Abstract
Backgroud: Both alcohol consumption and the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene polymorphism modulate serum lipid levels, but their interactions on serum lipid profiles are still unknown. The present study was undertaken to detect the interactions of PCSK9 E670G polymorphism and alcohol consumption on serum lipid levels.
Methods: Genotypes of the PCSK9 E670G in 1352 unrelated subjects (785 non-drinkers and 567 drinkers) were determined by polymerase chain reaction and restriction fragment length polymorphism combined with gel electrophoresis, and then confirmed by direct sequencing. The interactions between PCSK9 E670G genotypes and alcohol consumption on serum lipid parameters were detected by using a factorial design covariance analysis after controlling for potential confounders.
Results: The levels of serum triglyceride, high-density lipoprotein cholesterol, apolipoprotein (Apo) A1, and the ratio of ApoA1 to ApoB were higher in drinkers than in non-drinkers (P < 0.01 for all), whereas the levels of total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C) and ApoB were lower in drinkers than in non-drinkers (P < 0.001 for all). The genotypic and allelic frequencies of PCSK9 E670G were not different between non-drinkers and drinkers (P > 0.05 for each). The subjects with AA genotype in non-drinkers had higher serum LDL-C levels than the subjects with AG genotype, whereas the subjects with AG genotype in drinkers had higher serum TC levels than the subjects with AA genotypes (P < 0.05 for each). The effects of alcohol consumption on TC and LDL-C levels depended upon genotypes, the subjects with AA genotype had lower serum TC and LDL-C levels in drinkers than in non-drinkers.
Conclusions: Alcohol consumption can modify the effects of the PCSK9 E670G polymorphism on serum TC and LDL-C levels. The subjects with AA genotype of the PCSK9 E670G benefit more from alcohol consumption than the subjects with AG genotype in decreasing serum TC and LDL-C levels.
Keywords: proprotein convertase subtilisin/kexin type 9 gene, polymorphism, alcohol consumption, lipids, interaction.
Introduction
Light to moderate alcohol consumption has been shown to protect against the development of coronary artery disease (CAD) and mortality 1-3. The dose-response relation between alcohol consumption and risk of CAD is J- or U-shaped 4,5. The protective effects of regular, light to moderate alcohol consumption on CAD have been attributed to high serum high-density lipoprotein cholesterol (HDL-C) and apolipoprotein (Apo) A1 levels 6-10. However, alcohol in doses > 30 g/day in both sexes can augment serum triglyceride (TG) levels 8. The alcohol intake of 60 g/day increases the TG levels by about 0.19 mg/dl per 1 gram of alcohol consumed 11. Although the effects of alcohol consumption on low-density lipoprotein cholesterol (LDL-C) appear to vary by specific patient types or patterns of alcohol intake, and perhaps by population and sex, this topic has been the focus of much recent research 12. A recent study in older Italian subjects (65-84 years) has found that alcohol consumption increases serum LDL-C levels 13. Another recent study of Turks also found increases in LDL-C, as well as in ApoB and TG, with alcohol in men, while women had decreased TG and no change in LDL-C or ApoB with alcohol 14. A decrease in LDL-C with increased alcohol intake has also been reported in some studies, but this effect is less consistent and probably depends on the combination of one or more unmeasured factors 15.
Proprotein convertase subtilisin-like kexin type 9 (PCSK9, OMIM 607786) is a newly discovered serine protease that plays a key role in LDL-C homeostasis by mediating LDL receptor (LDLR) breakdown through a post-transcriptional mechanism 16-19. PCSK9 may also regulate ApoB-containing lipoprotein production and ApoB secretion 20,21, and promote production of nascent very low-density lipoprotein in the fasting state 22. Human PCSK9 gene is approximately 22 kb long, comprising the promoter region and 12 exons, and it is located on chromosome 1p32. The gene produces a mRNA of 3636 bp encoding a 692-amino acid glycoprotein. This protein, also called neural apoptosis regulated convertase, is a serine protease belonging to the protease K subfamily of subtilases. It is a subfamily of proteases largely involved in the processing of inactive precursor proteins to the active product and seems to be involved in the inactivation and degradation of LDLR 23-25. Adenoviral-mediated over-expression of human PCSK9 in mice promotes the accumulation of LDL-C in the plasma but this response is absent in LDLR-deficient animals 18,19,26. Recent studies show that PCSK9 binds directly to the extracellular domain of the LDLR 27,28 and increases its degradation 27. PCSK9 is expressed most abundantly in the liver, kidney, and small intestine 29. Several studies have found that missense mutations increasing the activity of PCSK9 (i.e., gain-of-function mutations) result in an increase of LDL-C levels and CAD 21,30-32 whereas nonsense mutations reducing PCSK9 activity (i.e., loss-of-function mutations) have the opposite effect, lowering LDL-C levels and reducing risk of CAD 33-35. These findings reveal that PCSK9 activity is a major determinant of plasma levels of LDL-C in humans and make it an attractive therapeutic target for LDL-C lowering. Among the genetic variants of the PCSK9 gene, a common single nucleotide polymorphism (SNP), E670G (rs505151), in exon 12 deserved greater scrutiny, as it was responsible for an amino acid change that could potentially be associated with altered PCSK9 activity. The PCSK9 E670G SNP in humans has been found to be associated with modifications of serum LDL-C levels in some studies 36-41 but not in others 35,42-44. The frequency of PCSK9 E670G SNP also varied significantly among different populations. For example, the frequency of G allele is rare in whites but present in approximately 24.8% of blacks 44. The minor-allele frequency (670G) in the Dallas Heart Study (DHS) was 3.6% in whites, 4.2% in Hispanics, and 26.0% in blacks 35. However, little is known about the interactions of PCSK9 E670G SNP and alcohol consumption on serum lipid concentrations. Therefore, the aim of the present study was to determine the interactions of PCSK9 E670G SNP and alcohol consumption on serum lipid levels.
Materials and methods
Subjects
A total of 1352 unrelated subjects were randomly selected from our previous stratified randomized cluster samples. Detailed description of the sampling in this cohort has been published 45,46. Briefly, twenty villages were selected randomly from Lihu (12 villages) and Baxu (15 villages) communities (10 villages for each). Then, sexes and ages (15-20, 20-29, 30-39, 40-49, 50-59, 60-69, ≥ 70 years) in each village were stratified and the subjects were determined from the local population registers. The response rate of the subjects was 97.0% 46. The age of the subjects in this study ranged from 19 to 80 years, with an average age of 46.42 ± 13.97 years. There were 785 non-drinkers and 567 drinkers. All of the subjects were rural agricultural workers. The subjects with a history or evidence of diseases related to atherosclerosis, CAD and diabetes or fasting blood glucose ≥ 7.0 mmol/L determined by glucose meter have been excluded. None of them had been treated with β-adrenergic blocking agents and lipid-lowering drugs such as statins or fibrates. The present study was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University. Informed consent was obtained from all subjects after they received a full explanation of the study.
Epidemiological survey
Epidemiological data including demographics, socioeconomic status, personal and family history of disease, current pharmacotherapy, alcohol consumption, cigarette smoking, and physical activity were collected using standardized questionnaires. Each participant was asked if he/she has consumed alcohol in the past 12 months. If yes, the average daily/weekly number of drinks and “Liangs”(traditional Chinese unit, about 50 mL) consumed over the past year for rice wine, corn wine, rum, beer, or liquor was recorded. As a matter of fact, 90% of the wine drunk by the drinkers was corn wine, rice wine, and rum, in which the alcohol content is about 15%, 30%, and 10% (v/v); respectively. Average daily alcohol intake (grams of alcohol per day) was then calculated. Alcohol consumption was categorized into groups of grams of alcohol per day: 0 (non-drinker), < 25 and ≥ 25. Smoking status was categorized into groups of cigarettes per day: 0 (non-smoker), < 20 and ≥ 20. At the physical examination, several parameters, such as height, weight, and waist circumference were measured. Body mass index (BMI) was calculated. Sitting blood pressure was measured three times with the use of a mercury sphygmomanometer after the subjects had a 5-minute rest.
Measurements of serum lipid levels
Venous blood samples were obtained from all subjects after at least 12 hours of fasting. The levels of serum total cholesterol (TC), TG, HDL-C, and LDL-C in samples were determined by enzymatic methods with commercially available kits. Serum ApoA1 and ApoB levels were detected by the immunoturbidimetric immunoassay using a commercial kit.
DNA amplification and genotyping
Genomic DNA was isolated from peripheral blood leukocytes using the phenol-chloroform method as previously described 41. The extracted DNA was maintained at 4°C until analysis. Genotyping of the PCSK9 E670G SNP was performed by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP). PCR amplification was performed using 5'-CACGGTTGTGTCCCAAATGG-3' and 5'-GAGAGGGACAAGTCGGAACC-3' (Sangon, Shanghai, People's Republic of China) as the forward and reverse primer pairs; respectively. Each amplification reaction was performed using 100 ng of genomic DNA in 25 μL of reaction mixture consisting of 25 μmol/L of each primer, 200 μmol/L of each deoxynucleotide triphosphate, 2.5 μL of 10 × PCR buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 20 mM MgCl2, 1% Triton), and 2 units of Taq polymerase. After initial denaturizing at 94 °C for 5 min, the reaction mixture was subjected to 35 cycles of 30 s denaturation at 94 °C, 30 s annealing at 58 °C and extension 30 s at 72 °C, followed by a final 5 min extension at 72 °C. After electrophoresis on a 1.2% agarose gel with 0.5 μg/mL ethidium bromide, the amplification products were visualized under ultraviolet light. Then 2 U of Eam 1104I restriction enzyme was added directly to the PCR products (10 μL) and digested at 37 °C overnight. After restriction enzyme digestion of the amplified DNA, the genotypes were identified by electrophoresis on 2% agarose gels and visualized with ethidium-bromide staining ultraviolet illumination. Genotypes were scored by an experienced reader blinded to epidemiological data and serum lipid levels. Six samples (AA and AG genotypes in three; respectively) detected by the PCR-RFLP were also confirmed by direct sequencing. The PCR products were purified by low melting point gel electrophoresis and phenol extraction, and then the DNA sequences were analyzed in Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., People's Republic of China.
Normal values of serum lipid levels
The normal values of serum TC, TG, HDL-C, LDL-C, ApoA1 and ApoB levels, and the ratio of ApoA1 to ApoB in our Clinical Science Experiment Center were 3.10-5.17, 0.56-1.70, 0.91-1.81, 2.70-3.20 mmol/L, 1.00-1.78, 0.63-1.14 g/L, and 1.00-2.50; respectively 45,46.
Statistical analysis
Continuous variables are expressed as mean ± standard deviation (serum TG levels are presented as medians and interquartile ranges). Categorical variables are presented as percentages. Allele frequency was determined via direct counting, and the standard goodness-of-fit test was used to test the Hardy-Weinberg equilibrium. Differences in genotype distribution between the groups were estimated by using the chi-square test. The difference in general characteristics between non-drinkers and drinkers was tested by the Student's unpaired t-test (TG by the Wilcoxon-Mann-Whitney test). The association of genotypes with serum lipid parameters was tested by analysis of covariance (TG by the Wilcoxon-Mann-Whitney test). The co-variables include sex, age, BMI, hypertension, and cigarette smoking. The interactions between genotypes and alcohol consumption were assessed by using a factorial design covariance analysis after controlling for potential confounders including sex, age, BMI, hypertension, and cigarette smoking. In order to evaluate the association of serum lipid levels with genotypes (AA = 1, AG = 2), multiple linear regression analysis was also performed in the non-drinkers and drinkers; respectively. A P value of less than 0.05 was considered statistically significant.
Results
General characteristics between non-drinkers and drinkers
The general characteristics of the two groups are shown in Table 1. The levels of body height, weight, the ratio of male to female, and the percentages of subjects who smoked cigarettes were higher in drinkers than in non-drinkers (P < 0.001 for all), whereas the levels of mean age and diastolic blood pressure were lower in drinkers than in non-drinkers (P < 0.01 for each). There were no significant differences in the levels of BMI, systolic blood pressure and pulse pressure between the two groups (P > 0.05 for all).
Table 1.
General characteristics and serum lipid levels between non-drinkers and drinkers.
| Parameter | Non-drinker (n = 785) |
Drinker (n = 567) |
t (χ2) | P |
|---|---|---|---|---|
| Male/female | 212/573 | 456/111 | 375.798 | 0.000 |
| Age (years) | 47.36 ± 14.34 | 45.13 ± 13.35 | 2.904 | 0.004 |
| Height (cm) | 152.65 ± 8.59 | 158.01 ± 7.65 | -11.847 | 0.000 |
| Weight (kg) | 54.21 ± 10.85 | 57.45 ± 8.77 | -5.861 | 0.000 |
| Body mass index (kg/m2) | 23.23 ± 4.19 | 22.95 ± 2.55 | 1.413 | 0.158 |
| Systolic blood pressure (mmHg) | 131.28 ± 20.21 | 130.56 ± 16.27 | 0.700 | 0.484 |
| Diastolic blood pressure (mmHg) | 89.32 ± 11.76 | 82.89 ± 11.23 | 10.109 | 0.000 |
| Pulse pressure (mmHg) | 59.96 ± 16.03 | 47.67 ± 14.89 | 14.329 | 0.220 |
| Cigarette smoking [n (%)] | ||||
| Non-smoker | 685 (87.2) | 250 (44.1) | ||
| < 20 cigarettes/day | 17 (2.2) | 148 (26.1) | ||
| ≥ 20 cigarettes/day | 83 (10.6) | 169 (29.8) | 308.608 | 0.000 |
| Alcohol consumption [n (%)] | ||||
| Non-drinker | 785 (100.0) | - | ||
| < 25 g/day | - | 372 (65.6) | ||
| ≥ 25 g/day | - | 195 (34.4) | ||
| Total cholesterol (mmol/L) | 4.82 ± 1.05 | 4.61 ± 1.01 | 3.687 | 0.000 |
| Triglyceride (mmol/L) | 1.02 (0.72) | 1.23 (0.91) | -5.226 | 0.000 |
| HDL-C (mmol/L) | 1.73 ± 0.44 | 1.82 ± 0.49 | -3.538 | 0.000 |
| LDL-C (mmol/L) | 2.92 ± 0.80 | 2.55 ± 0.82 | 8.304 | 0.000 |
| Apolipoprotein (Apo) A1 (g/L) | 1.35 ± 0.29 | 1.52 ± 0.42 | -8.802 | 0.000 |
| ApoB (g/L) | 0.92 ± 0.22 | 0.85 ± 0.25 | 5.450 | 0.000 |
| ApoA1/ApoB | 1.55 ± 0.49 | 1.66 ± 0.92 | -2.839 | 0.005 |
HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol. Data are expressed as percentage or mean ± SD [the values of triglyceride were presented as median (interquartile range)].
Serum lipid levels between non-drinkers and drinkers
Serum lipid levels between non-drinkers and drinkers are also shown in Table 1. The levels of TG, HDL-C, ApoA1, and the ratio of ApoA1 to ApoB were higher in drinkers than in non-drinkers (P < 0.01 for all), whereas the levels of TC, LDL-C and ApoB were lower in drinkers than in non-drinkers (P < 0.001 for all).
Results of electrophoresis and genotyping
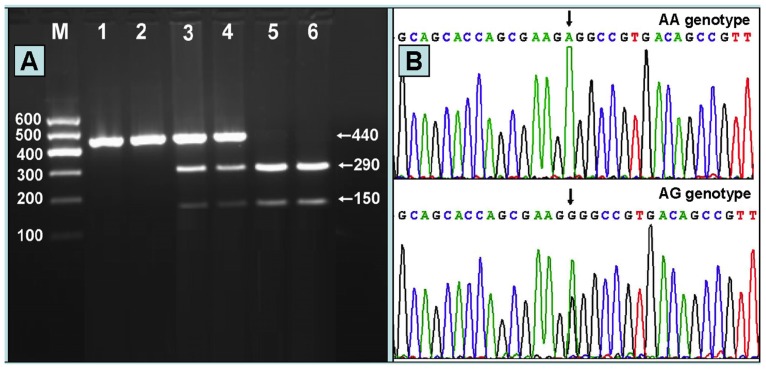
After the genomic DNA of the samples was amplified by PCR and imaged by 2% agarose gel electrophoresis, the PCR product of 440 bp nucleotide sequences could be seen in the samples (Figure 1A). The genotypes identified were named according to the presence or absence of the enzyme restriction sites, when an A to G transversion at 670 locus of the PCSK9 gene. The presence of the cutting site indicates the A allele, while its absence indicates the G allele (cannot be cut). Thus, AG genotype is heterozygote for the absence and presence of the site (bands at 440-, 290- and 150-bp), and AA genotype is homozygote for the presence of the site (bands at 290- and 150- bp; Figure 1A). The GG genotype was not detected in our study population. The genotypes detected by PCR-RFLP were also confirmed by sequencing; respectively (Figure 1B).
Figure 1.
(A) Genotyping of the PCSK9 E670G SNP. Lane M, 100 bp marker ladder; lanes 1 and 2, PCR products of the samples (440 bp); lanes 3 and 4, AG genotype (440-, 290- and 150-bp); and lanes 5 and 6, AA genotype (290- and 150-bp). The GG genotype was not detected in our study population. (B) A part of the nucleotide sequence of the PCSK9 E670G SNP. AA and AG genotypes.
Genotypic and allelic frequencies between non-drinkers and drinkers
The genotypic and allelic frequencies of PCSK9 E670G in non-drinkers and drinkers are shown in Table 2. The distribution of genotypes followed the Hardy-Weinberg equilibrium. The frequencies of A and G alleles were 97.4% and 2.6% in non-drinkers, and 97.9% and 2.1% in drinkers (P > 0.05); respectively. The frequencies of AA and AG genotypes were 94.8% and 5.2% in non-drinkers, and 95.8% and 4.2% in drinkers (P > 0.05); respectively. There was no significant difference in the genotypic and allelic frequencies between males and females, or between < 25 and ≥ 25 g/day drinkers.
Table 2.
Genotypic and allelic frequencies of the PCSK9 E670G SNP between non-drinkers and drinkers [n (%)].
| Group | n | Genotype | Allele | |||
|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||
| Non-drinker | 785 | 744 (94.8) | 41 (5.2) | 0 (0.0) | 1529 (97.4) | 41 (2.6) |
| Drinker | 567 | 543 (95.8) | 24 (4.2) | 0 (0.0) | 1110 (97.9) | 24 (2.1) |
| χ2 | - | 0.935 | 0.037 | |||
| P | - | 0.627 | 0.847 | |||
| Non-drinker | ||||||
| Male | 212 | 199 (93.9) | 13 (6.1) | 0 (0.0) | 411 (96.9) | 13 (3.1) |
| Female | 573 | 545 (95.1) | 28 (4.9) | 0 (0.0) | 1118 (97.6) | 28 (2.4) |
| χ2 | - | 0.613 | 0.000 | |||
| P | - | 0.736 | 0.994 | |||
| Drinker | ||||||
| Male | 456 | 433 (95.0) | 23 (5.0) | 0 (0.0) | 889 (97.5) | 23 (2.5) |
| Female | 111 | 110 (99.1) | 1 (0.9) | 0 (0.0) | 221 (99.5) | 1 (0.5) |
| χ2 | - | 5.857 | 2.791 | |||
| P | - | 0.053 | 0.095 | |||
| < 25 g/day | 372 | 355 (95.4) | 17 (4.6) | 0 (0.0) | 727 (97.7) | 17 (2.3) |
| ≥ 25 g/day | 195 | 188 (96.4) | 7 (3.6) | 0 (0.0) | 383 (98.2) | 7 (1.8) |
| χ2 | - | 4.733 | 3.235 | |||
| P | - | 0.094 | 0.072 | |||
Genotypes and serum lipid levels
The levels of LDL-C in non-drinkers were different between the two genotypes (P < 0.05), the subjects with AA genotype had higher LDL-C levels than the subjects with AG genotype. The levels of TC in drinkers were different between the two genotypes (P < 0.05), the subjects with AG genotype had higher TC levels than the subjects with AA genotypes. There was no significant difference in the levels of TG, HDL-C, ApoA1, ApoB, and the ratio of ApoA1 to ApoB between AA and AG genotypes in the both groups (P > 0.05 for all).
Interactions between genotypes and alcohol consumption on serum lipid parameters
The interactions between genotypes of the PCSK9 E670G and alcohol consumption on serum TC and LDL-C levels are also shown in Table 3. The subjects with AA genotype had lower serum TC and LDL-C levels in drinkers than in non-drinkers. There were no interactions between PCSK9 E670G genotypes and alcohol consumption on the levels of TG, HDL-C, ApoA1 and ApoB, and the ratio of ApoA1 to ApoB (P > 0.05 for all).
Table 3.
Genotypes of the PCSK9 E670G SNP and serum lipid levels between non-drinkers and drinkers and the interactions between genotypes and alcohol consumption on serum lipid parameters.
| Group | Genotype | n | TC (mmol/L) |
TG (mmol/L) |
HDL-C (mmol/L) |
LDL-C (mmol/L) |
ApoA1 (g/L) |
ApoB (g/L) |
ApoA1/ApoB |
|---|---|---|---|---|---|---|---|---|---|
| Non-drinker | AA | 744 | 4.84±1.07 | 1.02(0.73) | 1.74±0.44 | 2.94±0.81 | 1.35±0.29 | 0.92±0.22 | 1.55±0.49 |
| AG | 41 | 4.55±0.64 | 1.10(0.58) | 1.63±0.50 | 2.69±0.44 | 1.28±0.31 | 0.89±0.21 | 1.55±0.56 | |
| F | - | - | 3.849 | 0.081 | 2.523 | 4.547 | 2.521 | 1.090 | 0.008 |
| P | - | - | 0.050 | 0.935 | 0.113 | 0.033 | 0.113 | 0.297 | 0.928 |
| Drinker | AA | 543 | 4.59±0.99↓ | 1.23(0.92) | 1.82±0.49 | 2.55±0.82↓ | 1.48±0.32 | 0.85±0.25 | 1.96±1.10 |
| AG | 24 | 5.01±1.25 | 1.09(0.78) | 1.86±0.51 | 2.68±0.86 | 1.44±0.28 | 0.89±0.32 | 2.06±1.56 | |
| F | - | - | 4.415 | 0.212 | 0.110 | 0.692 | 0.353 | 0.634 | 0.240 |
| P | - | - | 0.036 | 0.832 | 0.741 | 0.406 | 0.553 | 0.426 | 0.625 |
| F1 | - | - | 4.518 | 2.839 | 1.908 | 4.644 | 0.249 | 0.256 | 1.002 |
| P1 | - | - | 0.032 | 0.092 | 0.167 | 0.029 | 0.618 | 0.613 | 0.317 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B. All data are expressed as mean ± SD [the values of TG are presented as median (interquartile range)]. The values of F1 and P1 of the drinkers are the interactions between the PCSK9 E670G genotypes and alcohol consumption on serum lipid parameters. ↓, genotype and alcohol consumption interactions to decrease serum lipid levels.
Correlation between genotypes and serum lipid parameters
Multiple linear regression analysis showed that the levels of TC and LDL-C in non-drinkers were negatively correlated with the genotypes of PCSK9 E670G (P < 0.05 for each), whereas the levels of TC and TG in drinkers were positively associated with the genotypes of PCSK9 E670G (P < 0.05 for each; Table 4).
Table 4.
Correlation of serum lipid parameters and genotypes between non-drinkers and drinkers.
| Lipid | Factor | Unstandardized coefficient | Standard error | Standardized coefficient | t | P |
|---|---|---|---|---|---|---|
| Non-drinker | ||||||
| TC | Genotype | -0.297 | 0.148 | -0.063 | -2.009 | 0.045 |
| LDL-C | Genotype | -0.260 | 0.116 | -0.072 | -2.250 | 0.025 |
| Drinker | ||||||
| TC | Genotype | 0.406 | 0.196 | 0.082 | 2.076 | 0.038 |
| TG | Genotype | 0.607 | 0.303 | 0.082 | 2.003 | 0.046 |
TC, total cholesterol; TG, triglyceride; LDL-C, low-density lipoprotein cholesterol.
Discussion
The results of the present study show that the levels of TG, HDL-C, ApoA1, and the ratio of ApoA1 to ApoB were higher in drinkers than in non-drinkers, whereas the levels of TC, LDL-C and ApoB were lower in drinkers than in non-drinkers. The subjects with AA genotype of PCSK9 E670G had lower serum TC and LDL-C levels in drinkers than in non-drinkers. The interactions between PCSK9 E670G genotypes and alcohol consumption on serum TC and LDL-C levels were also detected by using a factorial design covariance analysis after controlling for potential confounders. These findings suggest that the effect of alcohol consumption on serum TC and LDL-C levels may depend on the PCSK9 E670G genotypes. To the best of our knowledge, the interaction between PCSK9 E670G SNP and alcohol consumption on serum lipid levels has not been previously explored.
In the present study, we showed that the frequencies of G allele and AG genotype of PCSK9 E670G SNP was 2.6% and 5.2% in non-drinkers, and 2.1% and 4.2% in drinkers (P > 0.05 for each); respectively. There was no significant difference in the genotypic and allelic frequencies between males and females in both groups. Several previous studies showed that the minor-allele frequency (670G) was rare in whites but presented in approximately 24.8% of blacks 35,44. The frequency of the G allele in patients selected from Universitätsklinikum Hamburg-Eppendorf Martinistrasse, Hamburg, Germany was 5% 37 which lies between that observed in the TexGen population, 4.4% and that reported for the Lipoprotein Coronary Atherosclerosis Study (LCAS), 7.4% by Chen et al. 36 in their original study. There was no statistical significant difference in the frequency of the G allele in patients with LDL-C below the 50th percentile for age and sex, those with LDL-C between the 50th and 95th percentiles, and those with LDL-C above the 95th percentile 37. The 670G carrier in Chinese Taiwanese was identified less frequently in patients with CAD than in controls (9.9% vs. 11.9%), but the difference was not significant in a multivariable logistic regression analysis 39. These results indicate that the prevalence of the G allele variation of PCSK9 E670G may have a racial/ethnic specificity.
The association of PCSK9 E670G SNP and serum lipid levels is still controversial. Chen et al. 36 showed that PCSK9 E670G was an important determinant of plasma LDL-C levels, accounting for 3.5% of its variability (F = 14.6, P < 0.001). Accordingly, LDL-C levels were higher in those with the GG, intermediate in those with the AG, and lowest in those with the AA genotypes. Plasma TC, ApoB, and lipoprotein (a) levels were also associated with the PCSK9 E670G SNP. Evans and Beil 37 found that the PCSK9 E670G SNP in a European population was associated with increased LDL-C in men but not in women. Norata et al. 38 also showed that the 670G carriers were associated with increased plasma TC, LDL-C, and ApoB levels in the general population. Contradictory to these previous results, Hsu et al. 39 showed a significantly lower level of LDL-C in 670G carriers in Chinese Taiwanese than in non-carriers among 614 unrelated controls. However, the association between the PCSK9 E670G SNP and plasma LDL-C levels or CAD risk was not confirmed in other studies 35, 42-44. In the present study, we showed that the levels of LDL-C in non-drinkers were different between the two genotypes, the subjects with AA genotype had higher LDL-C levels than the subjects with AG genotype. The levels of TC in drinkers were different between the two genotypes, the subjects with AG genotype had higher TC levels than the subjects with AA genotypes. Multiple linear regression analysis showed that the levels of serum TC and LDL-C in non-drinkers were negatively correlated with the genotypes of PCSK9 E670G, whereas the levels of TC and TG in drinkers were positively associated with the genotypes of PCSK9 E670G. The reason for this discrepancy is most likely due to different gene-environmental interactions.
There are several potential limitations in the current study. First, we did not match drinkers and non-drinkers in terms of sex; therefore, the drinkers had a much higher proportion of males than did the non-drinkers. Second, the sample size of drinkers is a little small. In addition, the subject with GG genotype is not detected in both drinkers and non-drinkers. Third, the levels of body height, weight, and the percentages of subjects who smoked cigarettes were higher in drinkers than in non-drinkers, whereas the levels of mean age and diastolic blood pressure were lower in drinkers than in non-drinkers. Although sex, age, BMI, hypertension, and cigarette smoking have been adjusted for the statistical analysis, we could not completely eliminate the potential effects of these factors on serum lipid levels in both groups. Fourth, the use of questionnaire may not be a reliable measure for alcohol intake in our population because 90% of the wine drunk by the drinkers was corn wine, rice wine, and rum, in which the alcohol content is very low. Fifth, it is well known that serum lipid levels are modulated by multiple environmental and genetic factors, and their interactions. Although we have detected the interactions of PCSK9 E670G SNP and alcohol consumption on serum lipid levels in this study, there are still many unmeasured environmental and genetic factors and their interactions. Thus, the interactions of environment-environment, environment-gene, and gene-gene on serum lipid levels remain to be determined. Finally, the number of 670G allele carriers in our study populations is rather small, the findings in our current study should be confirmed in the other populations.
Conclusion
The present study shows that there were no significant differences in the genotypic and allelic frequencies of PCSK9 E670G SNP between non-drinkers and drinkers. The subjects with AA genotype in non-drinkers had higher serum LDL-C levels than the subjects with AG genotype. The individuals with AG genotype in drinkers had higher TC levels than the individuals with AA genotypes. The effects of alcohol consumption on TC and LDL-C levels depended upon genotypes, the subjects with AA genotype had lower serum TC and LDL-C levels in drinkers than in non-drinkers. These results suggest that the subjects with AA genotype of the PCSK9 E670G benefit more from alcohol consumption than the subjects with AG genotype in decreasing serum TC and LDL-C levels.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No: 30660061).
References
- 1.Doll R, Peto R, Hall E. et al. Mortality in relation to consumption of alcohol. BMJ. 1994;309:911–8. doi: 10.1136/bmj.309.6959.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camargo CA Jr, Hennekens CH, Gaziano JM. et al. Prospective study of moderate alcohol consumption and mortality in US male physicians. Arch Intern Med. 1997;157:79–85. [PubMed] [Google Scholar]
- 3.Grønbaek M, Deis A, Becker U. et al. Alcohol and mortality: is there a U-shaped relation in elderly people? Age Ageing. 1998;27:739–44. doi: 10.1093/ageing/27.6.739. [DOI] [PubMed] [Google Scholar]
- 4.Kiechl S, Willeit J, Rungger G. et al. Alcohol consumption and atherosclerosis: what is the relation? Prospective results from the Bruneck Study. Stroke. 1998;29:900–7. doi: 10.1161/01.str.29.5.900. [DOI] [PubMed] [Google Scholar]
- 5.McElduff P, Dobson AJ. How much alcohol and how often? Population based case-control study of alcohol consumption and risk of a major coronary event. BMJ. 1997;314:1159–64. doi: 10.1136/bmj.314.7088.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Criqui MH, Cowan LD, Tyroler HA. et al. Lipoproteins as mediators for the effects of alcohol consumption and cigarette smoking on cardiovascular mortality: results from the Lipid Research Clinics Follow-up Study. Am J Epidemiol. 1987;126:629–37. doi: 10.1093/oxfordjournals.aje.a114702. [DOI] [PubMed] [Google Scholar]
- 7.Choudhury SR, Ueshima H, Kita Y. et al. Alcohol intake and serum lipids in a Japanese population. Int J Epidemiol. 1994;23:940–7. doi: 10.1093/ije/23.5.940. [DOI] [PubMed] [Google Scholar]
- 8.Rimm EB, Williams P, Fosher K. et al. Moderate alcohol intake and lower risk of coronary heart disease: Meta-analysis of effects on lipids and haemostatic factors. BMJ. 1999;319:1523–8. doi: 10.1136/bmj.319.7224.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Oliveira E Silva ER, Foster D, McGee Harper M. et al. Alcohol consumption raises HDL cholesterol levels by increasing the transport rate of apolipoproteins A-I and A-II. Circulation. 2000;102:2347–52. doi: 10.1161/01.cir.102.19.2347. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal DP. Cardioprotective effects of light-moderate consumption of alcohol: a review of putative mechanisms. Alcohol Alcohol. 2002;37:409–15. doi: 10.1093/alcalc/37.5.409. [DOI] [PubMed] [Google Scholar]
- 11.Stampfer MJ, Kraussm RM, Ma J. et al. A prospective study of trigliceryde level, low-density lipoprotein particle diameter, and risk of myocardial infarction. JAMA. 1996;276:882–8. [PubMed] [Google Scholar]
- 12.Brinton EA. Effects of ethanol intake on lipoproteins and atherosclerosis. Curr Opin Lipidol. 2010;21:346–51. doi: 10.1097/MOL.0b013e32833c1f41. [DOI] [PubMed] [Google Scholar]
- 13.Perissinotto E, Buja A, Maggi S. et al. Alcohol consumption and cardiovascular risk factors in older lifelong wine drinkers: the Italian Longitudinal Study on Aging. Nutr Metab Cardiovasc Dis. 2010;20:647–55. doi: 10.1016/j.numecd.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Onat A, Hergenc G, Dursunoglu D. et al. Associations of alcohol consumption with blood pressure, lipoproteins, and subclinical inflammation among Turks. Alcohol. 2008;42:593–601. doi: 10.1016/j.alcohol.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Savolainen MJ, Kesaniemi YA. Effects of alcohol lipoproteins in relation to coronary heart disease. Curr Opin Lipidol. 1995;6:243–50. doi: 10.1097/00041433-199508000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell KN, Breslow JL. Adenoviral-mediated expression of PCSK9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA. 2004;101:7100–5. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjannet S, Rhainds D, Essalmani R. et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279:48865–75. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 18.Park SW, Moon YA, Horton JD. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J Biol Chem. 2004;279:50630–8. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 19.Lalanne F, Lambert G, Amar MJ. et al. Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells. J Lipid Res. 2005;46:1312–9. doi: 10.1194/jlr.M400396-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Ouguerram K, Chetiveaux M, Zair Y. et al. Apolipoprotein B100 metabolism in autosomal-dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler Thromb Vasc Biol. 2004;24:1448–53. doi: 10.1161/01.ATV.0000133684.77013.88. [DOI] [PubMed] [Google Scholar]
- 21.Sun XM, Eden ER, Tosi I. et al. Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet. 2005;14:1161–9. doi: 10.1093/hmg/ddi128. [DOI] [PubMed] [Google Scholar]
- 22.Lambert G, Jarnoux AL, Pineau T. et al. Fasting induces hyperlipidemia in mice overexpressing proprotein convertase subtilisin kexin type 9: lack of modulation of very-low-density lipoprotein hepatic output by the low-density lipoprotein receptor. Endocrinology. 2006;147:4985–95. doi: 10.1210/en.2006-0098. [DOI] [PubMed] [Google Scholar]
- 23.Zhou A, Webb G, Zhu X. et al. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274:20745–8. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell KN, Breslow JL. Proprotein convertase subtilisin kexin 9: the third locus implicated in autosomal dominant hypercholesterolemia. Curr Opin Lipidol. 2005;16:167–72. doi: 10.1097/01.mol.0000162321.31925.a3. [DOI] [PubMed] [Google Scholar]
- 25.Poirier S, Mayer G, Benjannet S. et al. The proprotein convertase PCSK9 induces the degradation of LDLR and its closest family members VLDLR and APOER2. J Biol Chem. 2008;283:2363–72. doi: 10.1074/jbc.M708098200. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell KN, Fisher EA, Breslow JL. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc Natl Acad Sci USA. 2005;102:2069–74. doi: 10.1073/pnas.0409736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang DW, Lagace TA, Garuti R. et al. Binding of PCSK9 to EGFA repeat of LDL receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–12. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham D, Danley DE, Geoghegan KF. et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14:413–9. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- 29.Seidah NG, Benjannet S, Wickham L. et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci USA. 2003;100:928–33. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abifadel M, Varret M, Rabès JP. et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–6. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 31.Timms KM, Wagner S, Samuels ME. et al. A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum Genet. 2004;114:349–53. doi: 10.1007/s00439-003-1071-9. [DOI] [PubMed] [Google Scholar]
- 32.Leren TP. Mutations in the PCSK9 gene in Norwegian subjects with autosomal dominant hypercholesterolemia. Clin Genet. 2004;65:419–22. doi: 10.1111/j.0009-9163.2004.0238.x. [DOI] [PubMed] [Google Scholar]
- 33.Cohen JC, Boerwinkle E, Mosley TH Jr. et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 34.Cohen J, Pertsemlidis A, Kotowski IK. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–5. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 35.Kotowski IK, Pertsemlidis A, Luke A. et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–22. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen SN, Ballantyne CM, Gotto AM Jr. et al. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am Coll Cardiol. 2005;45:1611–9. doi: 10.1016/j.jacc.2005.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans D, Beil FU. The E670G SNP in the PCSK9 gene is associated with polygenic hypercholesterolemia in men but not in women. BMC Med Genet. 2006;7:66. doi: 10.1186/1471-2350-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norata GD, Garlaschelli K, Grigore L. et al. Effects of PCSK9 variants on common carotid artery intima media thickness and relation to ApoE alleles. Atherosclerosis. 2010;208:177–82. doi: 10.1016/j.atherosclerosis.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Hsu LA, Teng MS, Ko YL. et al. The PCSK9 gene E670G polymorphism affects low-density lipoprotein cholesterol levels but is not a risk factor for coronary artery disease in ethnic Chinese in Taiwan. Clin Chem Lab Med. 2009;47:154–8. doi: 10.1515/CCLM.2009.032. [DOI] [PubMed] [Google Scholar]
- 40.Ding K, Kullo IJ. Molecular population genetics of PCSK9: a signature of recent positive selection. Pharmacogenet Genomics. 2008;18:169–79. doi: 10.1097/FPC.0b013e3282f44d99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aung LH, Yin RX, Miao L. et al. The proprotein convertase subtilisin/kexin type 9 gene E670G polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2011;10:5. doi: 10.1186/1476-511X-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scartezini M, Hubbart C, Whittall RA. et al. The PCSK9 gene R46L variant is associated with lower plasma lipid levels and cardiovascular risk in healthy U.K. men. Clin Sci (Lond) 2007;113:435–41. doi: 10.1042/CS20070150. [DOI] [PubMed] [Google Scholar]
- 43.Polisecki E, Peter I, Robertson M. et al. Genetic variation at the PCSK9 locus moderately lowers low-density lipoprotein cholesterol levels, but does not significantly lower vascular disease risk in an elderly population. Atherosclerosis. 2008;200:95–101. doi: 10.1016/j.atherosclerosis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang CC, Fornage M, Lloyd-Jones DM, Wei GS. et al. Longitudinal association of PCSK9 sequence variations with low-density lipoprotein cholesterol levels: the Coronary Artery Risk Development in Young Adults Study. Circ Cardiovasc Genet. 2009;2:354–61. doi: 10.1161/CIRCGENETICS.108.828467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruixing Y, Qiming F, Dezhai Y. et al. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 2007;48:2673–81. doi: 10.1194/jlr.M700335-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Ruixing Y, Dezhai Y, Shuquan L. et al. Hyperlipidaemia and its risk factors in the Guangxi Bai Ku Yao and Han populations. Public Health Nutr. 2009;12:816–24. doi: 10.1017/S1368980008003273. [DOI] [PubMed] [Google Scholar]