Abstract
In this study, we explored the regulation and the role of up-regulated microRNAs in idiopathic pulmonary fibrosis (IPF), a progressive interstitial lung disease of unknown origin. We analyzed the expression of microRNAs in IPF lungs and identified 43 significantly up-regulated microRNAs. Twenty-four of the 43 increased microRNAs were localized to the chromosome 14q32 microRNA cluster. We validated the increased expression of miR-154, miR-134, miR-299–5p, miR-410, miR-382, miR-409–3p, miR-487b, miR-31, and miR-127 by quantitative RT-PCR and determined that they were similarly expressed in embryonic lungs. We did not find evidence for differential methylation in this region, but analysis of transcription factor binding sites identified multiple SMAD3-binding elements in the 14q32 microRNA cluster. TGF-β1 stimulation of normal human lung fibroblasts (NHLF) caused up-regulation of microRNAs on chr14q32 that were also increased in IPF lungs. Chromatin immunoprecipitation confirmed binding of SMAD3 to the putative promoter of miR-154. Mir-154 was increased in IPF fibroblasts, and transfection of NHLF with miR-154 caused significant increases in cell proliferation and migration. The increase in proliferation induced by TGF-β was not observed when NHLF or IPF fibroblasts were transfected with a mir-154 inhibitor. Transfection with miR-154 caused activation of the WNT pathway in NHLF. ICG-001 and XAV939, inhibitors of the WNT/β-catenin pathway, reduced the proliferative effect of miR-154. The potential role of miR-154, one of multiple chr14q32 microRNA cluster members up-regulated in IPF and a regulator of fibroblast migration and proliferation, should be further explored in IPF.
Keywords: idiopathic pulmonary fibrosis, microRNA, TGF-β, WNT/β-catenin, developmental pathways
Clinical Relevance
Idiopathic pulmonary fibrosis (IPF) is a lethal and untreatable disease. The expression of microRNAs, small noncoding RNAs that regulate gene and protein expression, is altered in the IPF lung. Understanding how dysregulated microRNAs affect fibrosis-related pathways may have significant importance for the development of new interventions. In this manuscript we identify 43 significantly up-regulated microRNAs in IPF lungs and demonstrate one of these microRNAs, mir-154, as a regulator of fibroblast fibrosis–related phenotypes.
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and usually lethal disease of uncertain etiology (1). The lung phenotype in IPF is characterized by aberrant remodeling and profound changes in the phenotypes of alveolar epithelial cells and lung fibroblasts as well as activation of developmental pathways (2–4). IPF lungs are characterized by the histological appearance of usual interstitial pneumonia, including the presence of fibroblastic foci that are an aggregation of fibroblasts and myofibroblasts where fibroblasts proliferate, migrate, and contribute to excessive production of extracellular matrix (5).
Although the etiology of IPF is not well understood, there has been significant progress in understanding the molecular mechanisms that underlie the lung phenotype in IPF. Transforming growth factor (TGF)-β1 is considered a key regulator of lung fibrosis, an inducer of fibroblast proliferation, and a regulator of gene and microRNA expression in various cell types (6–9). Aberrant recapitulation of developmental programs and activation of WNT/β-catenin pathway has also been recently reported as one of the main phenotypic characteristics of the IPF lung (10–12). Inhibition of WNT/β-catenin/CBP driven transcription may have clinical significance in cancers and prevents and reverses established fibrosis in mice (13–15).
Recently, a role for microRNAs, small noncoding RNAs that regulate gene expression, has been suggested in lung fibrosis (9, 16). MicroRNAs are endogenous RNAs of approximately 19 to 25 nucleotides and are processed from primary transcripts in sequential steps by the RNase III endonucleases, Drosha in the nucleus (17) and Dicer in the cytoplasm. They negatively regulate target mRNAs by imperfectly matching the pairing to their 3′ untranslated regions (3′UTR) of mRNA targets (18). Differential expression of microRNAs is directly associated with developmental processes and with the initiation and progression of cancer, diabetes, cardiovascular disease, and lung disease (16, 19–22). Many microRNA are located in genomic clusters (23). The largest microRNA cluster in the human genome spans 100 kb at human chromosome 19q13.41 and comprises 46 microRNA genes exclusively expressed in the placenta, and the second largest cluster is on human chromosome14q32 (24, 25).
In this manuscript we focus on up-regulated microRNAs in IPF. We demonstrate that most of them are localized to the chromosome 14q32 microRNA cluster and that many belong to the miR-154 family. We study their regulation and downstream effects and demonstrate that they are TGF-β1 target microRNAs and provide preliminary evidence that mir-154 may be a regulator of disease-relevant fibroblast behavior. Some of the results presented in this manuscript have been previously reported as an abstract (26).
Materials and Methods
Sample Population
Tissues from 13 patients with IPF and 12 normal lung histology samples obtained from the University of Pittsburgh Health Sciences Tissue Bank as previously described (27) were used for microarrays, and 32 IPF samples and 28 normal histology controls from the Lung Tissue Research Consortium were used for quantitative RT-PCR (qRT-PCR). Fetal lung tissues were obtained from the University of Pittsburgh Tissue Bank. The University of Pittsburgh Institutional Review Board approved all experiments.
Cell Culture and Transfection
Primary normal human lung fibroblasts (NHLF) were purchased from Lonza Ltd. (Basel, Switzerland), and IPF fibroblasts were obtained from explanted lungs as previously described (28). All fibroblasts were between passages 3 through 8. Wherever indicated, cells were stimulated with recombinant TGF-β1 (R&D, Minneapolis, MN). Before stimulation or transfection, cells were cultured overnight in 1% FBS. Cells were transfected with pre–miR-154, mir-154 inhibitor, or negative controls (Ambion, Austin, TX). For cotransfection experiments, a TCF Reporter Plasmid Kit (EMD Millipore, Jaffrey, NH) was used with a Renilla luciferase reporter (Promega, Madison, WI) to normalize for transfection efficiency.
RNA Isolation and qRT-PCR
RNA from tissues and cells was isolated using the miRNeasy Mini kit (Qiagen, Valencia, CA), and quality was determined by an Agilent Bioanalyzer 2100. TaqMan MicroRNA and gene expression assays (ABI, Foster City, CA) were used for qRT-PCR.
Microarrays
RNA was labeled and hybridized to 8X15K Agilent microRNA or 4X44k whole human genome Agilent microarrays as described by us (9, 29). Microarray data were deposited in Gene Expression Omnibus (datasets GSE27430 and GSE27156).
Analysis of 14q32 Cluster and Chromatin Immunoprecipitation
Genomic coordinates of all transcripts were obtained from the UCSC Genome Browser. Putative Smad-binding elements were found by the Footer algorithm (30). Chromatin immunoprecipitation analyses were performed according to the manufacturer of the EZ ChIP protocol (Millipore, Billerica, MA)
MassARRAY EpiTYPER Analysis
Quantitative analysis of CpG dinucleotide methylation was performed using MassARRAY EpiTYPER analysis (31).
Proliferation Assays
NHLF were cultured in round-bottom, 96-well plates for 24 hours. After transfection with miR-154, the cells were pulsed with [3H]-thymidine (1 μCi/well) (PerkinElmer, Waltham, MA), and the [3H]-thymidine incorporation was measured. For FACS, NHLF were pulsed with 10 μM BrdU for 16 hours and then stained with 7-AAD for cell cycle analysis.
Migration Assays
Cell migration was determined by counting the number of cells that migrated through Matrigel inserts with 8-μm pores (Becton Dickinson, Woburn, MA).
ICG-001 and XAV939 Treatment
Human lung fibroblasts were transfected in a 96-well plate with 50 nM of pre–miR-154. Six hours later, the cultures were replaced with fresh medium, and 10 μM of ICG-001, a selective inhibitor of the CBP–β-catenin interaction (14, 32), or 10 μM of XAV939, a tankyrase inhibitor that inhibits WNT signaling (33). Proliferation was measured 24 hours after transfection using CellTiter 96 AQueous (Promega).
Statistical Analysis of Data
MicroRNA and mRNA microarray data analysis was done using GeneSpring v11, and visualization was performed with Java Treeview. Group comparisons were made using an unpaired, two-tailed Student’s t test, and significant enrichment of overexpression of microRNAs in specific chromosomal locations was determined using Fisher’s exact test.
Detailed methods are provided in the online supplement.
Results
The Majority of Increased MicroRNAs in IPF Localize to Chromosome 14q32 and Are Enriched with Members of the mir-154 Family
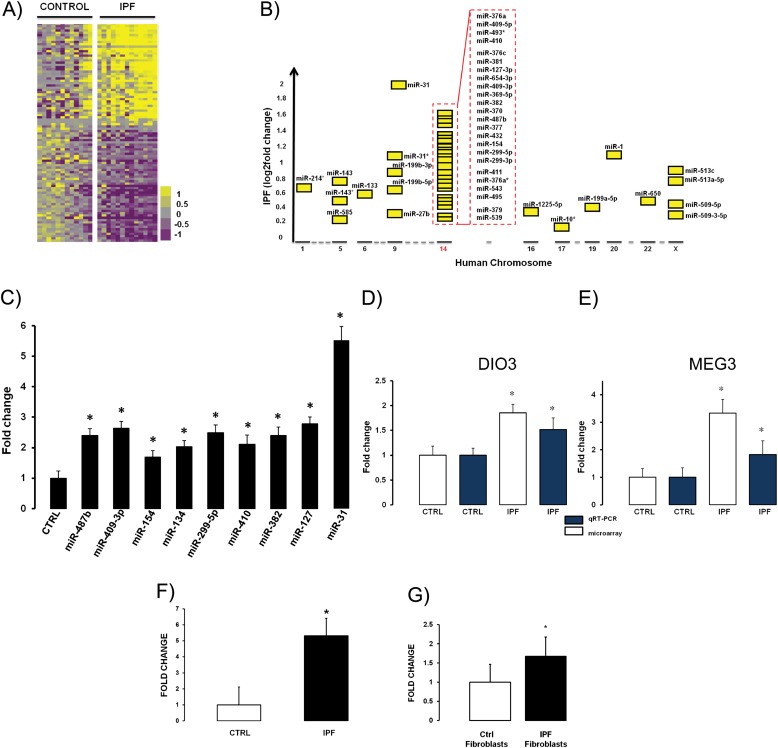
Ninety-four microRNAs were significantly differentially expressed (P < 0.05), and 43 of them were higher in IPF (Figure 1A; see Table E1 in the online supplement). The complete microRNA microarray data have been deposited in the Gene Expression Omnibus (GSE27430) and are publicly available. The majority of up-regulated microRNAs (n = 24) were localized to chromosome 14q32 (Figure 1B), and this enrichment was statistically significant (Figure E2). In fact, in silico analysis of the 14q32 microRNAs demonstrated that, when taken globally, the transcripts located at chromosome 14 are significantly transcriptionally activated (Figures E3A and E3B; Table 5E) (a detailed description is provided in the online supplement), suggesting that they operate as a transcriptional module as previously described (34). Twelve of the 14q32 microRNAs (miR-369–5p, miR-299–3p, miR-409–3p, miR-299–5p, miR-409–5p, miR-410, miR-377, miR-382, miR-487b-5p, miR-154, miR-539, and miR-493*) belong to the miR-154 family. The miR-154 family was the most abundant microRNA family within increased microRNAs expression in IPF.
Figure 1.
The majority of increased microRNAs in idiopathic pulmonary fibrosis (IPF) localize to chromosome 14q32 and are enriched with members of the mir-154 family. (A) microRNA microarray expression profiles were analyzed using RNA from 12 control and 13 pulmonary fibrosis lung tissues. The heat map represents statistically significant (P ≤ 0.05), differentially expressed microRNAs. Up-regulated microRNAs are shown in yellow, and down-regulated microRNAs are shown in purple. Columns represent individual samples; selected microRNAs are represented in rows. The names of the all differentially expressed microRNAs are provided in the Table E1. (B) Chromosomal distribution of significantly up-regulated microRNAs in IPF. The x axis represents human chromosomes, and the y axis represents fold change in log2 scale. The black lines on the x axis represent human chromosomes with significantly up-regulated miRs, and the gray lines represent chromosomes without significant differential expression of microRNAs. The members of miR-154 family located on chr14q32 are miR-369–5p, miR-299–3p, miR-409–3p, miR-299–5p, miR-409–5p, miR-410, miR-377, miR-382, miR-487b-5p, miR-154, miR-539, and miR-493*. (C) Quantitative RT-PCR (qRT-PCR) confirmation of the microarray results demonstrates significant changes in all qRT-PCR–measured microRNAs. Results are presented as mean ± SEM. DIO3 (D) and MEG3 (E) are increased in IPF lungs in comparison to controls. DIO3 and MEG3 mRNA levels in total lung homogenate were measured by microarray (white column) and qRT-PCR (blue column) (n ≥ 16 were used for microarray, and n = 5 for qRT-PCR). (F) qRT-PCR confirmation of the microarray results on the independent cohort of patients demonstrates significant (P = 2.7E-12) up-regulation of miR-154 in IPF samples (n = 32) compared with normal histology controls (n = 28). (G) qRT-PCR analysis of miR-154 is significantly increased in IPF fibroblasts compared with normal human lung fibroblasts (NHLF) (n = 4 each group; P < 0.05). Results are presented as mean ± SEM (P < 0.05).
We confirmed the microarray results of nine microRNAs (miR-154, miR-134, miR-299–5p, miR-410, miR-382, miR-409–3p, miR-487b, miR-31, and miR-127) by qRT-PCR (Figure 1C). Up-regulated microRNAs on chromosome 14q32 are part of the imprinted domain DLK1-DIO3, which contains paternally imprinted genes DLK1 and DIO3 and the maternally imprinted MEG3 gene (Figure 2A). DIO3 and MEG3 were increased in our previously published microarray data (29), and qRT-PCR confirmed their up-regulation in IPF lungs (Figures 1D and 1E).
Figure 2.
TGF-β1 partially regulates expression of microRNAs on chr14q32. (A) The regional physical map of the human chromosome 14q32. Genes are shown in red, up-regulated microRNAs in green, differentially methylated regions (DMR) in blue, and Smad-binding elements (SBEs), predicted by the Footer algorithm, in purple. DIO3 (B) and MEG3 (C) mRNA levels were analyzed by qRT-PCR using RNA from lung fibroblast stimulated with TGF-β1 for 24 hours (n = 6). Results are presented as mean ± SEM (P < 0.05). (D) Sequenom EpiTYPER quantitative methylation analysis of CpG dinucleotide was performed at the methylated regions (DMRs) on chromosome 14q32 on the following locations: DMR1 (100244848–100246120), DMR2 (100262504–100263352), DMR3 (100360176–100360721), and DMR4 (100601296–100602237). The primer sequences used in the Sequenom EpiTYPER study are shown in Table E4. (E) Direct comparison of differentially expressed microRNAs in IPF lungs and lung fibroblast stimulated with TGF-β1 for 2 hours (5 ng/μl). The seven underlined microRNAs indicate miR-154 family members. The color codings outlined in the inserted box indicate human chromosomes. (F) SMAD3 chromatin immunoprecipitation assay discovered an association with the predicted promoters (P1 and P2) of miR-154. Only P1 showed an association with SMAD3 under stimulation of TGF-β1. P1 and P2 are 391 and 322 bp, respectively, upstream from the pre–miR-154 gene. L = ladder; NC = negative control (normal mouse IgG); PC = positive control (anti-RNA polymerase II).
The overexpression of miR-154, as a representative of the most abundant up-regulated microRNA family, was confirmed in an additional cohort of 32 patients with IPF (Figure 1F). Expression of miR-154 was also significantly increased in IPF fibroblasts (Figure 1G).
TGF-β1 Drives Expression of MicroRNAs in the 14q32 Cluster and Induces Changes Similar to Those Seen in IPF
To resolve whether the coordinated increase in the microRNAs and whether gene expression on chromosome 14q32 was determined by the result of changes in methylation, we analyzed the methylation status of CpG islands on the putative promoters of DLK1 (100244848–100246120 and 100262504–10063352), MEG3 (100360176–100360721), and the CpG islands within the microRNA cluster (100601296–100602237) using CpG island arrays recently published by us (35) and validated with MassArray Epityper comparing fibrotic lungs with the controls. All four CpG islands were similarly methylated, and there was no significant difference detected between those two groups (Figure 2D).
To find alternative transcriptional regulatory mechanisms, we searched for transcription bindings sites of putative promoters upstream of microRNAs on chr14q32 using the Footer algorithm (30). We found 16 Smad (3 and/or 4) binding elements 5 kb upstream of up-regulated miRs on chr14q32 (Figure 2A; Table E3), nine of which were detected upstream of miR-154 family members. We also found on the same region putative transcription factor binding sites of p53, c-Myc, and E2F1.
To address the question of whether TGF-β regulates the mRNAs and microRNAs in this region, we stimulated NHLF with TGF-β1. TGF-β1 induced significant increases in MEG3 but not in DIO3 (Figures 2B and 2C). MicroRNA microarray analysis of the same NHLF revealed 84 differentially expressed microRNAs (Table E2); of these, 52 were increased and 32 were decreased. Of the 13 microRNAs localized to Chr14q32, seven microRNAs from the miR-154 family were increased by TGF-β1.
To determine whether TGF-β1–induced changes in microRNA expression in NHLF were reflective of those seen in IPF, we compared the microRNA changed in IPF to controls with NHLF stimulated with TGF-β (Figure 2E). We found 14 microRNAs that were commonly up-regulated in both groups; among them were 12 microRNAs from chr14q32, and of these seven microRNAs were miR-154 family members, suggesting that TGF-β1 is responsible for some of the increases in chromosome 14q32 microRNAs in IPF. Real-time PCR confirmed that TGF-β1 treatment significantly induced mir-154 in NHLF but not in IPF fibroblasts (data not shown).
We used chromatin immunoprecipitation to validate the binding of SMAD3 to the two closest Smad-binding elements of the miR-154 gene (Figure 2A; Table E3). Our data revealed binding of SMAD3 to position 391 bp upstream of the miR-154 gene in TGF-β1–stimulated cells (Figure 2F, lane P1) but not in the nonstimulated cells. In contrast, the binding of SMAD3 at the 322 bp site (Figure 2F, lane P2) upstream of pre–miR-154 was evident with or without TGF-β1 stimulation.
In summary, these findings suggest that TGF-β1 is a positive inducer of genes and microRNAs on chromosome 14q32. Because miR-154 is increased in IPF and induced by TGF-β1 and because many of its family members are induced in IPF, we focused on it as a representative member of the mir-154 family.
miR-154 Increases Migration and Proliferation of Lung Fibroblasts and Inhibits p15
To determine the downstream effects of increased expression of miR-154, we transfected NHLF with miR-154 and analyzed changes in migration, proliferation, and cell cycle. Transfection of NHLF with miR-154 induced a significant increase in proliferation of adult and fetal lung fibroblasts as measured by [3H] thymidine incorporation (Figures 3A and 3B) and a significantly higher percentage of BrdU-FITC labeled DNA of S-phase access compared with the scrambled controls, suggesting miR-154 promotes S-phase entry (Figure 3D) and a significant increase in cell migration (Figure 3C). The cell cycle inhibitor CDKN2B (p15), a CDK4 inhibitor, is a TargetScan-predicted target of miR-154. Western blot analysis revealed significantly decreased protein levels of p15 (Figure 3E) in NHLF transfected with miR-154 in comparison to control, suggesting that in part the effects of miR-154 on cell cycle were mediated through p15. TGF-β1–induced increases in proliferation were significantly reduced in the presence of a mir-154 inhibitor in NHLF and IPF fibroblasts (Figure 3F).
Figure 3.
miR-154 increases migration and proliferation of fibroblasts and decreases protein levels of cell cycle inhibitor p15. (A) NHLF transfected with pre–miR-154 or a negative (scrambed) control (SCR) for 24 hours demonstrate an increase in proliferation measured by [3H] thymidine incorporation assay. Data shown are means of n ≥ 5, ± SEM (y axis: CPM). (B) miR-154 increases proliferation of fetal fibroblasts transfected with pre–miR-154 or a scrambled control (SCR) for 24 and 48 hours, and the increase in proliferation was measured by [3H]thymidine incorporation assay. To each well, 1 μCi of [3H] was added for the final 18 hours of incubation. Data shown are means of n ≥ 5, ± SEM (y axis: CPM). (C) NHLF transfected by miR-154 exhibit significant increase in migration as measured by Boyden chamber assay; data are presented as a mean ± SEM of two independent experiments (n ≥ 5). (D) Flow cytometric analysis of a cell cycle comparing cells transfected for 24 hours with pre–miR-154 and the negative control (SCR) using BrdU incorporation. Lung fibroblasts were cultured with 10 μM BrdU for 12 to 16 hours, and BrdU incorporation was measured (with FITC–anti-BrdU) over total DNA content stained with 7-AAD. The results are shown as mean of n = 5 ± SEM. (E) p15 proteins levels were determined in total protein lysate from fibroblasts transfected with pre–miR-154 and scrambled control (SCR) for 24 hours using Western blot analysis. β-Actin was used as a loading control. (F) Anti–miR-154 reduced the proliferative effect of TGF-β1 in fibrotic and control fibroblasts. Fibrotic and normal fibroblasts were transfected with 30 nM of anti–miR-154 or scrambled control and 6 hours later stimulated with 5 ng/ml of TGF-β1. Transfection of anti–miR-154 was compared with scrambled. Proliferation was measured 48 hours after transfection using CellTiter 96 AQueous.
miR-154–Induced Changes in Proliferation Are Mediated through Activation of the WNT/β-Catenin Pathway
To examine whether microRNAs increased in IPF were potential regulators of lung development, we compared the expression of miR-487b, miR-409–3p, miR-154, miR-154*, miR-134, miR-299–5p, miR-410, miR-382, miR-377, and miR-296 in patients with IPF, in control subjects, and in fetal lungs by qRT-PCR. microRNAs increased in IPF, including miR-154, were also highly expressed in fetal lungs (Figure 4A), suggesting that the increased microRNA profile of IPF lungs represented reversal of lung differentiation. To determine whether at least part of the effect of miR-154 on NHLF was mediated through dysregulation of the WNT/β-catenin pathway implicated in IPF, we assessed mRNA levels of 84 WNT pathway–related genes using a WNT signaling PCR array in NHLF 24 hours after transfection with miR-154 and determined that 10 WNT pathway genes were significantly changed. The WNT pathway inhibitors DKK2, DIXDC1, and PPP2CA (36–38) were significantly decreased (Figure 4B), whereas activators of the pathway, such as the WNT receptors FZD 4/5/6, LRP, and KREMEN1, as well as β-catenin and WISP1 were increased (Figure 4B). Total protein levels of β-catenin were also significantly increased (Figures 4C and 4D), and MiR-154 transfection significantly induced activity of the TOPFLASH reporter assay (Figure 4E). Taken together, these results suggest that transfection of NHLF with miR-154 leads to activation of the WNT/β catenin pathway. Finally, to determine whether the effects of miR-154 on fibroblast proliferation were mediated through activation of the WNT/β catenin pathway, we used two inhibitors of the pathway: ICG-001, an inhibitor of β-catenin/CBP-mediated transcription that specifically binds to CBP (14), and XAV939, which inhibits Wnt/β-catenin signaling by increasing axin protein levels (33). We transfected NHLF with miR-154 and, 6 hours after transfection, treated the cells with 10 μM ICG-001 or XAV939. Treatment with either inhibitor eliminated the miR-154–induced proliferation (Figures 4F and 4G), suggesting that miR-154 induces fibroblast proliferation through its permissive effect on the WNT/β-catenin pathway.
Figure 4.
Mir-154 is a regulator of the WNT/β-catenin pathway. (A) microRNA qRT-PCR expression profiles were analyzed using RNA from 10 control subjects and from 10 fetal and 10 pulmonary fibrosis lung tissues. The heat map represents statistically significant (P ≤ 0.05), up-regulated microRNAs, which are shown in the yellow. The purple represents down-regulated miRs, and I represents nonexpressed miRs; the fetal and fibrotic tissues are normalized to the controls. Each column represents individual samples; selected microRNAs are represented in rows. (B) WNT superarray analysis was done on primary lung fibroblasts transfected with the negative control or pre–miR-154, and mRNA levels of 10 WNT signaling genes were found to be significantly elevated (*). The results are shown as mean of n = 4 ± SEM (*P ≤ 0.05). (C) Protein levels of β-catenin were determined in fibroblasts transfected with the precursor of miR-154 or negative control (24 h) using Western blot analysis. β-actin was used as a loading control. (D) Densitometry confirms changes in proteins. Relative intensity is calculated by dividing density of target protein by the density of the loading control for every lane as determined using ImageJ software (n = 3, *P ≤ 0.05). (E) NHLF cells were transfected with TOPFLASH reporter or its mutant, FOPFLASH, followed by transfection of pre–miR-154 or scrambled for 24 hours. Luciferase activity was normalized to Renilla luciferase activity (n = 4; *P < 0.05). (F) ICG-001 reduced the proliferative effect of miR-154 in human fibroblasts. Scrambled (SCR) and SCR+ICG-001 were used as a control for pre–miR-154 and miR-154+ICG-001 transfections, respectively (n ≥ 4, *P ≤ 0.05). Proliferation was measured 24 hours after transfection using CellTiter 96 AQueous. The results are shown as the percentage relative to scrambled. (G) XAV939 reduced the proliferative effect of miR-154 in human fibroblasts. Scrambled (SCR) and SCR+XAV939 were used as a control for pre–miR-154 and miR-154+XAV939 transfections, respectively (n ≥ 4, *P ≤ 0.05). Proliferation was measured 24 hours after transfection using CellTiter 96 AQueous. Results are shown as optical density at 490 nm.
Discussion
In the current study, we identified 94 microRNAs that were differentially expressed in IPF. Of the 43 increased microRNAs, 24 were localized to the microRNA cluster on chromosome 14q32, and 13 of them were members of the miR-154 family. Methylation analysis did not provide evidence for epigenetic regulation of this region in IPF, but transcription factor binding site analysis revealed enrichment of SMAD-binding sites. Indeed, 14 microRNAs from this region were induced by TGF-β1 stimulation of NHLF, including seven miR-154 family microRNAs. Transfection with miR-154 enhanced proliferation and migration of fibroblasts and repressed protein expression of cell cycle inhibitor p15; inhibited WNT pathway repressors DKK2, DIXDC1, and PPP2CA; and increased FZD 4/5/6, LRP, KREMEN1, β-catenin, and WISP1, leading to an overall activation of the WNT/β-catenin pathway. The miR-154–induced proliferation was reversed by inhibition of the WNT/β-catenin pathway, suggesting that the effects of miR-154 on lung fibroblasts were at least in part mediated through this pathway.
One of the most intriguing findings in our study is that the majority of up-regulated microRNAs in IPF can be localized to a single chromosomal cluster on chromosome 14q32. This is a relatively well described microRNA cluster that has been implicated in several diseases such as inflammatory bowel disease, schizophrenia, and cancer (39–43). The majority of these studies assumed structural changes such as amplifications, deletions, or changes in epigenetic regulation as the cause for the dysregulation. Indeed, the microRNA cluster on chr14q32 has been shown to be regulated by a differentially methylated region (IG-DMG) located approximately 200 kb upstream of the microRNA cluster (24). We analyzed this differentially methylated region on chr14q32 in patients with IPF and found no difference in methylation when compared with control subjects and thus looked for another potential explanation: shared transcriptional regulation. The discovery that the Chr14q32 microRNA cluster is enriched with SMAD binding sites and may be coordinately regulated by TGF-β1 is novel and suggests a different model by which this pluripotent growth factor exerts global effects on cellular phenotypes. Although we did not provide direct evidence for SMAD binding for every one of these SMAD-binding elements, we did provide evidence in silico (see Figures E3A and E3B for an analysis of gene expression across the region) that the Chr14q32 microRNA cluster behaved as a transcriptional module, and we confirmed in vitro the binding of SMAD3 to the putative promoter of mir-154 and demonstrated a significant increase in microRNAs in this cluster after TGF-β1 stimulation. The role of TGF-β1 as a regulator of single microRNA expression in IPF has been described by us and by other groups (16, 44), but this finding highlights the extent to which TGF-β1 exerts its effects on cellular phenotypes, considering that these microRNAs have multiple targets. Considering that the microRNAs in this cluster are usually up-regulated during lung development (21), it may be speculated that at least some of the recapitulation of developmental pathways observed in IPF is a result of chronic cellular exposure to TGF-β1.
In the context of its role in lung development, the impact of miR-154 on the WNT/β-catenin pathway may provide an important clue to the profound phenotypic changes observed in IPF. Recapitulation of developmental programs, particularly WNT signaling, has been implicated in IPF (2, 3, 10, 13, 45), but the mechanisms are not completely known. It is possible that at least part of the aberrant activation of the WNT/β-catenin pathway is caused by the permissive effects of miR-154 through inhibition of inhibitors of the pathways, as is gleaned from the down-regulation of miR-154 predicted targets; the WNT inhibitors DKK2, DIXDC1, and PPP2CA; and a concomitant increase in pathway activators FZD4/5/6, LRP6, KREMEN1, and intracellular molecules WISP1 and β-catenin. Regulation of WNT/β-catenin through targeting its inhibitors has recently been demonstrated in 293T cells (46). The downstream effects of miR-154 on fibroblast proliferation were completely abolished by the use of either ICG-001, a selective inhibitor of WNT/β-catenin/CBP-driven transcription previously shown to attenuate bleomycin-induced lung fibrosis in mice (13, 14), or by XAV939, an inhibitor that stabilizes axin and inhibits Wnt signaling (33), suggesting that the effects of mir-154 on fibroblast proliferation are at least partially mediated through aberrant dysregulation of the WNT/β-catenin pathway. A frequent conceptual challenge when interpreting microRNA experiments is the integration of all the parallel and intersecting effects that microRNAs exhibit. In our case, the discovery that overexpression miR-154 leads also to inhibition of the cyclin-dependent kinase inhibitor p15 (CDKN2B) suggests that mir-154 induces proliferation through more than one pathway. P15, a well known TGF-β1 responsive gene (47, 48), is a known inhibitor of cell cycle, and loss of p15 expression has been associated with several cancer types (49). Taken together, our observations suggest that overexpression of miR-154 may be associated with a general overproliferative state in lung fibroblasts.
There are several limitations of our work. First, the evidence for a potential role of mir-154 is derived from profiling of human lungs and from experiments performed in vitro in human lung fibroblasts. In vivo experiments that involve perturbations of mir-154 expression in animal models of fibrosis are required to prove that mir-154 plays a profibrotic role in vivo. Second, although we provide compelling data about the effects of mir-154 in NHLF and some limited observations in IPF fibroblasts, we did not perform a detailed analysis of the regulation and effects of mir-154 in IPF fibroblasts. A detailed analysis that includes a large number of primary lines from patients with IPF and potentially freshly harvested cells as recently suggested (50) will be required to determine whether mir-154 is consistently overexpressed in IPF fibroblasts and whether its inhibition alters their phenotypes. Third, although we provide evidence that mir-154 is induced by TGF-β1 and that it may be a regulator of the WNT/β-catenin, we do not provide a detailed mechanistic analysis of its role in the intersection of these critically important pathways. Last, although our results present a compelling view of a chromosomal location potentially regulated by TGF-β1 and then focus on the downstream profibrotic effects of mir-154, we do not address other potential mechanisms that may activate mir-154. As an example, in addition to multiple SMAD3/4 binding elements on chr14q32, we also found some predicted binding sides for p53, c-Myc, and E2F1 in the same region. We did not validate the binding of these factors or determine their effects on the expression of the cluster, but future studies may indicate whether they have an inhibitory or a coactivator role. However, the current finding of the regulation of mir-154 and the 14q32.31 cluster by TGF-β1 is novel and, in the context of fibrosis, is likely to be important. It may also suggest that mir-154 should be investigated as a potential previously unrecognized link between the TGF-β1 and the WNT/β-catenin pathways.
MicroRNAs have emerged as key regulators of fibrotic processes in the lung. In this manuscript we provide evidence for a regionally coordinated up-regulation of microRNAs potentially in response to TGF-β1 and highlight that the microRNA profile of IPF lungs is similar to that of the embryonic human lung and suggests activation of programs that were suppressed during lung differentiation. We then focused on mir-154 and demonstrate a role for this microRNA in the regulation of fibroblast proliferation and migration probably through the inhibitory effect on CDKN2B and a permissive effect on the global activation of the WNT signaling pathway. Our results suggest that increases in mir-154 may allow the activation of fibrotic transcriptional programs in lung fibroblasts and, thus, assessing the potential therapeutic value of in vivo inhibition of mir-154 in animal models of disease and detailed mechanistic analysis of its effects would be the logical next steps.
Supplementary Material
Acknowledgments
The authors thank Dr. Carol Feghali-Bostwick at the Simmons Center and the Scleroderma Center at the University of Pittsburgh for generously providing the IPF fibroblasts, Dr. Yingze Zhang for technical support on reporter assays, and Joseph Stahovic Jr. for outstanding intellectual and technical support.
Footnotes
This work was supported by National Institutes of Health grants NIH RO1HL07374530, RO1HL095397, R01LM009657, RC2HL101715, and RO1HL073722 and by the Dorothy P. and Richard P. Simmons Endowed Chair for Pulmonary Research.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2011-0377OC on October 4, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N Engl J Med 2001;345:517–525 [DOI] [PubMed] [Google Scholar]
- 2.Konigshoff M, Eickelberg O. Wnt signaling in lung disease: a failure or a regeneration signal? Am J Respir Cell Mol Biol 2010;42:21–31 [DOI] [PubMed] [Google Scholar]
- 3.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med 2008;5:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White ES, Thannickal VJ, Carskadon SL, Dickie EG, Livant DL, Markwart S, Toews GB, Arenberg DA. Integrin alpha4beta1 regulates migration across basement membranes by lung fibroblasts: a role for phosphatase and tensin homologue deleted on chromosome 10. Am J Respir Crit Care Med 2003;168:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coward WR, Saini G, Jenkins G. The pathogenesis of idiopathic pulmonary fibrosis. Ther Adv Respir Dis 2010;4:367–388 [DOI] [PubMed] [Google Scholar]
- 6.Pottier N, Maurin T, Chevalier B, Puissegur MP, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, et al. Identification of keratinocyte growth factor as a target of microrna-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS ONE 2009;4:e6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Konigshoff M, Jayachandran A, Handley D, Seeger W, Kaminski N, Eickelberg O. Transgelin is a direct target of tgf-beta/smad3-dependent epithelial cell migration in lung fibrosis. FASEB J 2008;22:1778–1789 [DOI] [PubMed] [Google Scholar]
- 8.Willis BC, Borok Z. Tgf-beta-induced emt: mechanisms and implications for fibrotic lung disease. Am J Physiol 2007;293:L525–L534 [DOI] [PubMed] [Google Scholar]
- 9.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010;182:220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vuga LJ, Ben-Yehudah A, Kovkarova-Naumovski E, Oriss T, Gibson KF, Feghali-Bostwick C, Kaminski N. Wnt5a is a regulator of fibroblast proliferation and resistance to apoptosis. Am J Respir Cell Mol Biol 2009;41:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konigshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS ONE 2008;3:e2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, et al. Wnt1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest 2009;119:772–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson WR, Jr, Chi EY, Ye X, Nguyen C, Tien YT, Zhou B, Borok Z, Knight DA, Kahn M. Inhibition of wnt/beta-catenin/creb binding protein (cbp) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci USA 2010;107:14309–14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Kim HY, Moon SH, et al. A small molecule inhibitor of beta-catenin/creb-binding protein transcription [corrected]. Proc Natl Acad Sci USA 2004;101:12682–12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YM, Ma H, Oehler VG, Gang EJ, Nguyen C, Masiello D, Liu H, Zhao Y, Radich J, Kahn M. The gamma catenin/cbp complex maintains survivin transcription in beta-catenin deficient/depleted cancer cells. Curr Cancer Drug Targets 2011;11:213–225 [DOI] [PubMed] [Google Scholar]
- 16.Pandit KV, Milosevic J, Kaminski N. Micrornas in idiopathic pulmonary fibrosis. Transl Res 2011;157:191–199 [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear rnase iii drosha initiates microrna processing. Nature 2003;425:415–419 [DOI] [PubMed] [Google Scholar]
- 18.Krol J, Loedige I, Filipowicz W. The widespread regulation of microrna biogenesis, function and decay. Nat Rev Genet 2010;11:597–610 [DOI] [PubMed] [Google Scholar]
- 19.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. Microrna-21 contributes to myocardial disease by stimulating map kinase signalling in fibroblasts. Nature 2008;456:980–984 [DOI] [PubMed] [Google Scholar]
- 20.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. Mir-21 is an egfr-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA 2009;106:12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted micrornas are differentially expressed during mouse and human lung development. Dev Dyn 2007;236:572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nana-Sinkam SP, Croce CM. Micrornas as therapeutic targets in cancer. Transl Res 2011;157:216–225 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhang R, Su B. Diversity and evolution of microRNA gene clusters. Sci China C Life Sci 2009;52:261–266 [DOI] [PubMed] [Google Scholar]
- 24.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microrna gene cluster at the mouse dlk1-gtl2 domain. Genome Res 2004;14:1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. The primate-specific microrna gene cluster (c19mc) is imprinted in the placenta. Hum Mol Genet 2010;19:3566–3582 [DOI] [PubMed] [Google Scholar]
- 26.Milosevic J, Pandit KV, Jacobs R, Benos PV, Yakhini Z, Chensny L, Kaminski N. The role of mir-154 microRNA family in IPF [abstract]. Am J Respir Crit Care Med 2009;109:A2732 [Google Scholar]
- 27.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali-Bostwick CA. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol 2005;166:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, Bisceglia M, Gilbert S, Yousem SA, Song JW, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2009;180:167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corcoran DL, Feingold E, Benos PV. Footer: A web tool for finding mammalian DNA regulatory regions using phylogenetic footprinting. Nucl Acids Res 2005;33:W442–W446. [DOI] [PMC free article] [PubMed]
- 31.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA 2005;102:15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMillan M, Kahn M. Investigating wnt signaling: a chemogenomic safari. Drug Discov Today 2005;10:1467–1474 [DOI] [PubMed] [Google Scholar]
- 33.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes wnt signalling. Nature 2009;461:614–620 [DOI] [PubMed] [Google Scholar]
- 34.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microrna expression atlas based on small rna library sequencing. Cell 2007;129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabinovich EI, Kapetanaki MG, Steinfeld I, Gibson KF, Pandit KV, Yu G, Yakhini Z, Kaminski N. Global methylation patterns in idiopathic pulmonary fibrosis. PLoS ONE 2012;7:e33770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao B, Niehrs C. Kremen2 modulates dickkopf2 activity during wnt/lrp6 signaling. Gene 2003;302:179–183 [DOI] [PubMed] [Google Scholar]
- 37.Wong CK, Luo W, Deng Y, Zou H, Ye Z, Lin SC. The dix domain protein coiled-coil-dix1 inhibits c-jun n-terminal kinase activation by axin and dishevelled through distinct mechanisms. J Biol Chem 2004;279:39366–39373 [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto H, Hinoi T, Michiue T, Fukui A, Usui H, Janssens V, Van Hoof C, Goris J, Asashima M, Kikuchi A. Inhibition of the wnt signaling pathway by the pr61 subunit of protein phosphatase 2a. J Biol Chem 2001;276:26875–26882 [DOI] [PubMed] [Google Scholar]
- 39.Benetatos L, Vartholomatos G, Hatzimichael E. Meg3 imprinted gene contribution in tumorigenesis. Int J Cancer 2011;129:773–779 [DOI] [PubMed] [Google Scholar]
- 40.Fasseu M, Treton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soule JC, Moreau R, et al. Identification of restricted subsets of mature microrna abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS ONE 2010;5:e13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ. Imprinted dlk1-dio3 region of 14q32 defines a schizophrenia-associated mirna signature in peripheral blood mononuclear cells. Mol Psychiatry 2012;17:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. Cdkn2a, nf2, and jun are dysregulated among other genes by mirnas in malignant mesothelioma: a mirna microarray analysis. Genes Chromosomes Cancer 2009;48:615–623 [DOI] [PubMed] [Google Scholar]
- 43.Luk JM, Burchard J, Zhang C, Liu AM, Wong KF, Shek FH, Lee NP, Fan ST, Poon RT, Ivanovska I, et al. Dlk1-dio3 genomic imprinted microrna cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem 2011;286:30706–30713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. Mir-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 2011;45:287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, et al. Aberrant wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol 2003;162:1495–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Huang T, Zhao X, Cheng L. Micrornas modulate the wnt signaling pathway through targeting its inhibitors. Biochem Biophys Res Commun 2011;408:259–264 [DOI] [PubMed] [Google Scholar]
- 47.Feng XH, Lin X, Derynck R. Smad2, smad3 and smad4 cooperate with sp1 to induce p15(ink4b) transcription in response to tgf-beta. EMBO J 2000;19:5178–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F, Nielsen LD, Lucas JJ, Mason RJ. Transforming growth factor-beta antagonizes alveolar type ii cell proliferation induced by keratinocyte growth factor. Am J Respir Cell Mol Biol 2004;31:679–686 [DOI] [PubMed] [Google Scholar]
- 49.Kool J, Uren AG, Martins CP, Sie D, de Ridder J, Turner G, van Uitert M, Matentzoglu K, Lagcher W, Krimpenfort P, et al. Insertional mutagenesis in mice deficient for p15ink4b, p16ink4a, p21cip1, and p27kip1 reveals cancer gene interactions and correlations with tumor phenotypes. Cancer Res 2010;70:520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emblom-Callahan MC, Chhina MK, Shlobin OA, Ahmad S, Reese ES, Iyer EP, Cox DN, Brenner R, Burton NA, Grant GM, et al. Genomic phenotype of non-cultured pulmonary fibroblasts in idiopathic pulmonary fibrosis. Genomics 2010;96:134–145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.