Abstract
Background: Anorectal malformations (ARMs) represent a variety of congenital disorders that involve abnormal termination of the anorectum. This study was to reveal relation between CDX1 and human ARMs phenotypes.
Methods: 108 Chinese patients and 120 Chinese controls were included in this study. We analyzed the relation between two by PCR, qRT-PCR, western blot and immunofluorescence.
Results: Four heterozygous mutations in CDX1 gene were identified in ARMs patients (3.7%, 4/108), no found in controls. CDX1 protein expression was significantly decreased in the ARMs compared with the control anorectum. All samples analyzed in ARMs group exhibited down-regulated CDX1 mRNA expression in comparison to matched normal group, demonstrated significant differences statistically.
Conclusion: The findings represented the relation between CDX1 mutations and CDX1 genotype. Furthermore, it was suggested that the downregulation of CDX1 might be related to the development of ARMs.
Keywords: Anorectal malformations, CDX1, mutation, children
Introduction
Anorectal malformations (ARMs) represents a variety of congenital disorders which involve abnormal termination of anorectum. The clinical spectrums of these anomalies range from anal stenosis to persistent cloaca. The overall incidence is approximately 1 in 5000 live births1. Besides non-syndromic forms, patients with ARMs often display syndromic features encompassing other congenital defects. Up to now, the etiology of ARMs remains unknown, although several evidences indicated that genetic component involved in the pathogenesis2. Recent studies in animal models with ARMs have greatly enhanced our knowledge in this multifaceted developmental process3,4, which has provided clues to genes and pathways that may be operating in human diseases and in most cases, mutations in the human orthologs give rise to similar or related phenotypes. During the essential period of hindgut development, considerable signaling molecules participated in the embryogenesis of the anorectum, including Wnt5a5,6, Tcf44, Sonic hedgehog (Shh)7,8, Fibroblast growth factor 10(Fgf10)9, Hoxa-13 and Hoxd-1310. Recently, Wong et al.11 reported that gene DKK4 may participate in the development of ARMs. Draaken et al.12 revealed that seven gene including WNT3A, WNT5A, WNT11, DACT1, FGF10, FGFR2 and the T gene mutations may nonetheless be a cause of ARMs in rare cases. Therefore, DNA alterations in any of the genes governing the development of the anorectum may result in the ARMs phenotype.
As ARMs is a multigenerational complex disease, ARMs may be caused by mutations in a variety of genes. At present, it is poorly understood and debatable whether there is a common molecular basis for ARMs2. Several genes, expressed during the stages of gastrulation, have helped in the elucidation of the molecular basis of this event. Many of them bear a common conserved sequence motif and homeobox. The homeobox gene recently isolated from Drosophila, mouse and human shown a very restricted expression pattern at the onset of gastrulation, suggesting an involvement in the formation of the digestive tract13,14. In previous studies, it was shown that CDX1, a member of the caudal-type homeobox gene family, is a transcription factor which is involved in establishing and maintaining differentiated patterns in embryo tissues and are required for anterior-posterior regional identity15. CDX1 might carry out a function in intestinal epithelium because its expression is specific to tissue and increases during development. In mutant animal model, it was found that homozygous CDX1 and heterozygous CDX2 knockout mice frequently develop dysplastic lesions, metaplasia and homeotic transformation of endoderm to a rostral phenotype in the colon16. It is suggested that CDX1 gene may be involved in the regulation of the proliferation and differentiation of the anorectal region development. Furthermore, the dynamic expression pattern of CDX1 along the developing has been proposed that CDX1 is involved in both the caudal region of the embryo and AP patterning17,18. On the other hand, it was reported that downregulation of CDX1 at the time of cloacal separation into rectum and urethra might be related to the development of ARMs19. Indeed, evidences from these studies have strengthened our observation that CDX1 plays an important role in human anorectal development and the pathogenesis of ARMs.
Only limited data concerning lineage specific expression of CDX1 in ARMs patients and normal children (controls) in Chinese Northeast area are available, hence, a large cohort of ARMs is necessary to further demonstrate CDX1 mutations. And more detailed understanding will potentially clarify the role of CDX1 in the etiology of ARMs. In view of the incomplete knowledge about the spectrum of ARMs relating to CDX1 mutations, we screened CDX1 mutations in a cohort of 108 Chinese ARMs cases. It is demonstrated that mutations and down regulation in CDX1 are associated with human ARMs phenotypes, suggesting a role for this newly identified pathway in human development and disease.
Methods
Patients and control samples
Total 108 Chinese patients from Northeast District and 120 Chinese controls were initially included into this study after having obtained informed consent from their parents or guiders.
Detection of CDX1 genotype
The CDX1 PCR primer sequences are in Table 1. PCR procedure as follow: denaturing at 95°C for 5 min, then 35 cycles of denaturing at 95°C for 30 s, annealing at 52-63°C for 30s and elongation at 72°C for 5 min, and finally incubation for 10 min at 72°C. The PCR products were purified by using an ABI PRISM Big Dye assay (Applied Biosystems, Foster City, CA). Direct sequencing was used to screen for DNA variants in coding sequence. Sequencing was performed using an ABI PRISM Big Dye Terminator v3.0 Cycle sequencing kit and an ABI3100 automated sequencer (Applied Biosystems, Foster City, CA). When a DNA alteration was found, PCR amplification and sequencing were repeated from the stock DNA or newly extracted DNA from blood of the patient bearing the DNA alteration.
Table 1.
Primers for the amplification of CDX1 exons.
| Exon | primer | Amplicon (bp) |
|---|---|---|
| 1 | F: 5'GTTCAGGTGAGCGGTTGC 3' | 552bp |
| R: 5' AGGGCGAGGGAAGGGT 3' | ||
| 2 | F: 5' AACCTGGAGTGTATGTCT 3' | 235bp |
| R: 5' GATGTAACGGCTGTAAT 3' | ||
| 3 | F: 5' GGGATAACAAGGAAGGC 3' | 1393bp |
| R: 5' AACGGGAAAGGAATACAC 3' |
Quantitative real-time RT-PCR
For real-time PCR amplification, total RNA of 0.05g anorectum were extracted using Trizol kit (Invitrogen, Carlsbad, CA). Single strand cDNA was prepared with SYBR PrimeR Script TM RT-PCR Kit (Takara, Dalian, China) as per manufacturers' instructions. The primers used for Real-time PCR were as follows: Forward Primer: 5'-CCG AAG ACG GGC AAG GAG T-3'; Reverse Primer: 5' -CCA GGA AAG ATG AGG AGG TG-3'. Relative quantitation by real-time PCR was carried out on LightCycler 2.0 instrument (Roche Diagnostics). The amplification protocol consisted of initial denaturation at 95°C for 10 s, 45 cycles of 5 s at 95°C for denaturation, 20 s at 60°C for annealing, the melting step was performed at 95°C for 10 s, 65°C for 15 s and 95°C for 10 s. A negative control was included in each run to access specificity of primers and possible contamination. A negative control was included in each run to access specificity of primers and possible contamination. Threshold cycle (Ct) was obtained by LightCycler software package. Quantification was done using the comparative Ct method, i.e. expression levels for the target genes were normalized to β-actin of each sample {2-△Ct=2-[Ct (target gene)-Ct (β-actin)]}. To ensure accuracy of the results all reactions were performed in triplicate and arithmetic means were represented for each sample.
Western blot analysis
Protein extract (50μg) was denatured, separated by SDS/PAGE, and transferred to PVDF membranes (Millipore, Billerica, MA, USA), blocked with 5% fat-free milk in Tris-buffered saline (1h, room temperature) and incubated overnight at 4°C in primary antibodies against CDX1(1:1000), TBP (1:1000). After washing, the membranes were incubated in secondary antibodies at room temperature for 1h. The membranes were washed and developed using a chemiluminescent substrate kit (SuperSignal West Pico, Pierce, Rockford, IL). For Western blot analysis, densitometric values were analyzed using the ECL Plus Western blot detection system.
Immunofluorescence
For immunocytochemical analysis, 4% paraformaldehyde-fixed tissue of anorectum was incubated at 4°C for 24 hr with mouse monoclonal antibodies to CDX1(1:200), rabbit polyclonal antibodies to Tcf4 (1:200). Alexa Fluor 488-labeled goat anti-mouse IgG and TRITC-conjugated goat anti-rabbit IgG were used as secondary antibodies.
Statistical analysis
All data were presented as mean ± SD. Significance of differences between all groups was evaluated by two-sample t test. Statistical analyses were performed using SPSS Software (V13.0, SPSS, USA), P values < 0.05 was considered to be statistically significant.
Results
CDX1 mutations in ARMs
Direct sequencing was performed on covering the coding regions of CDX1 gene of 108 patients with ARMs. Four heterozygous mutations in CDX1 gene were identified in ARMs patients (3.7%, 4/108), including 1 of 65 patients with type of perineal fistula (1.5%), 1 of 32 patients with vestibular fistula (3.1%), and 1 of 10 patients with rectourethral fistula (10%), and 1 in anal stenosis, who didn't have any other congenital afflictions. Likewise, three of the mutations were found in the homeodomain, of which two (K199N, Fig 1-A and R206S, Fig 1-B) were located in the DNA binding region, one (Q203H, Fig 1-C) did in the specific DNA base contact region. The other mutation (97-98insE, Fig 1-D) was located in the exon1. All the mutations have not been reported previously. (Table 2) However, the four types of mutations in CDX1 gene were not detected in 120 normal control DNA samples.
Figure 1.

CDX1 exons mutation in ARMs patients. Sequence electropherograms documenting the c.213-214Ins GAA mutation (arrow) found in exon 1 of CDX1 gene, the c.6G>C, c.27G>T, c.18A>C mutation (*) found in exon 3 of CDX1 gene.
Table 2.
Mutations in CDX1 gene identified in 108 ARMs.
| Exon | Coding change | Amino acid change | Associated phenotype | Number of individual |
|---|---|---|---|---|
| 1 | c.213-214Ins GAA | 96-98Ins E | anal stenosis | 1 |
| 3 | c.6G>C | K199N | perineal fistula | 1/65 |
| 3 | c.27G>T | R206S | vestibular fistula | 1/32 |
| 3 | c.18A>C | Q203H | rectourethral fistula | 1/10 |
Expression of CDX1 mRNA and protein
Tissue samples from 45 patients were included into the analysis.The16 patients exhibited high-type ARMs and other appeared low-type ARMs. The mean age of the patients was 1.5 months. As a control group, tissues from 12 patients without ARMs were obtained, including 6 patients who died of neonatal asphyxia, 4 died of intracranial hemorrhage and 2 died of septicaemia. The mean age of the control group was 1 month.
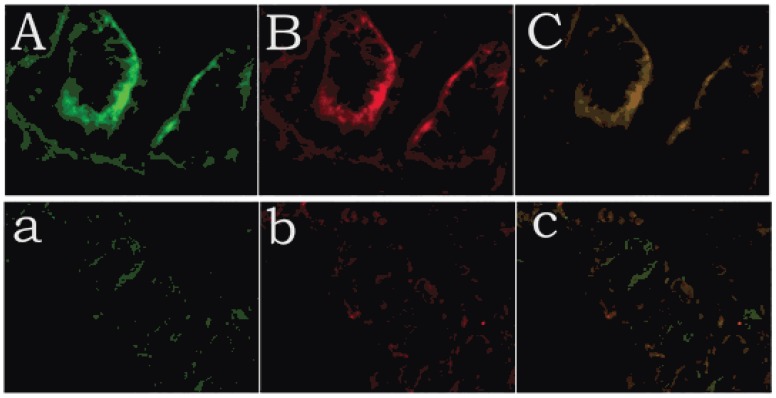
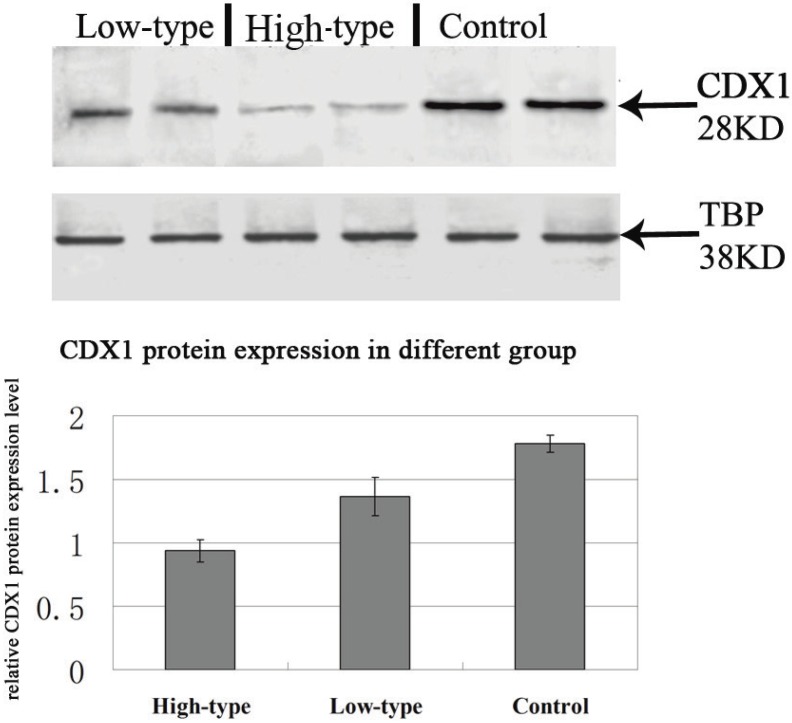
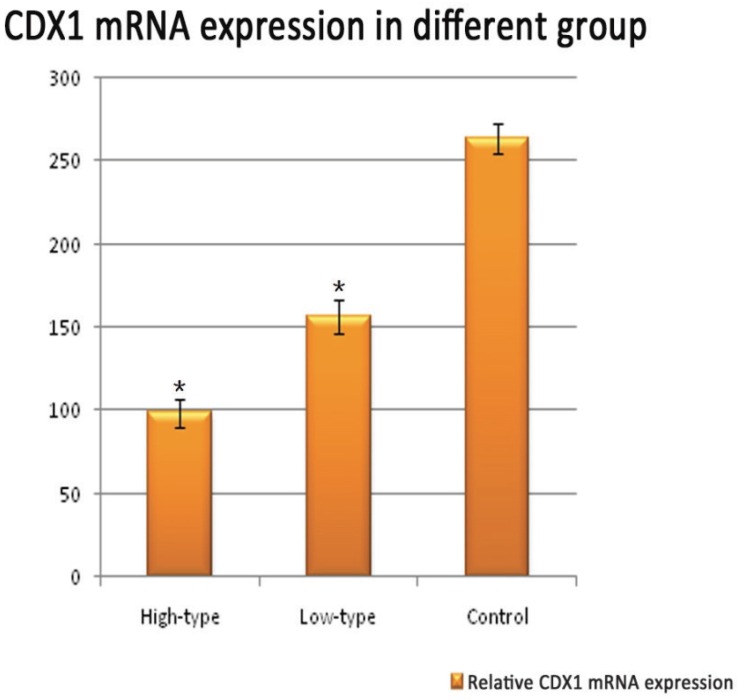
Positive immunolabelling was detected as green for CDX1 protein in the epithelium of the rectum and the intestine in the control. Expression of CDX1 was weaker in the high-type ARMs patients than that in the control group. (Fig 2) Western blot analysis specific for CDX1 was done to quantify the protein expression in the development of cloaca/hindgut. CDX1 was detected as an approximately 28-kD band on western blots of protein extracted from both the normal and ARMs tissue. Each protein band was normalized by a corresponding TBP band (Fig 3). CDX1 protein expression was significantly decreased in the ARMs patients compared with the control group (0.936±0.085 vs. 1.782±0.067, 1.365±0.15 vs. 1.782±0.067, P<0.05) (Table 3). The CDX1 mRNA and protein were found in ARMs and control group. All samples analysis in ARMs group exhibited significantly down-regulated CDX1 mRNA expression in comparison to matched normal group (98.67±8.3 vs. 263.81±8.92, 156.89±10.23 vs. 263.81±8.92, P<0.05). (Fig. 4, Table 4)
Figure 2.
Expression of CDX1 and Tcf4 of control and high-type ARMs, as revealed by immunofluorescence staining. Positive immunolabelling was detected as green for CDX1 protein in the histiocytic cytoplasm of the epithelium of the rectum and in the nuclei of intestinal villus in the control. Expression of CDX1 was weaker in the high-type ARMs patients than in the control group. A (CDX1), B (Tcf4), C (Merged): control; a (CDX1), b (Tcf4), c (Merged): ARMs.(original magnification×400).
Figure 3.
Representative Western blots showing CDX1 protein expression within the rectum of control and ARMs. β-Actin was used to confirm equal protein loading and transfer. Obvious differences in CDX1 protein expression between control and ARMs groups were found. *P<0.05 vs. the control.
Table 3.
Western blot results for CDX1 protein in the extremity of the rectum of groups of patients with high-type, low-type ARMs and in control group without ARMs.
| Group | NO. of sample | CDX1 expression level |
|---|---|---|
| High-type | 16 | 0.936±0.085a |
| Low-type | 29 | 1.365±0.15b |
| Control | 12 | 1.782±0.067 |
Data are mean ±SD of the ratio of the gray value of the protein to that of the gray of β-actin. aP<0.05 versus control group; bP<0.05 versus low-type ARMs group;
Figure 4.
Representative Quantitative real-time RT-PCR shown CDX1 mRNA expression within the rectum of control and ARMs. Obvious differences in CDX1 mRNA expression between control and ARMs groups were found. *P<0.05 vs. the control.
Table 4.
Real-time PCR analysis of CDX1 mRNA expression.
| Group | NO. of sample | CDX1 expression level |
|---|---|---|
| High-type | 16 | 98.67±8.3a |
| Low-type | 29 | 156.89±10.23b |
| Control | 12 | 263.81±8.92 |
The Value of Y-axis were obtained from2-△Ct×104. aP<0.05 versus control group; bP<0.05 versus low-type ARMs group;
Discussion
The familial clustering of ARMs suggests a role for genetic mutations but the rarity of large affected families, especially in China, which limits the pace of discovery by means of linkage analysis. The selection of candidate genes based on the phenotype observed in mutant mice is a common strategy for the identification of genes and has been applied to ARMs in this study20. Anorectal malformations represent a spectrum of congenital disorders which involve abnormal termination of the anorectum. The clinical presentation ranges from anal stenosis to ARMs with fistula to persistent cloaca. The clinical management of ARMs continues to be a challenge, and our current understanding for normal and abnormal development of the anorectum remains incomplete. Mutations in SHH and its downstream mediators included Gli2 and Gli321, have been shown to resemble various forms of ARMs, suggesting a role of their signaling in anorectal development.
The CDX1 gene, a member of the caudal-type homeobox gene family, has been shown to play crucial roles in developmental processes in both vertebrates and invertebrates. Studies in rodents have shown that CDX1 is essential for gut development, the caudal region of the embryo and AP patterning22,23. A previous study have shown the expression profile of CDX1 mRNA was first detected in the endoderm of the developing gut24. And then CDX1 were found throughout large and small intestine, the detailed study by James et al. showed that CDX1 was expressed at high levels in the large intestine, reaching a peak in the rectum25. The present study provides the evidence of the critical role of CDX1 in normal anorectal development. It is indicated that CDX1 is required for the normal development of the anorectum. In this study, we have investigated the expression pattern of CDX1, suggesting that the mRNA and protein expression levels of CDX1 were significantly higher in controls. In contrast, the CDX1 expression level in ARMs remained low and unchanging; in previous study, it was reported that in ethylenethiourea (ETU)-induced ARMs embryos, spatiotemporal expression of CDX1 was imbalance during the anorectal morphogenesis from GD13 to GD16, revealing that the downregulation of CDX1 at the time of cloacal separation into rectum and urethra might be related to the development of ARMs19. A network of signaling pathways, including Wnt, Shh, and FGF10, is involved in the proper development of the anorectum, which provides a powerful tool to enhance our knowledge in the pathogenesis of ARMs. In summary, it was suggested that CDX1 might play an essential role not only in the embryogenesis of the anoretum, but also in the development of ARMs.
Furthermore, we reasoned that the association of these genes with ARMs could have been missed due to the variability of the phenotype under study and, subsequent grouped patients according to additional characteristics such as presence/ absence of fistula. This strategy also failed to reveal any implication of this gene in ARMs, although it could be argued that sample classification was at the expense of statistical power. Garcia-Barceló et al. excluded the SHH gene as a potential responsible for ARMs21. However, so far, none of the genes involved in the Wnt pathway has been studied in CDX1 gene nonmutated ARMs cases. Consequently, all the caudal type homeobox gene proteins remain possible candidate genes19,26. As in many other congenital diseases, several genes acting in different tissues and may be involved in ARMs at different developmental stages. These genes are subjected to complex cis and trans regulations, hence mutations in regulatory elements may also contribute to disease. However, there were no similar reports regarding these mutations in the Chinese Han population. In our study, we found some novel CDX1 mutations in ARMs patients. There were some associations of CDX1 gene mutations with ARMs. This variation in the occurrence could be explained as a result of the difference in the mutation position or the difference in genetic modifier. This latter mutation results in the substitution of a His, which is highly conserved between different species and has been shown to be a site of in vivo phosphorylation, suggesting that it plays an important role in the receptor regulation and that its absence can result in the protein loss of function. Intraexonic silent mutations can alter mRNA processing, thus resulting in either altered mRNA levels or truncated proteins. The mutation demonstrated pleiotropic effects by the finding that non-enteric phenotypes were only found in mutated homozygotes whereas heterozygous mutation usually caused isolate enteric manifestation. Otherwise, we could not conclude a correlation between an allelic status of CDX1 mutation and the extent of the defect in the ARMs. It was interesting to us that CDX1 null genotypes may thus play a role in ARMs. Taken together, the overall mutation rate was 3.7%, indicating that CDX1 mutations are relatively uncommon in ARMs. Certainly, our findings expanded the knowledge about the phenotype spectrum and the prevalence of CDX1 mutations among Chinese ARMs patients. The mutations detected in this study were likely to be significant because they were not observed in otherwise healthy subjects. This study conveys the problematic assessment of the pathogenic role in disease of potentially causative but rare variants.
Obviously, DNA variations in the gene encoding the molecules are bound to account for the phenotypic variability of a given phenotype and for a low incidence of mutations in the candidate genes analyzed. Due to the high phenotypic heterogeneity, it is essential to identify a causative genetic alteration in some patient with ARMs. Information of mutation is valuable not only in terms of scientific knowledge, but also a meaningful genetic counseling for the affected family. In addition, not all the patients harbor CDX1 mutations. Exhaustive functional analysis of these variants would be out of the scope of this study and, indeed, due to variation in genetic background, further studies need to be conducted on different populations to confirm at this point. Further extension of the study to a larger population of related individuals may give a clearer picture.
Conclusions
Our findings are useful in understanding the occurrence of CDX1 mutations and the correlation between the CDX1 genotype and the ARMs phenotype in Chinese patients. Furthermore, it was suggested that the downregulation of CDX1 might be related to the development of ARMs. Therefore, our research result provides a new target for treatment of ARMs. However, further studies are required to determine the pre-clinical utility of this gene.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 81170578 and grant no. 30400473) and supported by the Outstanding Scientific Fund of Shengjing Hospital (grant no. m850).
Abbreviations
- ARMs
Anorectal malformations
- CDX1
caudal type homeobox gene-1
- qRT-PCR
quantitative reverse transcriptase-polymerase chain reaction
- ETU
ethylenethiourea.
References
- 1.van der Putte SC. Normal and abnormal development of the anorectum. J Pediatr Surg. 1986;21:434–40. doi: 10.1016/s0022-3468(86)80515-2. [DOI] [PubMed] [Google Scholar]
- 2.Holschneider AM, Hutson JM. Genetics, Pathogenesis and Epidemiology of Anorectal Malformations and Caudal Regression Syndrome. Anorectal malformations in children; 1st edition. New York: Springer; 2006. pp. 31–42. [Google Scholar]
- 3.Seifert AW, Harfe BD, Cohn MJ. Cell lineage analysis demonstrates an endodermal origin of the distal urethra and perineum. Dev Biol. 2008;318:143–52. doi: 10.1016/j.ydbio.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T, Bai YZ, Wang WL. et al. Spatiotemporal pattern analysis of transcription factor 4 in the developing anorectum of the rat embryo with anorectal malformations. Int J Colorectal Dis. 2009;24:1039–47. doi: 10.1007/s00384-009-0705-3. [DOI] [PubMed] [Google Scholar]
- 5.Tai CC, Sala FG, Bellusci S. et al. Wnt5a knock-out mouse as a new model of anorectal malformation. J Surg Res. 2009;156:278–82. doi: 10.1016/j.jss.2009.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakata M, Takada Y, Yoshida H. et al. Induction of Wnt5a-expressing mesenchymal cells adjacent to the cloacal plate is an essential process for its proximodistal elongation and subsequent anorectal development. Pediatr Res. 2009;66:149–54. doi: 10.1203/PDR.0b013e3181aa304a. [DOI] [PubMed] [Google Scholar]
- 7.Kimmel SG, Mo R, Kim PC. New mouse models of congenital anorectal malformations. J Pediatr Surg. 2000;35:227–30. doi: 10.1016/s0022-3468(00)90014-9. [DOI] [PubMed] [Google Scholar]
- 8.Mandhan P, Quan QB, Beasley S. et al. Sonic hedgehog, BMP4, and Hox genes in the development of anorectal malformations in Ethylenethiourea-exposed fetal rats. J Pediatr Surg. 2006;41:2041–5. doi: 10.1016/j.jpedsurg.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 9.Fairbanks TJ, De Langhe S, Burns RC. et al. Fibroblast growth factor 10 (Fgf10) invalidation results in anorectal malformation in mice. J Pediatr Surg. 2004;39:360–5. doi: 10.1016/j.jpedsurg.2003.11.034. [DOI] [PubMed] [Google Scholar]
- 10.Warot X, Fraulob V, Chambon P, Dollé P. et al. Gene dosage-dependent effects of the Hoxa-13 and Hoxd-13 mutations on morphogenesis of the terminal parts of the digestive and urogenital tracts. Development. 1997;124:4781–91. doi: 10.1242/dev.124.23.4781. [DOI] [PubMed] [Google Scholar]
- 11.Wong EH, Cui L, Ng CL. et al. Genome-wide copy number variation study in anorectal malformations. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds451. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Draaken M, Prins W, Zeidler C. et al. Involvement of the WNT and FGF signaling pathways in non-isolated anorectal malformations: Sequencing analysis of WNT3A, WNT5A, WNT11, DACT1, FGF10, FGFR2 and the T gene. Int J Mol Med. 2012;30:1459–64. doi: 10.3892/ijmm.2012.1124. [DOI] [PubMed] [Google Scholar]
- 13.Bonner CA, Loftus SK, Wasmuth JJ. Isolation, characterization, and precise physical localization of human CDX1, a caudal-type homeobox gene. Genomics. 1995;28:206–11. doi: 10.1006/geno.1995.1132. [DOI] [PubMed] [Google Scholar]
- 14.Silberg DG, Swain GP, Suh ER. et al. Cdx1 and Cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–71. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 15.van den Akker E, Forlani S, Chawengsaksophak K. et al. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–93. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- 16.Chawengsaksophak K, Beck F. Chromosomal localization of Cdx2, a murine homologue of the Drosophila gene caudal to mouse chromosome 5. Genomics. 1996;34:270–1. doi: 10.1006/geno.1996.0286. [DOI] [PubMed] [Google Scholar]
- 17.Freund JN, Domon-Dell C, Kedinger M. et al. The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 1998;76:957–69. doi: 10.1139/o99-001. [DOI] [PubMed] [Google Scholar]
- 18.Davidson AJ, Zon LI. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox gene expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Developmental Biology. 2006;292:506–18. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T, Bai YZ, Zhang D. et al. Temporal and spatial expression of caudal-type homeobox gene-1 in the development of anorectal malformations in rat embryos. J Pediatr Surg. 2009;44:1568–74. doi: 10.1016/j.jpedsurg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Beck F. The role of Cdx genes in the mammalian gut. Gut. 2004;53:1394–6. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Barcelo MM, Chi-Hang Lui V, Tam PK. et al. Mutational analysis of SHH and GLI3 in anorectal malformations. Birth Defects Res A Clin Mol Teratol. 2008;82:644–8. doi: 10.1002/bdra.20482. [DOI] [PubMed] [Google Scholar]
- 22.Allan D, Houle M, Bouchard N. et al. RARgamma and Cdx1 interactions in vertebral patterning. Dev Biol. 2001;240:46–60. doi: 10.1006/dbio.2001.0455. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian V, Meyer BI, Gruss P. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell. 1995;83:641–53. doi: 10.1016/0092-8674(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 24.Meyer BI, Gruss P. Mouse Cdx-1 expression during gastrulation. Development. 1993;117:191–203. doi: 10.1242/dev.117.1.191. [DOI] [PubMed] [Google Scholar]
- 25.James R, Kazenwadel J. Homeobox gene expression in the intestinal epithelium of adult mice. J Biol Chem. 1991;266:3246–51. [PubMed] [Google Scholar]
- 26.Lohnes D. The Cdx1 homeodomain protein: an integrator of posterior signaling in the mouse. Bioessays. 2003;25:971–80. doi: 10.1002/bies.10340. [DOI] [PubMed] [Google Scholar]