Abstract
The relationship between ecological variation and microbial genetic composition is critical to understanding microbial influence on community and ecosystem function. In glasshouse trials using nine native legume species and forty rhizobial strains, we find that bacterial rRNA phylotype accounts for 68% of inter-isolate variability in symbiotic effectiveness and 79% of host-specificity in growth response. We also find that rhizobial phylotype diversity and composition of soils collected from a geographic breadth of sites explains the growth responses of two acacia species. Positive soil microbial feedback between the two acacia hosts was largely driven by changes in diversity of rhizobia. Greater rhizobial diversity accumulated in association with the less responsive host species, Acacia salicina, and negatively affected the growth of the more responsive A. stenophylla. Together this work demonstrates correspondence of phylotype with microbial function, and demonstrates that the dynamics of rhizobia on host-species can feed back on plant population performance.
Keywords: Mutualism, Symbiosis, Rhizobia, Plant-microbe interactions, Plant ecology, Microbial ecology, Plant-soil feedback, Diversity-productivity, Molecular ecology, Specificity, Ecosystem function
Introduction
Advances in molecular methods have revealed a tremendous diversity of bacterial taxa in soil, with as many as 10,000 discrete taxa present per gram (Torsvik et al. 1990; Roesch et al. 2007). This bacterial diversity could be of great functional importance if genetically distinct taxa also have correspondingly different physiologies and ecologies. While differences in habitats, and spatial and temporal patterns support the ecological coherence of higher taxonomic ranks (von Mering et al. 2007; Philippot et al. 2010), examples confirming differing ecologies of phylogenetically distinct bacterial taxa (phylotypes) within a lineage are limited to a few systems involving intensive studies of tightly circumscribed groups (Palys et al. 2000; Johnson et al. 2006; Hunt et al. 2008). Thus, the generality of correspondence between genetic delineation of species and ecological function remains uncertain (Cohan 2006; Doolittle & Papke 2006; Konstantinidis et al. 2006), especially given the numerous examples of lateral transfer of genes for ecologically important traits within major soil microbial groups and across distantly related groups (Ochman et al. 2000; Koonin et al. 2001; Gogarten et al. 2002). This is particularly problematic because the causal link between genetics and ecology is a basic assumption underlying the rapidly growing fields of molecular ecology and environmental microbiology (von Mering et al. 2007; Philippot et al. 2010).
Soil microorganisms play fundamental roles in biogeochemical cycling and are major drivers of terrestrial ecosystem productivity and diversity (van der Heijden et al. 2008; Mangan et al. 2010). Experimental studies have identified that soil microbial diversity and composition can have large influences on plant productivity and diversity (Van der Heijden et al. 1998; Vogelsang et al. 2006). Moreover, the composition of microbial communities has been shown to be dynamic, changing rapidly with plant species (Bever 2002; Mitchell et al. 2010). This change in composition can generate feedbacks on plant fitness and recent work supports this primary role for soil biota in structuring plant communities (Kulmatiski et al. 2008; Bever et al. 2010; Mangan et al. 2010; Johnson et al. 2012). While soil-feedback inoculation studies provide phenomenological evidence of the importance of soil microbial change on plant ecology, the microbial drivers of these feedbacks are often obscure, with individual studies illustrating contributing roles of both soil pathogens and soil mutualists (Packer & Clay 2000; Bever 2002).
Here, we investigate the link between microbial genotype and function in the context of interactions between plants and their soil microbial communities, specifically targeting legumes and N2-fixing symbionts (i.e. rhizobia). This well-known interaction has the advantage that metrics of ecological function can be efficiently scored using assays of host growth. Specifically, average host growth promotion can represent the effect of the rhizobial community on terrestrial productivity and the specificity of host growth promotion underlies the potential impact of the rhizobial community on plant community dynamics (Bever et al. 1997; Mangan et al. 2010). Moreover, given that the genes coding for N-fixation within rhizobia are known to have spread laterally across taxonomic groups (Laguerre et al. 2001; Sprent 2001; Finan 2002; MacLean et al. 2007), studies of rhizobia provide a conservative test of the relationship between chromosomally defined genotypes and host growth promotion.
In particular, we take advantage of the extensive work on the ecology of the interactions between Australia acacias and their symbiotic rhizobia. Two recent studies of this system have demonstrated specificity of rhizobial effects on plant growth (Thrall et al. 2008) and the potential for positive plant-soil feedbacks (Thrall et al. 2007b). Here, we integrate information on rhizobial phylotype with phenotypic growth data from these two studies to test the relationship between rhizobial genotype (as assessed by rRNA markers) and ecological function. We first reanalyze the ecological variation measured within a comprehensive glasshouse inoculation study of individual isolates of rhizobia with a diverse set of ecologically important Acacia species (Thrall et al. 2008). We then integrate genetic analyses of rhizobial community composition and ecological function into a test of rhizobial mediation of plant-soil feedback between two widely distributed Acacia spp (Thrall et al. 2007b). Using these approaches we explicitly evaluate the extent to which rhizobial phylotypes and community structure can predict the ecological performance of their legume hosts.
Materials and Methods
Experiment 1: rRNA markers as predictors of ecological function
A full factorial replicated glasshouse trial involving nine Acacia species and 40 rhizobial isolates was conducted to assess variation in host specificity and rhizobial effectiveness at promoting plant growth (Thrall et al. 2008). The rhizobial strains were originally isolated from 17 Acacia host species from 22 locations across southeastern Australia; the strains used in the glasshouse study represented a haphazardly chosen subset of these. The set of host species used in the glasshouse trial included: A. brachybotrya, A. hakeoides, A. ligulata, A. mearnsii, A. pendula, A. pycnantha, A. rigens, A. salicina and A. stenophylla. These were selected to represent a broad range of geographic distributions and ecologies. Experimental details and data are provided in the Electronic Appendix and in Thrall et al (2008).
Characterization and identification of rhizobial phylotypes
Genomic DNA was extracted from purified single colony rhizobial isolates and the SSU was amplified. The SSU PCR product was digested with one of four restriction endonucleases (HhaI, HinfI, MspI and RsaI; New England Biolabs) and digested products were separated on agarose gels. Restriction profiles were used to identify genomic species as described by (Lafay & Burdon 1998). To determine the generic affiliations of isolates, 16s rDNA was sequenced from representatives of each newly identified phylotype as described in the Electronic Appendix and Hoque et al (2011).
Statistical Analysis
Of the forty isolates used in the glasshouse study, a total of 34 could be identified to phylotype and only these isolates were included in the analysis of phylotype. Log transformed plant dry weight was analyzed using Proc Mixed in SAS (SAS 1990) with plant species, the genus, phylotype and isolate of rhizobia and their interactions with plant species being random effects. Identifying rhizobial isolates and genera and acacia species as random effects allow us to generalize the extent to which taxonomic groupings based on the rRNA gene explain ecological variation in effectiveness of growth promotion of Australian acacias. Variance components were estimated using restricted maximum likelihood.
Experiment 2: Correlation of genotypic composition and ecological function in field soils
To examine patterns of genetic variation and adaptation in host and symbiont populations across geographic ranges, studies were conducted on multiple populations of two widespread native Australian Acacia spp. (A. salicina, A. stenophylla) and associated rhizobial bacteria. Both host species have broad distributions across the Murray Darling Basin in eastern Australia. Acacia stenophylla occurs on the western interior of the basin from the River Murray north while A. salicina occurs throughout the basin, extending more into the eastern flanking ranges than A. stenophylla. In total, 58 sites, including 28 with A. salicina and 30 with A. stenophylla, were characterized with regard to host abundance, symbiont population sizes, soil chemistry and environmental parameters (Thrall et al. 2007b).
Estimates of rhizobial abundance
The most probable number (MPN) plant infection test was used to enumerate rhizobia capable of nodulating seedlings of Acacia salicina and A. stenophylla (Thrall et al. 2007b). Measures of rhizobial density using these two species were log transformed and these values were strongly positively correlated (correlation = 0.80, P < 0.0001). We therefore separated these measures into two variables, one being the average of the two measures which approximated overall rhizobial density and the second being the difference in abundance of nodulation by the two hosts, which represents a measure of the host-specificity of association with the resident rhizobia.
Genetic characterisation of rhizobial communities
Across the 58 sites, a total of 1316 isolates (average of 22.7 per site) were obtained and classified into 119 phylotypes (Hoque et al. 2011). Of these isolates, 1285 (representing 109 of the phylotypes) could be reliably assigned to genera based on 16S rRNA sequence data (Hoque et al. 2011), as summarized in Table S1. Data on phylotype composition and generic affiliation were used to calculate community and species diversity indices.
Acacia growth assays
To examine variation in symbiont effectiveness at promoting plant growth, both Acacia species were grown in all 58 soils (data in electronic appendix), plus uninoculated N+ and N- controls. At harvest, plant roots were separated from the soil and scored for nodulation characteristics (Thrall et al. 2007b).
Statistical analyses
Responses of A. salicina and A. stenophylla to the individual soil communities were calculated as the average of log transformed final dry weights. These averages were analyzed with a two way analysis of variance (acacia species by soil origin) using Proc GLM in SAS (SAS 1990), with the variation in response to soil samples collected from individual species being the random effect. The sign and magnitude of the soil community interaction coefficient was used to measure soil community feedback (the difference in average growth in home soils from the average growth in each other’s soils), which was derived as a critical metric determining soil microbial effects on plant community dynamics (Bever et al. 1997). In this measure, growth in pots is assumed to be correlated with fitness in the field (e.g. (Pringle & Bever 2008; Mangan et al. 2010)). Tests of the relationship between Acacia growth response and rhizobial phylotype composition were performed using two approaches. First, we constructed a matrix of genetic similarities in rhizobial composition and a separate matrix of the difference in growth response of the individual Acacia species. We tested correlations between rhizobial phylotype composition and Acacia growth matrices using Mantel tests performed within GenAlEx 6 (Peakall & Smouse 2006).
The second approach to testing the dependence of Acacia growth response on the rhizobial soil community was to use regression and analysis of covariance. Metrics of rhizobial community composition were constructed using principal components analysis which included all isolates that occurred at least seven times within the sampling. The top ten axes of variation (Table S2), which together explained 75% of the total variation, were used as predictors of plant growth, along with measures of phylotype diversity of each sample (number of rhizobial phylotypes, evenness and diversity (H)), the proportion of Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium in each sample, the average density of rhizobia measured by the two Acacia species and the difference in rhizobial density measured by the two Acacia species. Rhizobial richness was log transformed prior to analysis to decrease the leverage of a few very diverse samples.
We constructed two sets of regression analyses. The first analysis tested predictors of the average growth response of the two Acacia species and the second tested predictors of the difference in growth response between A. salicina and A. stenophylla. The later corresponds to the plant*soil interaction term which measures the plant-soil feedback (Bever et al. 1997). Regression analyses were performed in Proc Reg in SAS. Within each set of analyses, candidate models were ranked by the Akaike’s Information Criterion corrected for small sample size (AICc). The Akaike weights (wi), which correspond to the likelihood that a given model is the best of those being considered (Burnham & Anderson 2002), were calculated and all models with Akaike weights of at least 5% were included (Tables S3 and S5). For predictors within this set of models, combined estimates of the regression coefficients were obtained by weighting the coefficients by the Akaike weights (Tables S4 and S6). The standard errors of the regression coefficients were calculated from the Akaike weighted average of individual model coefficients and from variation in coefficients between models (Burnham & Anderson 2002). In these analyses, a coefficient of zero was used for models which did not include a particular predictor. In order to evaluate the potential importance of individual predictors, we constructed t-tests of overall significance of individual predictors from the Akaike weighted averaged coefficients and standard errors. Significant relationships were depicted using partial regression plots (Figs 2b-2e) of the most likely model as judged by the Akaike weights.
Fig. 2.
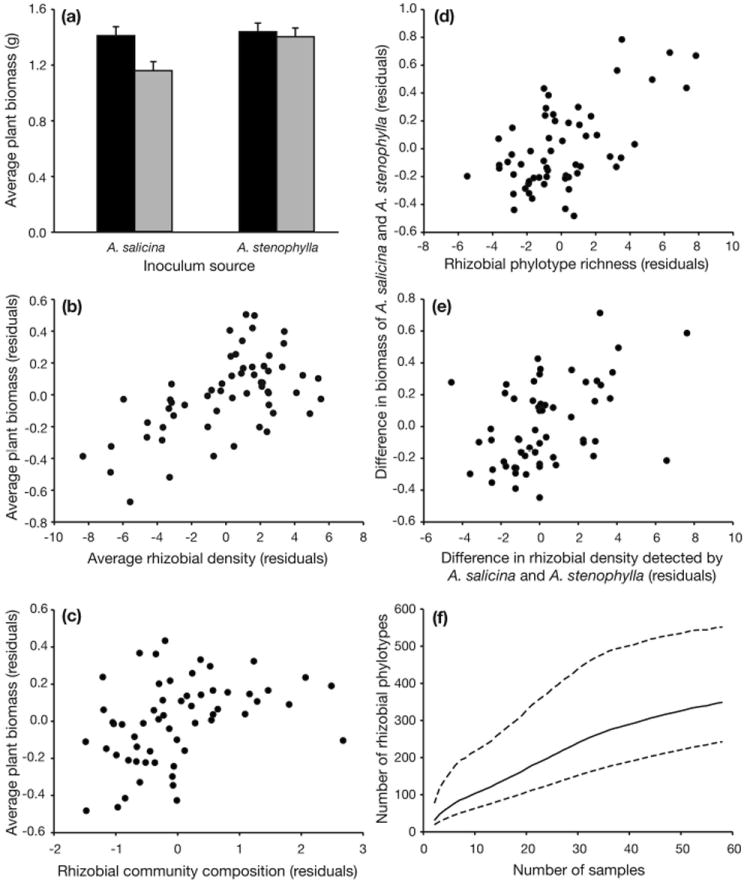
Fig 2a presents the growth of A. salicina (black bars) and A. stenophylla (grey bars) in soils sampled from A. salicina and A. stenophylla. On average, A. salicina and A. stenophylla growth increased with increasing rhizobial density (Fig. 2b) and values of the fifth principal component of rhizobial soil community composition (Fig. 2c). The difference in growth of A. salicina and A. stenophylla increased with phylotype richness of rhizobial communities (Fig. 2d) and with differences in rhizobial density measured by the two plant species (Fig. 2e). Figs. 2b-2e are partial regression plots which represent the slope and error after removal of other predictors in the best model. Fig. 2f presents the estimate and 95% CI for total phylotype diversity associated with A. salicina and A. stenophylla in southeastern Australia.
To test whether the significant rhizobial predictors of acacia growth mediate acacia response to the soils derived from A. salicina and A. stenophylla, we first evaluated whether these predictors differed between soils of these two origins using analysis of variance. The significant predictors that did differ between these two soil origins were then evaluated for their potential to explain the variation in plant response to soils derived from A. salicina versus A. stenophylla. We did this by using the significant rhizobial predictors as covariates in analyses of covariance of acacia growth response conducted in Proc GLM. Covariates that mediate the soil origin effect on acacia growth will reduce the sums of squares explained by soil origin. We constructed an F test for this mediation from the difference in sums of squared deviations due to soil origin without covariates and the sums of squared deviations with covariates (Tables S7 and S8).
Total rhizobial phylotype diversity was estimated using the mean of the bootstrapped estimates using Chao2 as generated by the program EsimateS (Colwell et al. 2012).
Results
Experiment 1: rRNA markers as predictors of ecological function
The 40 different rhizobial strains isolated from natural sites throughout southeastern Australia (Thrall et al. 2008) varied considerably in both their average ability to promote the growth of Acacia host plants (covariance estimate = 0.054, P = 0.0002) and in the specificity of their growth promotion across the nine Acacia host species (covariance estimate = 0.078, P < 0.0001). This substantial ecological variation between different isolates of rhizobia represents the majority of variation seen in this study.
We were able to characterize 34 of these isolates using restriction fragment length polymorphisms (RFLP) banding patterns and analysis of 16S rRNA sequences. These 34 isolates fall within thirteen different phylotypes across several rhizobial genera (Electronic Appendix).
Phylotype explains a remarkable amount of the variation in the ecological function of these rhizobial isolates. In fact, phylotype accounts for 62% of the variation in average growth promotion of the rhizobial strains (Z = 1.7, P = 0.04) and 74% of the variation in specificity of growth promotion (Z = 5.6, P = 0.0001). The four rhizobial genera explained 87% of the species variation in average growth promotion (Z = 0.9, ns) and 74% of the species variation in specificity of growth promotion (Z = 2.9, P = 0.001). Most Acacia spp. grew better with Bradyrhizobium than other genera of rhizobia (Fig. 1). However, A. stenophylla grew relatively poorly with phylotypes of Bradyrhizobium (Fig. 1b), while A. salicina was more consistently responsive to inoculation with a broader range of genera or phylotypes of rhizobia (Fig. 1c).
Fig. 1.
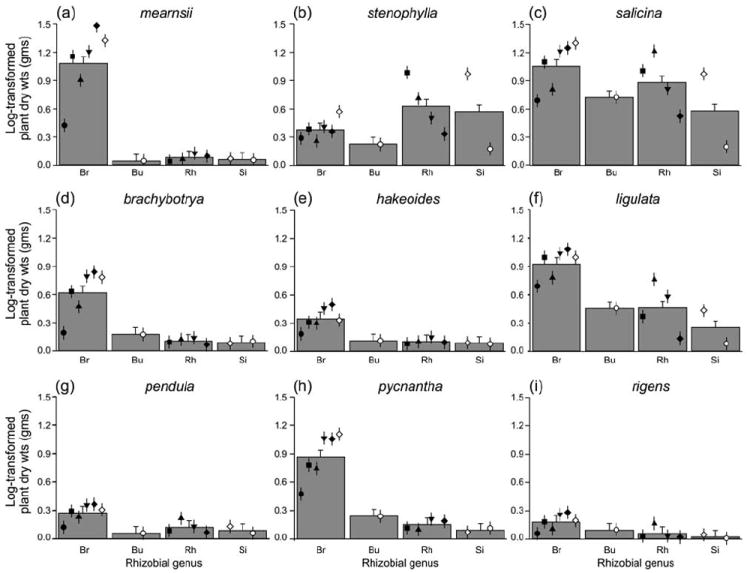
Growth of nine Acacia species was evaluated in association with 34 different isolates of rhizobia belonging to thirteen phylotypes and four genera. Rhizobial phylotypes showed consistent differences in their average growth promotion and the specificity of their growth promotion. While most Acacia species grew best with phylotypes of Bradyrhizobium, A. stenophylla (Fig. 1b) grew relatively poorly with phylotypes of Bradyrhizobium, and instead performed best with Rhizobium and Sinorhizobium. Acacia salicina was relatively insensitive to different rhizobial phylotypes (Fig. 1c). Symbols represent individual phylotypes and the histograms represent the means of the genera Bradyrhizobium, Burkholderia, Rhizobium and Sinorhizobium (= Ensifer), which are represented by Br, Bu, Rh and Si, respectively.
Experiment 2: Correlation of genotypic composition and ecological function in field soils
We found that on average both species of Acacia tended to grow larger when inoculated with soils derived from beneath adult plants of A. stenophylla than A. salicina (F1,56 = 2.89, P < 0.1). This was particularly true for A. stenophylla, which grew significantly better in soils derived from its own species compared to those of A. salicina (F1,56 = 7.66, P = 0.008, Fig. 2a), as previously reported in Thrall et al. (2007b). The differential response of A. stenophylla and A. salicina to their soils, with A. stenophylla growing relatively better with soil inocula derived from conspecific trees, generated a net pairwise positive soil community feedback (as derived in Bever et al. 1997).
To test whether this positive plant-soil feedback was mediated by changes in the rhizobial community, we characterized the rhizobial community composition for each of these soil samples, using the information presented in (Hoque et al. 2011). We then constructed a matrix of genetic similarities in rhizobial composition and a separate matrix of similarity in growth response of the individual Acacia species. We found a significant correlation between the growth of A. salicina and A. stenophylla and rhizobial community composition (Mantel test with 10,000 permutations, rM = 0.11, P = 0.02; rM = 0.081, P = 0.02, for A. salicina and A. stenophylla, respectively), affirming that acacias inoculated with soil communities containing similar rhizobial compositions had similar growth responses. This analysis provides a robust test confirming an overall correspondence between rhizobial community structure and acacia growth, but on its own is insufficient to identify rhizobial composition as the primary driver of plant-soil feedbacks.
We therefore tested potential causal relationships mediating the plant-soil feedback using regression approaches. Phylotype richness, diversity and evenness, measures of rhizobial community composition constructed from principal component analysis of the phylotype composition of the soils, and estimates of relative rhizobial density from each soil were evaluated as predictors of plant growth. Our analysis showed that the average growth response of the two Acacia species to a given soil inoculum is strongly positively related to measures of overall rhizobial density from that soil (t = 3.56, P = 0.0008, Fig. 2b, Tables S3, S4). Measures of the phylotype composition of the rhizobial communities were also important predictors, as acacias responded positively to increasing values of the fifth principal component of rhizobial community composition (t = 2.48, P = 0.02, Fig. 2c, Table S4). Acacia growth also tended to decline with increasing rhizobial species richness (t = -1.71, P = 0.09, Fig. 2d, Table S4). Of these three predictors, rhizobial density and species richness were dependent on the species of Acacia from which the soils originated. Rhizobial density was higher in soils derived from A. stenophylla (F1,57 = 2.95, P < 0.1), while rhizobial species richness was higher in soils derived from A. salicina (F1,57 = 8.25, P =0.006). Together these changes in rhizobial density and species richness explained 98% of the decline in average acacia growth in A. salicina soils (Fig. 2a), thus mediating the weak effect of soil origin (F1,57 = 2.81, P < 0.1, Fig. 3, Table S7).
Fig. 3.
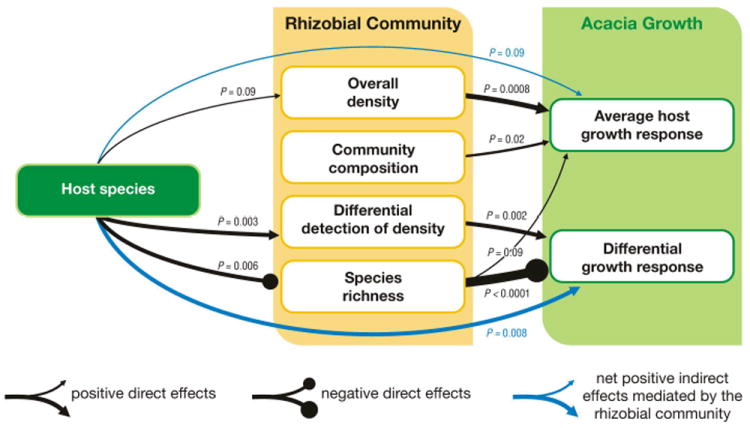
Path diagram showing rhizobial controls on average acacia growth and the difference in response of A. salicina and A. stenophylla. Soils from A. stenophylla generally have higher overall densities of rhizobia, particularly as measured by A. stenophylla, and lower phylotype richness. The increase in overall densities of rhizobia results in an increase in average acacia growth. The differential increase in rhizobia that colonize A. stenophylla but not A. salicina, and the reduction in rhizobial phylotype richness increase the growth of A. stenophylla relative to that of A. salicina, resulting in positive soil community feedback.
Soil community feedback is driven by the difference in growth response of the plant species (Bever et al. 1997), and we found that the differential response of the two Acacia species across the 58 soils assayed (Fig. 2a) was also determined by measures of rhizobial phylotype composition. Phylotype richness was the strongest predictor, and had a stronger negative effect on the growth of A. stenophylla than on that of A. salicina (t = 4.94, P < 0.0001, Figs. 2d, S1). This effect was strikingly robust to the inclusion of other predictors in the model (Tables S5, S6). The difference in soil rhizobial density detected using the two species was also a significant predictor, as A. stenophylla grew better in soils in which A. stenophylla test plants detected higher densities of nodulating bacteria relative to A. salicina (t = 3.21, P = 0.002, Fig. 2e, Table S6). As rhizobial phylotype richness was significantly higher in soils from A. salicina than soils from A. stenophylla (F1,57 = 8.25, P = 0.006), this phylotype effect contributes to the reduced growth of A. stenophylla when inoculated with soil communities present in A. salicina sites (Fig. 3, Table S8). A similar dependence on host origin was found for the differential detection of rhizobial bacteria between the two host species (F1,57 = 9.96, P = 0.003), reflecting a higher density of compatible rhizobia in soils of conspecific acacias. Together, these two predictors explain 99.5% of the difference in growth response between these two species to inoculation with conspecific versus heterospecific field soils (F1,57 = 7.59, P = 0.008, Fig. 3, Table S8). This result confirms that changes in the rhizobial community generated the positive soil feedback observed in this system.
Phylogenetic diversity of Rhizobia on Acacia salicina and stenophylla
Bootstrapped estimates put the number at nearly 400 (95% confidence limits of 247-610) different rhizobial phylotypes within part of the range of only two host species (Fig. 2f). However, this number is likely to be an underestimate given the high number of phylotypes that were unique to one site as reflected in the steady rise of the extrapolated total diversity with increasing sample size.
Discussion
We find that rhizobial isolates vary in their effects on plant productivity and in the host-specificity of these effects. Together these results are consistent with rhizobial community structure contributing to both terrestrial productivity and host community dynamics, extending similar findings for mycorrhizal fungi (Van der Heijden et al. 1998; Vogelsang et al. 2006). Our study is unique in demonstrating concordance between ecological function and rRNA phylotype across a broad range of bacterial isolates, and in demonstrating that changes in rhizobial community composition can drive feedbacks on plant populations.
Relationship of rRNA genotype and ecological function
Our work strongly supports the largely untested hypothesis that rhizobial groups delineated by rRNA gene sequences (i.e. phylotypes) represent ecologically distinct species. In both the factorial manipulation of plant and acacia isolates and an assay of acacia response to field soils, we find that measures of rhizobial composition explain the majority of the observed variation in acacia growth promotion and in the specificity of acacia response. The strong correspondence we observed between rRNA phylotype and rhizobial ecology is particularly surprising, given that genes encoding for their symbiosis with legumes are carried on relatively mobile plasmids or symbiosis islands. As such, rhizobia have been prominent examples of promiscuous exchange of ecologically relevant genes among phylogenetically distant taxa (Laguerre et al. 2001; Sprent 2001; Finan 2002; MacLean et al. 2007). Evidence of such exchanges has fueled expectations of facile shifts in ecologically relevant genes and rapid evolutionary adaptation of microbes to their local environment. With this expectation, local mixing and adaptation may obscure any ecological signal of the diversity or composition of genes on bacterial chromosomes. While we cannot say whether horizontal mixing of functional genes alters ecological function of these rhizobia, we can conclude that the level of horizontal mixing in these populations was not so frequent as to obscure a strong signal of the rRNA gene with ecological function. We find that rRNA gene sequence reliably predicts average growth promotion and the specificity of that growth promotion by independently isolated rhizobial isolates. Moreover, the growth promotion of field soils is predicted by the local diversity and composition of rhizobial isolates in those soils as measured from the rRNA gene. This is impressive given the many soil organisms besides rhizobia, from mycorrhizal fungi to soil pathogens (Bever et al. 2012), that can impact plant growth response.
Our studies also provide support for functional redundancy of rhizobial species, as, nine out of the top ten axes of variation in the rhizobial community composition did not contribute to an explanation of growth response of A. salicina and A. stenophylla. We note, however, that our assay of rhizobial function was limited to two dimensions (average and differential effects on these two species), while many other aspects of rhizobial biology could be important in other contexts (e.g. effect on other legume hosts or salinity tolerance (Thrall et al. 2008)). We therefore suggest that given the support we found for groupings based on phylogenetic analyses of rRNA representing functionally distinct bacterial species, there is likely to be a tremendous amount of functional diversity in microbial systems. Even within our limited sampling, we estimate the presence of nearly 400 (95% confidence limits of 247-610) different rhizobial phylotypes within part of the range of only two host species (Fig. 2f). Given the existence of more than 1,900 species of native legumes in Australia alone and the breadth of environments in which they occur, we predict a staggering level of ecologically relevant species diversity in symbiotic rhizobia associated with these plant species. Extrapolating to other microbial groups, our results suggest that the extremely high phylogenetic diversity observed in soils likely translates to a correspondingly high level of variation in the broad array of ecological functions they represent (e.g. ammonia oxidization, denitrification, free-living N2-fixation).
Rhizobial Diversity as a Driver of Plant-Soil Feedbacks
The acacia species performed relatively better with soil communities derived from conspecifics, a positive pairwise feedback dynamic that could contribute to physical separation of monomorphic patches of the two species (Bever et al. 1997; Molofsky & Bever 2002). While changes in density and composition of beneficial soil symbionts have been observed to generate feedback in other systems (Bever 2002; Dickie et al. 2005; Vogelsang & Bever 2009), this is the first demonstration of positive plant-soil feedback mediated by changes in symbiont diversity. The direction of this effect is surprising, given that A. stenophylla growth declines with increasing species richness of rhizobial communities (p=0.0005, Fig. S1), a result that contrasts with results of glasshouse demonstrations of increases in productivity with increasing symbiont diversity (Van der Heijden et al. 1998; Vogelsang et al. 2006).
The decline in productivity with increasing rhizobial diversity supports general expectations from evolutionary ecology. More diverse communities are expected to have greater likelihood of inclusion of less beneficial symbionts, which can proliferate and decrease the average efficiency of the microbial mutualism (Sachs & Simms 2006; Thrall et al. 2007a; Bever et al. 2009). While observations of partner choice and host sanctions in legumes (Kiers et al. 2003; Simms et al. 2006; Heath & Tiffin 2009) suggest that non-beneficial symbionts should decrease over time, weaker sanctions have been observed in less responsive legume hosts (Kiers et al. 2007). Consistent with this work, we observed greater rhizobial diversity in soils associated with A. salicina, the less selective and less responsive host (Thrall et al. 2008). Together, our work suggests that A. salicina, by permitting the proliferation of less effective mutualists, indirectly inhibits the success of the highly symbiont-dependent A. stenophylla. While more work is required to confirm that the prevalence of non-beneficial rhizobia is correlated with rhizobial diversity, this study potentially represents the first field evidence of ecological consequences of dynamic tensions between host sanctions and the proliferation of cheaters during the evolutionary maintenance of microbial mutualisms (Kiers & Denison 2008; Bever et al. 2009).
A growing body of evidence identifies microbial dynamics as important determinants of plant community dynamics and plant community structure (Bever et al. 2010; Inderjit & van der Putten 2010; Mangan et al. 2010). While plant-soil feedback tests have contributed to our understanding of the role of soil organisms in plant communities, relatively few studies have identified the microbial drivers of these feedbacks (Bever et al. 2012). Our work demonstrates that integration of molecular ecology characterizations with phenomenological plant-soil feedback experiments can facilitate the identification of the microbial agents and processes generating feedback on plant populations.
Supplementary Material
Acknowledgments
We were encouraged to pursue these issues by J. Burdon, J. Sprent, P. Marschner and J. Brockwell. We thank C. Eliasson and S. Hoque for technical support. This work was supported by grants from the NSW Environmental Trust (LMB, PHT) and the National Action Plan for Salinity and Water Quality (PHT). JDB was supported through a Fulbright Fellowship, NSF DEB-0919434, NSF DEB-1050237 and NIH-5 R01 GM092660. Earlier versions of this manuscript were read and greatly improved by A. Bissett, J. Sprent, L. Barrett, S. Mangan, G. Velicer, C. Fuqua, U. Schuette, P. Zee, A. Larimer, T. Platt and A.L. Laine.
Footnotes
Supporting Information
Additional Supporting Information may be downloaded via the online version of this article at Wiley Online Library (www.ecologyletters.com). As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organised for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Statement of authorship: PHT designed the individual studies, LMB collected the molecular data, and JDB performed the statistical analyses. JDB wrote the first draft of the manuscript, and PHT and LMB contributed substantially to revisions.
References
- Bever JD. Negative feedback within a mutualism: Host-specific growth of mycorrhizal fungi reduces plant benefit. Proceedings of the Royal Society of London, B. 2002;269:2595–2601. doi: 10.1098/rspb.2002.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M, Rillig MC, Stock WD, Tibbett M, Zobel M. Rooting theories of plant community ecology in microbial interactions. Trends in Ecology & Evolution. 2010;25:468–478. doi: 10.1016/j.tree.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Platt TG, Morton ER. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annual Review of Microbiology. 2012;66:265–83. doi: 10.1146/annurev-micro-092611-150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecology Letters. 2009;12:13–21. doi: 10.1111/j.1461-0248.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- Bever JD, Westover KM, Antonovics J. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. Journal of Ecology. 1997;85:561–573. [Google Scholar]
- Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information–Theoretic Approach. Springer Science; New York, NY: 2002. [Google Scholar]
- Cohan FM. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos Trans R Soc B Biol Sci. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell RK, Chao A, Gotelli NJ, Lin S-Y, Mao CX, Chazdon RL, Longino JT. Models and estimators linking individual-based and sample-based rarefaction, extrapolation, and comparison of assemblages. Journal of Plant Ecology. 2012;5:3–21. [Google Scholar]
- Dickie IA, Schnitzer SA, Reich PB, Hobbie SE. Spatially disjunct effects of co-occurring competition and facilitation. Ecology Letters. 2005;8:1191–1200. doi: 10.1111/j.1461-0248.2005.00822.x. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Papke RT. Genomics and the bacterial species problem. Genome Biology. 2006;7:116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM. Evolving Insights: Symbiosis Islands and Horizontal Gene Transfer. J Bacteriol. 2002;184:2855–2856. doi: 10.1128/JB.184.11.2855-2856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Molecular Biology and Evolution. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Heath KD, Tiffin P. Stabilizing mechanisms in a legure-rhizobium mutualism. Evolution. 2009;63:652–662. doi: 10.1111/j.1558-5646.2008.00582.x. [DOI] [PubMed] [Google Scholar]
- Hoque MS, Broadhurst LM, Thrall PH. Genetic characterization of root-nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across south-eastern Australia. Int J Syst Evol Microbiol. 2011;61:299–309. doi: 10.1099/ijs.0.021014-0. [DOI] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Inderjit, van der Putten WH. Impacts of soil microbial communities on exotic plant invasions. Trends in Ecology & Evolution. 2010;25:512–519. doi: 10.1016/j.tree.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Johnson DJ, Beaulieu WT, Bever JD, Clay K. Conspecific Negative Density Dependence and Forest Diversity. Science. 2012;336:904–907. doi: 10.1126/science.1220269. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Denison RF. Sanctions, Cooperation, and the Stability of Plant-Rhizosphere Mutualisms. Annu Rev Ecol Evol S. 2008;39:215–236. [Google Scholar]
- Kiers ET, Hutton MG, Denison RF. Human selection and the relaxation of legume defences against ineffective rhizobia. Proceedings of the Royal Society B-Biological Sciences. 2007;274:3119–3126. doi: 10.1098/rspb.2007.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Rosseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: Quantification and classification. Annual Review of Microbiology. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmatiski A, Beard KH, Stevens JR, Cobbold SM. Plant-soil feedbacks: a meta-analytical review. Ecology Letters. 2008;11:980–992. doi: 10.1111/j.1461-0248.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- Lafay B, Burdon JJ. Molecular diversity of rhizobia occurring on native shrubby legumes in southeastern Australia. Appl Environ Microbiol. 1998;64:3989–3997. doi: 10.1128/aem.64.10.3989-3997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 2001;147:981–993. doi: 10.1099/00221287-147-4-981. [DOI] [PubMed] [Google Scholar]
- MacLean AM, Finan TM, Sadowsky MJ. Genomes of the symbiotic nitrogen-fixing bacteria of legumes. Plant Physiol. 2007;144:615–622. doi: 10.1104/pp.107.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan SA, Schnitzer SA, Herre EA, Mack K, Valencia M, Sanchez E, Bever JD. Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature. 2010;466:752–755. doi: 10.1038/nature09273. [DOI] [PubMed] [Google Scholar]
- Mitchell RJ, Campbell CD, Chapman SJ, Cameron CM. The ecological engineering impact of a single tree species on the soil microbial community. Journal of Ecology. 2010;98:50–61. [Google Scholar]
- Molofsky J, Bever JD. A novel theory to explain species diversity in landscapes: positive frequency dependence and habitat suitability. Proceedings of the Royal Society of London, B. 2002;269:2389–2393. doi: 10.1098/rspb.2002.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Packer A, Clay K. Soil pathogens and spatial patterns of seedling mortality in a temperate tree. Nature. 2000;404:278–281. doi: 10.1038/35005072. [DOI] [PubMed] [Google Scholar]
- Palys T, Berger E, Mitrica I, Nakamura LK, Cohan FM. Protein-coding genes as molecular markers for ecologically distinct populations: the case of two Bacillus species. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1021–1028. doi: 10.1099/00207713-50-3-1021. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. The ecological coherence of high bacterial taxonomic ranks. Nature Reviews Microbiology. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- Pringle A, Bever JD. Analogous effects of arbuscular mycorrhizal fungi in the laboratory and a North Carolina field. New Phytologist. 2008;180:162–175. doi: 10.1111/j.1469-8137.2008.02537.x. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. Pyrosequencing enumerates and contrasts soil microbial diversity. Isme Journal. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Simms EL. Pathways to mutualism breakdown. Trends in Ecology & Evolution. 2006;21:585–592. doi: 10.1016/j.tree.2006.06.018. [DOI] [PubMed] [Google Scholar]
- SAS. SAS/STAT User’s Guide. SAS Institute Inc.; Cary, NC: 1990. [Google Scholar]
- Simms EL, Taylor DL, Povich J, Shefferson RP, Sachs JL, Urbina M, Tausczik Y. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proceedings of the Royal Society of London. 2006;273:77–81. doi: 10.1098/rspb.2005.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI. Nodulation in Legumes. Royal Botanical Gardens; Kew: 2001. [Google Scholar]
- Thrall PH, Bever JD, Slattery JF. Rhizobial mediation of Acacia adaptation to soil salinity: evidence of underlying trade-offs and tests of expected patterns. J Ecol. 2008;96:746–755. [Google Scholar]
- Thrall PH, Hochberg ME, Burdon JJ, Bever JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends in Ecology & Evolution. 2007a;22:120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Slattery JF, Broadhurst LM, Bickford S. Geographic patterns of symbiont abundance and adaptation in native Australian Acacia-rhizobia interactions. J Ecol. 2007b;95:1110–1122. [Google Scholar]
- Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Applied and Environmental Microbiology. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- Vogelsang KM, Bever JD. Mycorrhizal densities decline in association with nonnative plants and contribute to plant invasion. Ecology. 2009;90:399–407. doi: 10.1890/07-2144.1. [DOI] [PubMed] [Google Scholar]
- Vogelsang KM, Reynolds HL, Bever JD. Mycorrhizal fungal identity and richness determine the diversity and productivity of a tallgrass prairie system. New Phytologist. 2006;172:554–562. doi: 10.1111/j.1469-8137.2006.01854.x. [DOI] [PubMed] [Google Scholar]
- von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


