
Reagents
Chemicals and cell culture reagents were purchased from Sigma-Aldrich (St. Louis, MO) and Gibco BRL (Grand Island, NY). cAMP analogs, 8-chlorophenylthio-cAMP (CPT), N6-Phenyl-cAMP (N6), and 8-pCPT-2′-O-Me-cAMP (8ME) were purchased from the Biolog Life Science Institute (Bremen, Germany).
Cell Culture and Quantitative PCR
EL4 cells were purchased from ATCC (Manassas, VA). Murine primary CT4+ T cells were isolated from lung tissue. Cell suspensions were obtained after digestion of lung tissues in collagenase and DNase, and were then passed through a 70-μm and a 40-μm cell strainer. CD4+ T cells were isolated by magnetic-activated cell sorting system (Miltenyi Biotec Inc., Auburn, CA). Cells were treated with CPT (100 μM), forskolin (10 μM), isobutylmethylxanthine (200 μM), rolipram (10 μM), LPS (1 μg/ml), and rapamycin (0.1 μg/ml). Total RNA was isolated and quantitative PCR (qPCR) was performed. The primer pair for CTLA4 was described previously (21).
Cloning of the CTLA4 Promoter and Construction of Deletion and Linker Scanning Mutation Constructs
The murine CTLA4 promoter containing −1221 base pair (bp) from the transcription start site was cloned into the pXP2 basic vector (p1221) (1). The p1221 was used as a template for 5′ deletion constructs −343 (p343), −238 (p238), −167 (p167), and −64 (p64). Series of 20 bp linker scanning mutation (LSM) constructs were generated from −270 to −110 bp.
Transfections and Reporter Gene Assays
Transient transfections were performed using FuGENE 6 (Roche, Indianapolis, IN). After 12 hours of transfection, cAMP analogs and other stimuli were added at the indicated time, and luciferase activity was measured (Promega, Sunnyvale, CA).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay was performed using the Upstate ChIP kit (Billerica, MA). Cells were fixed and nuclei were sonicated using Bioruptor sonicator (Diagenode Inc., Denville, NJ). Immunoprecipitations were performed with a cAMP response element–binding (CREB) antibody (Cell Signaling, Danvers, MA). The DNA complex was reversed and analyzed by qPCR. Primers specific for CREB binding site are: 5′-CCTCAGAGGTGACTCGAAGCTTCA-3′ and 5′-ACGAGAAAGGAAGCCGTGGGT-3′.
Acute Lung Injury Model
BALB/c mice (8 wk old) were purchased from Harlan Breeders (Indianapolis, IN). The mice were maintained according to the guidelines of the University of California, San Diego Animal Care Program. LPS (100 μg) in 50 μl PBS was administered intratracheally (22). For the rapamycin group, mice were injected with rapamycin (50 μg) or vehicle (PBS) intraperitoneally 3 hours before LPS administration. Lungs were harvested at 48 hours after LPS exposure. Bronchoalveolar lavage (BAL) fluid and BAL cells were collected as previously described (22).
cAMP Accumulation [125I] Radioimmunoassay
BAL cells were divided into two tubes: one was treated with forskolin (10 μM) for 10 minutes to enhance the cAMP. Lung tissues were incubated with 125I-cAMP and anti-cAMP antibody. Radioactivity was counted, and the production of cAMP was normalized to the cell numbers or the amount of protein.
Statistical Analysis
ANOVA was performed using GraphPad Prism software (La Jolla, CA). Statistical analyses of data were performed using paired and unpaired Student’s t tests or using one-way ANOVA. Statistical significance is defined by a P value less than 0.05.
Results
cAMP Signaling Increases CTLA4 mRNA Levels in T Cells
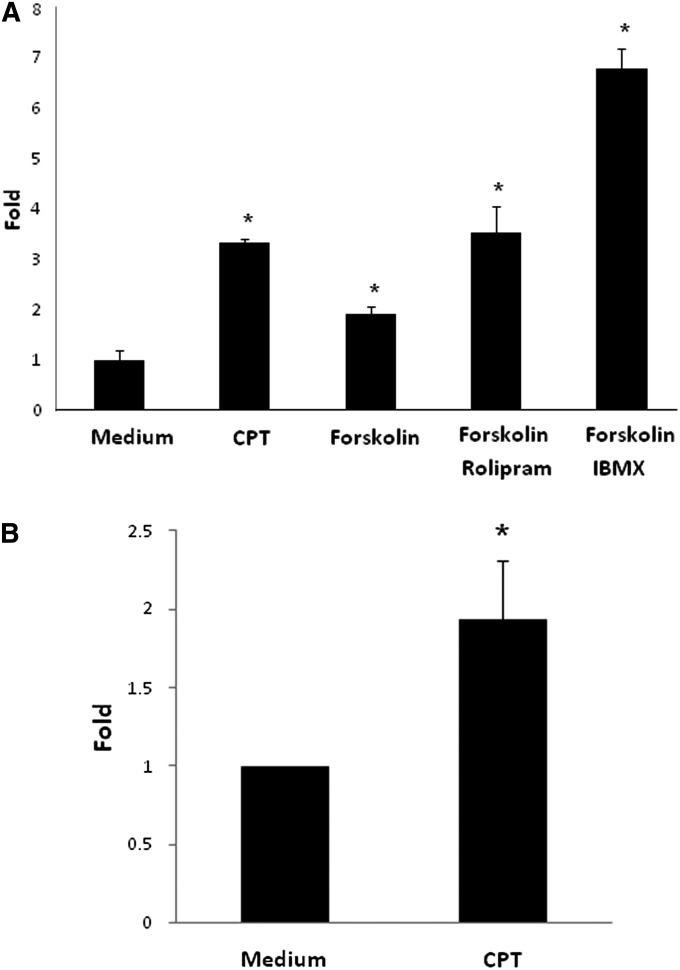
We investigated the impact of cAMP signaling on CTLA4 levels in T cells (EL4 cells) by analyzing pharmacological reagents that increase intracellular cAMP levels (Figure 1A). Treatment with the cAMP analog CPT and forskolin, a direct adenylyl cyclase activator, with and without the PDE4-specific inhibitor, rolipram (because PDE4 is the highest expressed PDE in T cells [23]), or the nonspecific PDE inhibitor, isobutylmethylxanthine, all increased CTLA4 mRNA expression (P < 0.05). Similar results were found in mouse primary lung CD4+ T cells treated with CPT (Figure 1B). These data indicate that cAMP increases CTLA4 mRNA levels in T cells.
Figure 1.
Cyclic AMP (cAMP) signaling increases cytotoxic T lymphocyte antigen (CTLA) 4 mRNA Levels. (A) T cells (EL4) were incubated with a cyclic AMP (cAMP) analog, 8-chlorophenylthio-cAMP (CPT; 100 μM), or forskolin (10 μM), with or without a phosphodiesterase (PDE) 4 inhibitor, rolipram (10 μM), or a nonspecific PDE inhibitor, isobutylmethylxanthine (IBMX; 200 μM) for 12 hours. (B) Mouse primary lung CD4+ T cells were incubated with CPT (100 μM) for 12 hours. CTLA4 mRNA levels were determined by quantitative PCR (qPCR) after normalizing with glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data are shown as means (±SD) (n = 3). *P < 0.05, compared with medium group.
cAMP Analogs Increase CTLA4 Promoter Activity through the PKA Pathway
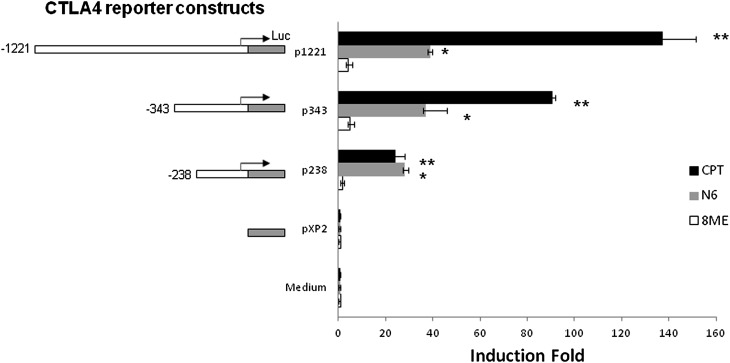
To determine whether the cAMP-mediated increase in CTLA4 mRNA levels is regulated at the transcriptional level, we analyzed CTLA4 luciferase reporter constructs. We tested the effects of pathway-specific cAMP analogs on the regulation of CTLA4 transcription by analysis with cAMP analogs that display different specificities toward cAMP downstream effectors. We examined the impact of CPT (which activates both PKA and Epac), N6 (PKA specific), and 8ME (Epac specific) on CTLA4 promoter activity. We tested three CTLA4 luciferase reporter constructs, 1,221 bp, 343 bp, and 238 bp, relative to the CTLA4 transcription start site (Figure 2). Both CPT and N6 increased the promoter activities of all three CTLA4 reporter constructs; however, this increase was not observed with 8ME treatment. These data suggest that cAMP increases CTLA4 promoter activity through the PKA, but not the Epac, pathway. As CPT induced greater CTLA4 promoter activity, we focused on analysis with CPT as the cAMP analog.
Figure 2.
CTLA4 promoter activity is increased by cAMP signaling through the protein kinase (PK) A, but not the exchange proteins activated by cAMP (Epac) pathway. Transient transfections of CTLA4 reporter constructs into T cells (EL4) were performed using FuGENE 6 transfection reagent for 12 hours. cAMP analogs, CPT (100 μM), N6-Phenyl-cAMP (N6; 50 μM), or 8-pCPT-2′-O-Me-cAMP (8ME; 50 μM) were added into the medium. After 12 hours of incubation, the cell lysates were measured for luciferase activity. Luciferase activity was normalized for transfection efficiency by β-galactosidase activity. The CTLA4 reporter constructs are −1,221 bp (p1,221), −343 bp (p343), and −238 bp (p238) relative to the CTLA4 transcription start site. pXP2 is the promoterless vector. Data are presented as the induction fold of relative luciferase activity (mean ± SD; n = 6). *P < 0.01, compared with N6 and pXP2 group; **P < 0.01, compared with CPT and pXP2 group.
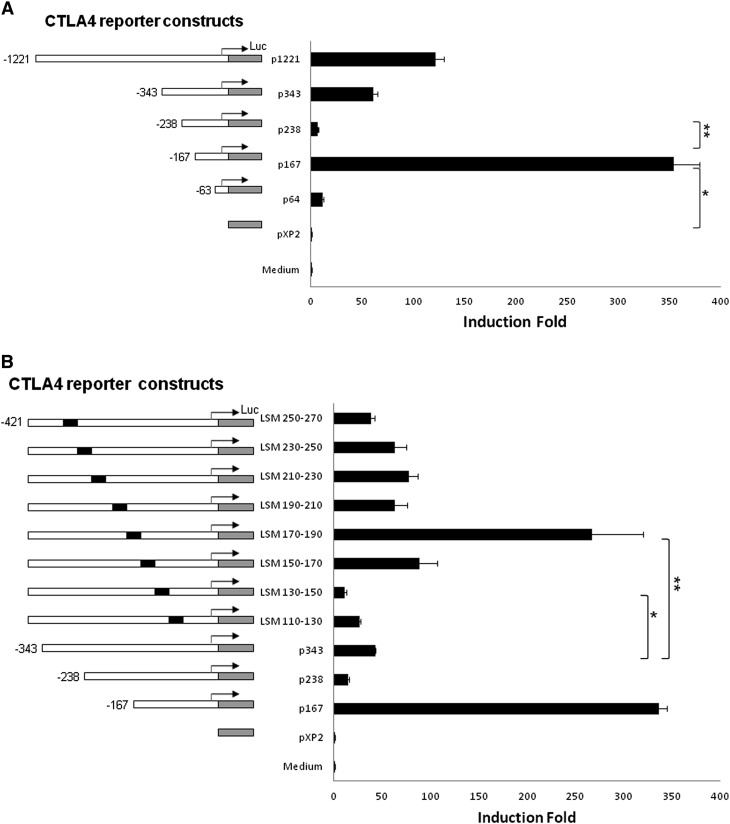
Localization of a cAMP-Responsive Region in the CTLA4 Promoter
We further analyzed the promoter activities to localize cAMP-responsive regions. T cells were stimulated with CPT after transfection of CTLA4 luciferase reporter constructs (including −1221, −343, −238, −167, and −63 bp relative to the CTLA4 transcription start site) (Figure 3A). The largest construct (p1221) showed significant CTLA4 promoter activity, which decreased with the smaller (p343 and p238) constructs. Analysis of a new construct, p167 (−167 bp), indicated a more than 200-fold increase in promoter activity (P < 0.01; Figure 3A, p167), suggesting the presence of an enhancer binding site in this region. The p238 CTLA4 promoter construct exhibits a significant decrease relative to the p167 construct (P < 0.05; Figure 3A, p238), suggesting a putative inhibitor binding site located between −238 and −167 bp. Next, we further defined cAMP-responsive regions through LSM from position −270 to −110 bp (Figure 3B, left panel). Multiple constructs consisting of 20-bp tandem mutations were analyzed. LSM 170–190, with mutations between −190 bp and −170 bp relative to the transcription start site, had high promoter activity similar to p167 (Figure 3B, right panel, LSM 170–190), indicating that a putative inhibitor binding site is located in the −190 to −170 bp region. In contrast, LSM 130–150 construct, with mutations between −150 and −130 bp, exhibited the lowest promoter activity (Figure 3B, right panel; LSM, 130–150), indicating that a putative enhancer binding site is located within the −150 to −130 bp region.
Figure 3.
Localization of cAMP inducible regulatory region(s) in the CTLA4 promoter. (A) After transient transfection of CTLA4 reporter constructs into T cells (EL4), cAMP analog, CPT (100 μM), was added into the medium. After 12 hours of incubation, the cell lysates were measured for luciferase activity. Five CTLA4 luciferase reporter constructs, spanning −1221 to −63 bp relative to the CTLA4 transcription start site, were analyzed. Data are presented as the induction fold of relative luciferase activity (mean ± SD; n = 6). *P < 0.01, compared with pXP2; **P < 0.01, compared with p167. (B) Eight linker scanning mutation (LSM) deletions (20 bp mutations, black) within −110 to −270 bp relative to the CTLA4 transcription start site were analyzed. Data are presented as the induction fold of relative luciferase activity (mean ± SD; n = 6). *P < 0.05, compared with LSM 130–150 and p343; **P < 0.01, compared with LSM 170–190 and p343.
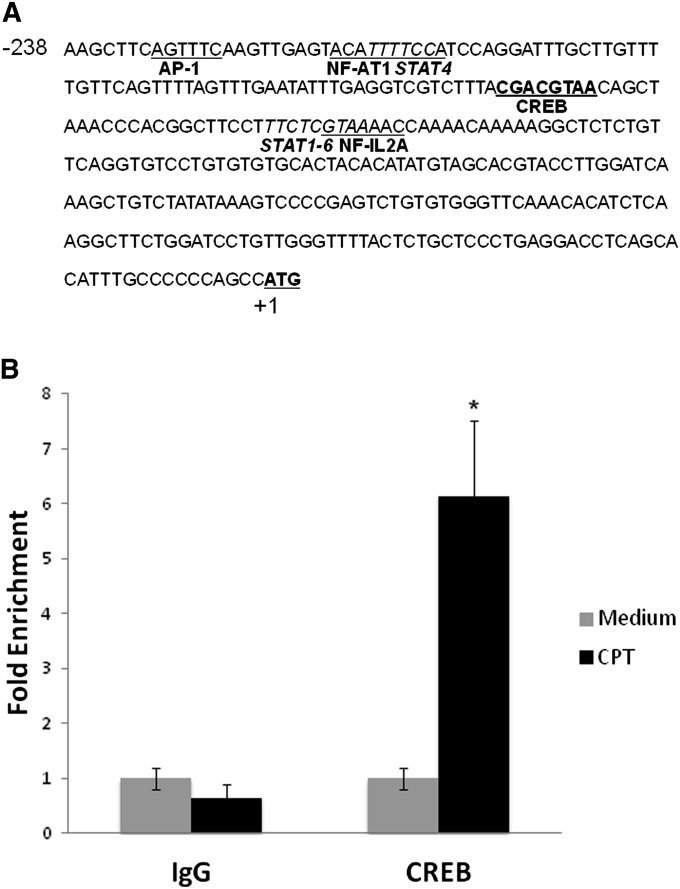
CTLA4 Promoter cis-Regulatory Element Analysis and ChIP Assay
Computer analysis of the CTLA4 promoter region (−238 to −62 bp) revealed potential DNA binding sites (Figure 4A). The promoter region from −150 to −130 bp contains a putative CREB binding site, a well known downstream transcription factor of cAMP signaling. To define whether the putative CREB binding site (−150 to −143 bp) is responsive to cAMP, we performed ChIP assay with CREB antibody in T cells, with or without CPT treatment. The DNA region flanking the putative CREB binding site and a proximal DNA region (as internal control) were amplified by qPCR. There was a sixfold enrichment of the CREB binding site in the CPT treatment group in T cells (Figure 4B). These data suggest that CREB is present and binds to the CTLA4 promoter after increases in cAMP.
Figure 4.
CTLA4 promoter analysis and chromatin immunoprecipitation (ChIP) assay. (A) Response elements in the CTLA4 promoter region from −238 to +1 bp relative to the transcription start site. ATG (bold with heavy underline) is the transcription start site. CREB (bold with heavy underline) is a putative cAMP response element–binding (CREB) binding site, located at −150 to −143 bp relative to transcription start site. (B) ChIP assay using an anti-CREB antibody. Nuclei from T cells (EL4) were prepared and sonicated, and immunoprecipitations were performed using an anti-CREB antibody. Immunoprecipitated DNA was purified and analyzed by qPCR. Folds of CREB binding site DNA enrichment were analyzed with primers specific for CREB binding site after normalizing with the proximal promoter region as an internal control. Data are presented as means (±SD) (n = 6). *P < 0.05, compared with IgG control group.
Rapamycin Decreases LPS- or cAMP-Induced CTLA4 Transcription
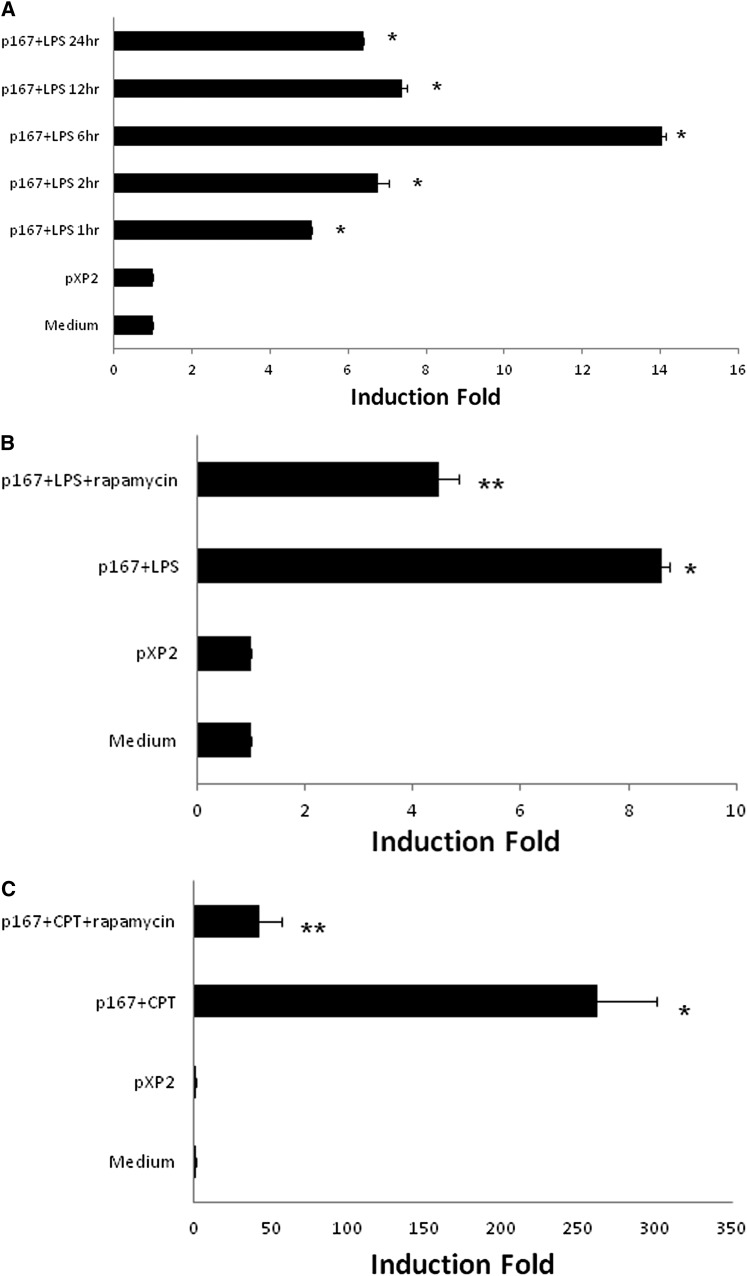
cAMP is activated by multiple stimuli, including response to microbial challenges or LPS. We previously showed that CTLA4 expression is increased in an LPS-induced model of acute lung injury (ALI), then tested whether LPS modifies CTLA4 at the level of transcription (22). We analyzed LPS stimulation of the CTLA4 promoters at different time points. At 6 hours after LPS administration, the promoter activity of p167 construct was significantly induced, albeit not as robustly as with cAMP stimulation (data not shown and Figure 5A). We have previously shown that the immunosuppressant, rapamycin, decreases adaptive immune signaling–induced CTLA4 transcription (1). Here, we tested whether rapamycin modified LPS- or cAMP-induced CTLA4 transcription in T cells. Rapamycin decreases LPS-induced CTLA4 promoter activity from 8-fold to 4-fold (Figure 5B), and cAMP-induced CTLA4 promoter activity from above 200-fold to 40-fold (Figure 5C).
Figure 5.
LPS increases CTLA4 transcription similar to cAMP and rapamycin decreases LPS- or cAMP-induced CTLA4 transcription in T cells. (A) After transient transfection of CTLA4 reporter construct (p167) into T cells (EL4), LPS (1 μg/ml) was added into medium. After 1, 2, 6, 12, and 24 hours of incubation, cell lysates were measured for luciferase activity. *P < 0.01, compared with p167 + LPS and pXP2 group. (B) After transient transfection of the CTLA4 reporter construct (p167), cAMP analog, CPT (100 μM), with or without rapamycin (0.1 μg/ml), was added into the medium. After 12 hours of incubation, cell lysates were measured for luciferase activity. Data are presented as the induction fold of relative luciferase activity (mean ± SD; n = 6). *P < 0.01, compared with pXP2 group; **P < 0.01, compared with p167 + CPT group.
Rapamycin Decreases Elevated cAMP in a Model of Acute Lung Injury
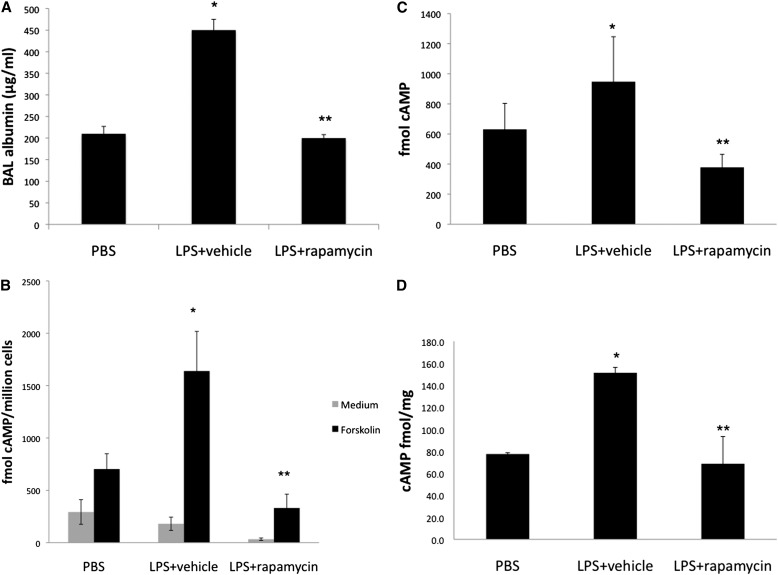
We have previously shown that CTLA4 levels are increased in vivo in a model of ALI (24); therefore, we tested whether cAMP levels are changed in a model of ALI. LPS induced ALI with increased inflammatory parameters (Figure 6A) (22). In addition, cAMP accumulation in BAL cells, BAL fluid, and lung tissues were increased (Figures 6B–6D). Rapamycin administration decreased inflammation and cAMP accumulation in ALI (Figures 6A–6D).
Figure 6.
Rapamycin decreases the elevated cAMP activity in LPS-induced acute lung injury (ALI). Rapamycin (100 μg) or vehicle was administered intraperitoneally 3 hours before LPS exposure. LPS (100 μg) or PBS was administered intratracheally to mice (BALB/c). Bronchoalveolar lavage (BAL) fluid was harvested 48 hours after LPS exposure. (A) BAL albumin levels were determined by ELISA. Data are shown as means (±SD) (n = 3). *P < 0.05, compare with PBS group; **P < 0.05, compared with vehicle group. (B) Total BAL cells were incubated with or without forskolin (10 μM) for 10 minutes. cAMP levels were measured by [125I] radioimmunoassay. (C) BAL fluid and (D) lung tissue cAMP levels were analyzed by [125I] radioimmunoassay. Data are shown as means (±SD) (n = 3). *P < 0.05, compared with PBS group; **P < 0.05, compared with LPS + vehicle group.
Discussion
Increased cAMP levels are associated with inhibition of the activation and proliferation of lymphocytes (25, 26). CTLA4 is a negative regulator of T cells modified by adaptive immune signaling (1). Tregs, a subset of T cells with potent immunoregulatory properties, express high levels of cAMP and transfer cAMP into activated target cells via gap junctions (7, 27), suggesting that cAMP is crucial for naturally occurring Treg-mediated suppression. Tregs can express CTLA4 constitutively (27, 28) to inhibit the development of autoreactive T cells (29). The pathways by which non–antigen-dependent signals modify CTLA4 are not well defined. In a previous study, we demonstrated that CD4+ T cells and the T cell pathways involving CTLA4 are involved in an LPS-induced model of ALI (22).
In this study, we define a cAMP inducible region located within the −167 bp region relative to the CTLA4 transcription start site. A putative inhibitor binding site was located in the region between −190 and −170 bp, which merits further investigation. Previously, we identified an adaptive signaling-responsive region on the CTLA4 promoter, located in the −335 bp region (1, 18), containing a nuclear factor of activated T cells binding site (21). The same nuclear factor of activated T cells binding site is functional in the human CTLA4 promoter as well (30). Interestingly, the cAMP-responsive region differs from the adaptive signaling-responsive region. In contrast, LPS increases CTLA4 promoter activity via the same region as that induced by cAMP, supporting the concept that immune signaling may be mediated through different pathways.
Because cAMP activates the downstream mediators, PKA and Epac, we tested their roles in cAMP-induced CTLA4 expression. Using analogs specific for each downstream mediator, together with analysis of CTLA4 promoter constructs containing the cAMP response regions, our data suggest that cAMP induction of CTLA4 is mediated through the PKA pathway, but not the Epac pathway. Further analysis of the DNA sequence of the cAMP-responsive region revealed a potential CREB transcription factor binding site. ChIP assay results indicate binding of CREB with the CTLA4 promoter. CREB is a transcription factor downstream of the PKA pathway. Taken together, our data suggest that cAMP induces CTLA4 expression via a PKA/CREB–dependent pathway. Because CREB is also important in the regulation of cytokine genes (reviewed in Ref. 31), cAMP induction of CTLA4 transcription suggests alternative pathways modulating immune suppression in the absence of adaptive signaling and full T cell activation. Other examples of cAMP/PKA–mediated pathways include modulation of LPS-induced expression of TNF-α, granulocyte colony-stimulating factor, and IL-10 (15). Similarly, Toll-like receptor 4 LPS-mediated expulsion of bacteria is mediated by the cAMP/PKA pathway (32–35).
We have previously shown that rapamycin decreases adaptive immune signaling–induced CTLA4 transcription (1). In the current study, rapamycin decreased both LPS- and cAMP-induced CTLA4 promoter activity via the same promoter region, indicating the potential cross-talk between the TLR and mammalian target of rapamycin pathways mediated by CTLA4.
We have previously shown that CTLA4 plays a role in decreasing inflammatory responses in allergy and ALI (22, 24). In LPS-induced ALI models, increased cAMP levels in BAL cells, BAL fluid, and lung tissues are found at 48 hours after LPS exposure, the same time point as the elevated CTLA4 levels and inflammatory parameters. Rapamycin decreases LPS-induced elevation of cAMP accumulation, in parallel with a decrease in inflammatory parameters.
Our data indicate that cAMP/PKA/CREB increases CTLA4 transcription in T cells via the same promoter region of LPS-induced CTLA4 transcription. These data suggest a potential mechanism by which cAMP regulates immune suppression and a potential link between innate and adaptive immune signaling. Rapamycin decreases LPS-induced elevation of cAMP accumulation in ALI. Together, these data suggest that cAMP may increase CTLA4 as a means of negatively regulating pulmonary inflammatory responses in vivo and in vitro.
Supplementary Material
Acknowledgments
The authors thank Ms. Kathryn Reed for assistance with manuscript preparation.
Footnotes
This work was supported by National Institutes of Health grants 5R01AI053878, 5R01AI075317, and T32HL098062.
Originally Published in Press as DOI: 10.1165/rcmb.2012-0155OC on September 28, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Finn PW, He H, Wang Y, Wang Z, Guan G, Listman J, Perkins DL. Synergistic induction of CTLA-4 expression by costimulation with TCR plus CD28 signals mediated by increased transcription and messenger ribonucleic acid stability. J Immunol 1997;158:4074–4081 [PubMed] [Google Scholar]
- 2.Krinzman SJ, De Sanctis GT, Cernadas M, Mark D, Wang Y, Listman J, Kobzik L, Donovan C, Nassr K, Katona I, et al. Inhibition of T cell costimulation abrogates airway hyperresponsiveness in a murine model. J Clin Invest 1996;98:2693–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linsley PS, Brady W, Urnes M, Grosmaire LS, Damle NK, Ledbetter JA. CTLA-4 is a second receptor for the B cell activation antigen B7. J Exp Med 1991;174:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med 1996;183:2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodor J, Fehervari Z, Diamond B, Sakaguchi S. ICER/CREM–mediated transcriptional attenuation of IL-2 and its role in suppression by regulatory T cells. Eur J Immunol 2007;37:884–895 [DOI] [PubMed] [Google Scholar]
- 6.Bodor J, Fehervari Z, Diamond B, Sakaguchi S. Regulatory T cell–mediated suppression: potential role of ICER. J Leukoc Biol 2007;81:161–167 [DOI] [PubMed] [Google Scholar]
- 7.Bopp T, Becker C, Klein M, Klein-Hessling S, Palmetshofer A, Serfling E, Heib V, Becker M, Kubach J, Schmitt S, et al. Cyclic adenosine monophosphate is a key component of regulatory T cell–mediated suppression. J Exp Med 2007;204:1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson KW, Davis BH, Smith KA. cAMP antagonizes interleukin 2–promoted T-cell cycle progression at a discrete point in early G1. Proc Natl Acad Sci USA 1988;85:6072–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J, Abraham SN. TLR-mediated immune responses in the urinary tract. Curr Opin Microbiol 2008;11:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aronoff DM, Carstens JK, Chen GH, Toews GB, Peters-Golden M. Short communication: differences between macrophages and dendritic cells in the cyclic AMP–dependent regulation of lipopolysaccharide-induced cytokine and chemokine synthesis. J Interferon Cytokine Res 2006;26:827–833 [DOI] [PubMed] [Google Scholar]
- 11.Koga K, Takaesu G, Yoshida R, Nakaya M, Kobayashi T, Kinjyo I, Yoshimura A. Cyclic adenosine monophosphate suppresses the transcription of proinflammatory cytokines via the phosphorylated c-fos protein. Immunity 2009;30:372–383 [DOI] [PubMed] [Google Scholar]
- 12.Ydrenius L, Majeed M, Rasmusson BJ, Stendahl O, Sarndahl E. Activation of cAMP-dependent protein kinase is necessary for actin rearrangements in human neutrophils during phagocytosis. J Leukoc Biol 2000;67:520–528 [DOI] [PubMed] [Google Scholar]
- 13.Aronoff DM, Canetti C, Peters-Golden M. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor–mediated increase in intracellular cyclic amp. J Immunol 2004;173:559–565 [DOI] [PubMed] [Google Scholar]
- 14.Serezani CH, Ballinger MN, Aronoff DM, Peters-Golden M. Cyclic AMP: master regulator of innate immune cell function. Am J Respir Cell Mol Biol 2008;39:127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall EA, Zavzavadjian JR, Chang MS, Randhawa B, Zhu X, Hsueh RC, Liu J, Driver A, Bao XR, Sternweis PC, et al. Suppression of LPS-induced TNF-alpha production in macrophages by cAMP is mediated by PKA-AKAP95-p105. Sci Signal 2009;2:ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Pouw Kraan TC, Boeije LC, Smeenk RJ, Wijdenes J, Aarden LA. Prostaglandin-E2 is a potent inhibitor of human interleukin 12 production. J Exp Med 1995;181:775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aronoff DM, Canetti C, Serezani CH, Luo M, Peters-Golden M. Cutting edge: macrophage inhibition by cyclic AMP (cAMP): differential roles of protein kinase A and exchange protein directly activated by cAMP-1. J Immunol 2005;174:595–599 [DOI] [PubMed] [Google Scholar]
- 18.Perkins D, Wang Z, Donovan C, He H, Mark D, Guan G, Wang Y, Walunas T, Bluestone J, Listman J, et al. Regulation of CTLA-4 expression during T cell activation. J Immunol 1996;156:4154–4159 [PubMed] [Google Scholar]
- 19.Vendetti S, Riccomi A, Sacchi A, Gatta L, Pioli C, De Magistris MT. Cyclic adenosine 5′-monophosphate and calcium induce CD152 (CTLA-4) up-regulation in resting CD4+ T lymphocytes. J Immunol 2002;169:6231–6235 [DOI] [PubMed] [Google Scholar]
- 20.Vendetti S, Patrizio M, Riccomi A, De Magistris MT. Human CD4+ T lymphocytes with increased intracellular cAMP levels exert regulatory functions by releasing extracellular cAMP. J Leukoc Biol 2006;80:880–888 [DOI] [PubMed] [Google Scholar]
- 21.Jen KY, Campo M, He H, Makani SS, Velasco G, Rothstein DM, Perkins DL, Finn PW. CD45RB ligation inhibits allergic pulmonary inflammation by inducing CTLA4 transcription. J Immunol 2007;179:4212–4218 [DOI] [PubMed] [Google Scholar]
- 22.Nakajima T, Suarez CJ, Lin KW, Jen KY, Schnitzer JE, Makani SS, Parker N, Perkins DL, Finn PW. T cell pathways involving CTLA4 contribute to a model of acute lung injury. J Immunol 2010;184:5835–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schudt C, Tenor H, Hatzelmann A. PDE isoenzymes as targets for anti-asthma drugs. Eur Respir J 1995;8:1179–1183 [DOI] [PubMed] [Google Scholar]
- 24.Lin KW, Jen KY, Suarez CJ, Crouch EC, Perkins DL, Finn PW. Surfactant protein D–mediated decrease of allergen-induced inflammation is dependent upon CTLA4. J Immunol 2010;184:6343–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minguet S, Huber M, Rosenkranz L, Schamel WW, Reth M, Brummer T. Adenosine and cAMP are potent inhibitors of the NF-kappa B pathway downstream of immunoreceptors. Eur J Immunol 2005;35:31–41 [DOI] [PubMed] [Google Scholar]
- 26.Bodor J, Bodorova J, Gress RE. Suppression of T cell function: a potential role for transcriptional repressor ICER. J Leukoc Biol 2000;67:774–779 [DOI] [PubMed] [Google Scholar]
- 27.Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak TW, Sakaguchi S. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte–associated antigen 4. J Exp Med 2000;192:303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonuleit H, Schmitt E, Stassen M, Tuettenberg A, Knop J, Enk AH. Identification and functional characterization of human CD4(+)CD25(+) T cells with regulatory properties isolated from peripheral blood. J Exp Med 2001;193:1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen S, Ding Y, Tadokoro CE, Olivares-Villagomez D, Camps-Ramirez M, Curotto de Lafaille MA, Lafaille JJ. Control of homeostatic proliferation by regulatory T cells. J Clin Invest 2005;115:3517–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson HM, Hedgcock CJ, Aufiero BM, Wilson AJ, Hafner MS, Tsokos GC, Wong HK. Induction of the CTLA-4 gene in human lymphocytes is dependent on NFAT binding the proximal promoter. J Immunol 2007;179:3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torgersen KM, Vang T, Abrahamsen H, Yaqub S, Tasken K. Molecular mechanisms for protein kinase A–mediated modulation of immune function. Cell Signal 2002;14:1–9 [DOI] [PubMed] [Google Scholar]
- 32.Song J, Duncan MJ, Li G, Chan C, Grady R, Stapleton A, Abraham SN. A novel TLR4-mediated signaling pathway leading to IL-6 responses in human bladder epithelial cells. PLoS Pathog 2007;3:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J, Bishop BL, Li G, Duncan MJ, Abraham SN. TLR4-initiated and cAMP-mediated abrogation of bacterial invasion of the bladder. Cell Host Microbe 2007;1:287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop BL, Duncan MJ, Song J, Li G, Zaas D, Abraham SN. Cyclic AMP–regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat Med 2007;13:625–630 [DOI] [PubMed] [Google Scholar]
- 35.Song J, Bishop BL, Li G, Grady R, Stapleton A, Abraham SN. TLR4-mediated expulsion of bacteria from infected bladder epithelial cells. Proc Natl Acad Sci USA 2009;106:14966–14971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.