Abstract
Studies in model organisms suggest that aged cells can be functionally rejuvenated, but whether this concept applies to human skin is unclear. Here we apply 3′-end sequencing for expression quantification (“3-seq”) to discover the gene expression program associated with human photoaging and intrinsic skin aging (collectively termed “skin aging”), and the impact of broadband light (BBL) treatment. We find that skin aging was associated with a significantly altered expression level of 2,265 coding and noncoding RNAs, of which 1,293 became “rejuvenated” after BBL treatment; i.e., they became more similar to their expression level in youthful skin. Rejuvenated genes (RGs) included several known key regulators of organismal longevity and their proximal long noncoding RNAs. Skin aging is not associated with systematic changes in 3′-end mRNA processing. Hence, BBL treatment can restore gene expression pattern of photoaged and intrinsically aged human skin to resemble young skin. In addition, our data reveal, to our knowledge, a previously unreported set of targets that may lead to new insights into the human skin aging process.
Introduction
Aging is under complex genetic and environmental control. Aging is associated with large-scale changes in gene expression, and how such changes may be modulated for healthful benefits in human beings is not clear. Numerous single-gene mutations have been identified that can extend the lifespan of model organisms (Partridge, 2010; de Magalhaes et al., 2012), and dietary restriction can slow the rate of aging, even if applied late in life (Partridge, 2010). More recently, several interventions have been shown to confer features of youthfulness to aged cells or tissues, demonstrating a remarkable plasticity of the aging process. For instance, heterochronic parabiosis between young and old mice enables circulatory factors to restore the functions of aged muscle stem cells (Liu and Rando, 2011). Similarly, inducible blockade of the transcription factor NF-κB in aged murine epidermis can abrogate cellular senescence and restore the global gene expression program of old skin to resemble that of young skin (Adler et al., 2007). An important question is whether similar plasticity exists in human skin, where aging occurs over decades rather than over months or years as seen in model organisms. Defining clinically viable strategies to unlock the plasticity of human aging is a critical challenge.
An ideal technology to test this concept is broadband light (BBL), also known as intense pulse light, a commonly available and popular treatment to “rejuvenate” the skin. According to the American Society for Aesthetic Plastic Surgery, over $215 million dollars were spent in the United States in 2009 on these procedures. Unlike ablative light-based treatments that improve the overall appearance of aged skin through thermal destruction and regrowth of the epidermis and superficial dermis, BBL uses a broad band of noncoherent light waves, ranging from 560 to 1,200 nm, that are absorbed by a number of components in the skin. Currently, BBL procedures are used to decrease the appearance of fine rhytides, dyspigmentation, erythema, and elastosis (Bitter Jr, 2000; Negishi et al., 2001). Nevertheless, the molecular changes that are induced by this treatment are unclear.
“Rejuvenation” is a term that has been used by many investigators and the lay public with different meanings, and thus needs to be carefully defined. Here we define “rejuvenation” as the restoration of characteristics of youthfulness to aged cells and tissues. After BBL treatment, is the skin truly “rejuvenated” at a molecular level, i.e., more closely resembles younger skin, or is the treatment merely inducing a wounding or scarring response that differs fundamentally from uninjured youthful skin?
Histologically, BBL has been reported to diminish melanin deposition in the dermis and reduce telangiectasias (Bitter Jr, 2000; Prieto et al., 2002), with some reports also reporting an increase in new upper papillary dermal collagen formation at 3 weeks after treatment (Negishi et al., 2001). However, this neocollagen formation may be a more variable or short-term effect, as ultrastructural analyses of skin 3 months after treatment have not shown any collagen or elastin fiber effects (Prieto et al., 2002). We examine the molecular basis of the BBL treatment response by defining the global gene expression programs of photoaged and intrinsically aged human skin and response to BBL. The intent is to capture the broadest spectrum of changes in RNA induced by aging and BBL, including alterations in gene expression (coding and noncoding) and gene regulation.
Results
Clinical and histologic changes after BBL treatment
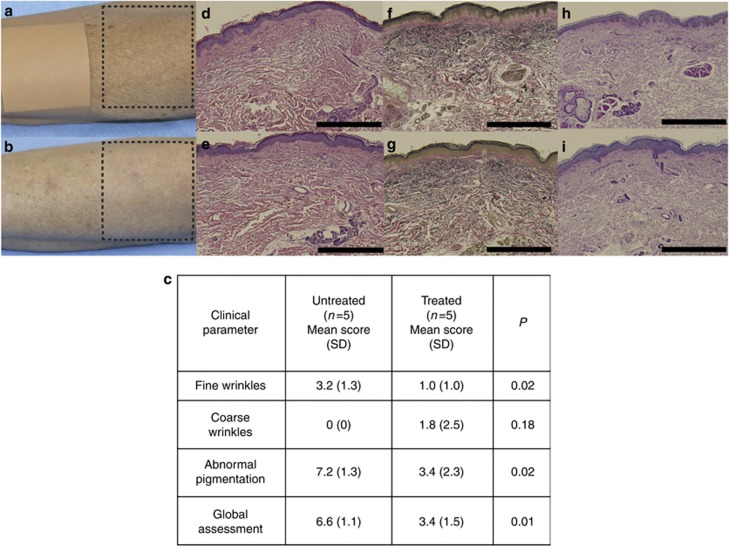
To gain insights into the gene expression program associated with skin aging and BBL treatment, we used skin biopsies from young female volunteers (age <30 years, n=5) and site-matched untreated and treated skin of aged female volunteers (age >50 years, n=5), the latter after three courses of monthly BBL treatment (n=5; Figure 1a). The treated subjects were healthy older females with moderate to severe photodamage on the forearms, and resided in the Santa Clara or San Jose, California metropolitan area, where on average there are 257 sunny days out of 365 days, with the average UV Index being 5.1 (average UV Index in the United States is 4.3; source: www.bestplaces.net, accessed 25 April 2012). Tanning beds, topical retinoids, or any other skin treatments on the arms were prohibited for 1 month before enrollment and during the study. During the study, the participants were instructed to sun-protect their arms with a broad-spectrum sunscreen and long-sleeved clothing, as well as avoid prolonged sun exposure. The untreated young subjects had the same inclusion criteria, but did not have evidence of photoaging on the arms.
Figure 1.
Clinical and histologic effects of broadband light (BBL) treatment. (a) Arm of a 73-year old female before BBL treatment (dashed box indicates area to be treated and bandage indicates untreated skin). (b) The same forearm after three BBL treatments with reduced fine wrinkling, hyperpigmentation, and erythema in the treated area (dashed box) compared with the untreated area. (c) Skin aging parameters show significant decreases in fine wrinkling, abnormal pigmentation, and global skin aging assessment after BBL treatment. The P-value by two-sided t-test. (d) Histology of skin before BBL treatment shows elastosis (original magnification × 200, hematoxylin and eosin (H&E) stain) and (e) reduced elastosis (original magnification × 200, H&E stain) after BBL treatment. (f) Before treatment, elastosis is prominent (original magnification × 200, von Giesen stain). (g) After treatment, elastosis is less distinct (original magnification × 200, von Giesen stain). (h) Before treatment, collagen fibers appear attenuated and disordered (original magnification × 200, periodic acid–Schiff (PAS) stain). (i) After treatment, collagen fibers are more uniform (original magnification × 200, PAS stain). Bars=1 mm each.
After three BBL treatments, arm skin showed improvements in clinical ratings of intrinsic and extrinsic skin aging parameters: fine wrinkling (P=0.03), abnormal pigmentation (P=0.02), and global skin aging assessment (P=0.01; Figure 1a–c). On histologic examination, the elastotic fibers in the treated aged samples were found to be diminished and less distinct compared with those in untreated aged samples (Figure 1d–g). The periodic acid–Schiff stain showed no obvious changes in collagen quantity in the dermis between treated and untreated aged samples, although they did appear less disordered after treatment (Figure 1h and i). The treated aged samples also displayed subjective increases in epidermal thickness (Figure 1e, g and i) compared with untreated aged samples (Figure 1d, f and h).
Expression program of coding and noncoding RNAs in aging skin
Although gene expression programs of aging in several tissues have been previously examined by microarray hybridization, we used 3′-end sequencing for expression quantification (3-seq), an efficient strategy of deep sequencing of RNA 3′ ends (Tariq et al., 2011). The potential advantages of 3-seq include accurate quantification of transcript levels not obscured by cross-hybridization, an ability to determine alterations in RNA termination and processing, and the ability to discover previously unannotated genes, such as long noncoding RNAs (lncRNAs). We generated 6.5–12.4 million uniquely mappable reads for each sample, and identified differentially expressed transcripts using DESeq algorithm (see Materials and Methods).
To rigorously define aging in molecular terms, we first identified transcript alterations associated with aging by comparing untreated young with untreated aged samples, and then tested how BBL treatment to aged skin affected these parameters.
Comparison of mRNA transcript levels in untreated young versus untreated aged, as well as untreated aged versus treated aged, samples revealed a consistent significant change in the expression level in 3,530 genes (Figure 2a). The directionality of the gene expression change with BBL treatment is shown in Figure 2a, with blue indicating a 2-fold decrease and yellow indicating a 2-fold increase. Genes whose transcript levels changed significantly between untreated young and untreated aged (n=2,265) are shown in Supplementary Table S1 online.
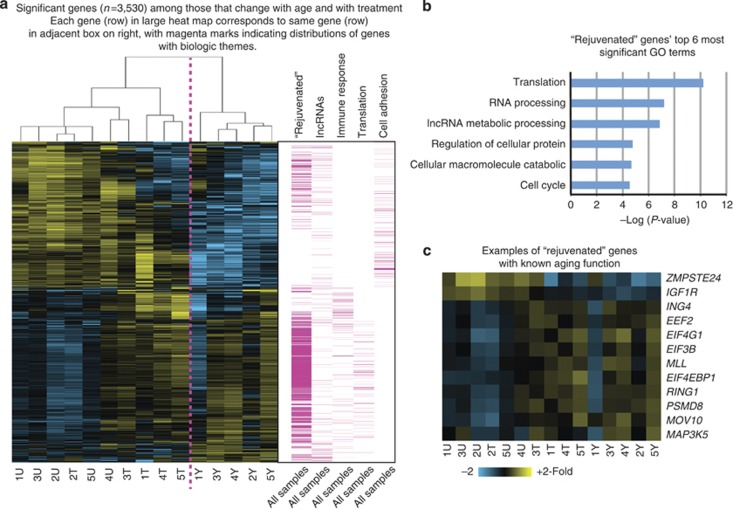
Figure 2.
Effects of broadband light (BBL) treatment on coding and noncoding RNAs in aging skin. (a) Gene expression clustering of treated aged samples is intermediate between untreated young and untreated aged samples. Transcript levels that significantly change with untreated young versus untreated aged samples, as well as untreated aged versus treated aged samples (n=3,530 total transcripts), are shown. Columns indicate single subject sample and rows indicate gene. T, aged treated; U, aged untreated; Y, young untreated. Magenta columns are visual representations of the gene distributions on the large heat map (left) as grouped by biological function. For instance, “immune response” and “translation” related genes are on the lower half of the heat map, with yellow indicating increased levels (or “up”) in treated aged and untreated young groups; the corresponding location in the large heat map for immune response and translation are blue (or “down”) in the untreated aged group. Distributions on the large heat map of rejuvenated genes (RGs; n=1,293) and “long noncoding RNAs” (lncRNAs) are shown in the first and second magenta columns, respectively. (b) The top six most significant Gene Ontology (GO) terms among RGs. (c) Examples of RGs with known aging function.
To visually display the locations of significant genes on the large heat map (Figure 2a), we have provided columns (in magenta) to the right of the large heat map that represent biological themes, according to Gene Ontology (GO) terms. For instance, the “rejuvenated genes” (RGs) and lncRNAs are distributed on both the upper and lower parts of the large heat map. In contrast, the “immune response” genes and “translation” genes are located on the lower half of the heat map. The “cell adhesion” genes are located on the upper half of the large heat map and are decreased in the untreated young group, increased in the untreated aged group, and intermediate in the treated aged group. The magenta columns hence provide a general sense of what biological function is altered and in what direction (increased (yellow) or decreased (blue)), enabling comparison between untreated aged, treated aged, and untreated young in the large heat map. For instance, both the treated older samples and the young untreated samples show increased transcript levels in “immune response” and “translation,” as both these groups are “up” (yellow). In contrast, the untreated aged group shows decreased transcript levels, or “down” (blue) in “immune response” and “translation” genes compared with the other two groups.
The gene programs associated with aging are multifaceted, and are enriched for several biological themes. The top five most significant GO terms that are increased in the aged untreated compared with young untreated group included translation (P=4.7 × 10−12), translational elongation (P=5.1 × 10−7), macromolecular complex assembly (P=7.5 × 10−6), ncRNA metabolic processing (P=6.2 × 10−6), and RNA processing (P=2.5 × 10−6). The top five GO terms that decreased in the aged untreated group compared with the young untreated group were genes encoding functions related to cell adhesion (P=1.5 × 10−17), biological adhesion (P=1.7 × 10−17), homophilic cell adhesion (P=7.8 × 10−8), skeletal system development (P=3.2 × 10−7), and enzyme-linked receptor protein signaling pathway (P=5.2 × 10−6). These gene sets are reminiscent of gene expression changes associated with aging in other tissues and organisms. For instance, translation-related genes or regulation of translation affects aging in Caenorhabditis elegans (Long et al., 2002) and Drosophila melanogaster (Kirby et al., 2002). In addition, translation is believed to underlie the important role of the TOR (target of rapamycin) pathway in stem cell aging (Chen et al., 2009; Nelson et al., 2009; Liu and Rando, 2011; Serrano, 2011).
BBL treatment promotes the gene expression pattern of young skin
Genes whose average expression level in aged treated skin was closer to young untreated skin than aged untreated skin were defined as RGs. Specifically, mean gene expression levels in the treated aged group were subtracted from mean gene expression levels in the untreated young group as well as from the untreated aged group. If the difference in gene expression level was less with the untreated young group compared with the difference with the untreated aged group, the gene was operationally defined as “rejuvenated”. A total of 1,293 transcripts qualified as RGs (Supplementary Table S2 online). Hierarchical clustering showed that the gene expression pattern of treated aged skin more closely resembled that of untreated young skin than untreated aged skin from the same individuals (Figure 2a). The RGs reflect coherent biological themes and include genes that fall under the following top six most significant GO terms: translation (P=5.8 × 10−11), RNA processing (P=6.3 × 10−8), ncRNA metabolic processing (P=1.4 × 10−7), regulation of cellular protein metabolic process (P=1.6 × 10−5), cellular macromolecular catabolic process (P=2.1 × 10−5), and cell cycle (=2.4 × 10−5; (Figure 2b, upper right).
A closer inspection of genes with expression patterns that were “rejuvenated” by BBL treatment revealed several key regulators known to control organismal aging (Figure 2c). These include ZMPSTE24, a metalloproteinase that processes lamin A, the gene defective in the dramatic premature aging syndrome, Hutchinson-Guilford progeria. In addition, the IGF1R receptor was one of the RGs identified, and this gene product is directed linked to aging and longevity in human beings, mice, and other model organisms (Liang et al., 2011; Tazearslan et al., 2011), as well as in other model organisms. Other RGs include EIF4G1 and EIF4EBP1, which are associated with increased lifespan in C. elegans (Curran and Ruvkun, 2007). MLL is a transcription regulator that associates with telomeres (Caslini et al., 2009), and methylates H3K4, which is required for normal lifespan in C. elegans (Greer et al., 2010). MAP3K5 (ASK10) regulates kinase activity in response to oxidative stress in a Klotho aging mouse model (Hsieh et al., 2010). PSMD8 is a proteasome component, and proteasome malfunction has been reported to contribute to aging in human skin (Hwang et al., 2007). RING1 and MOV10 are in the Polycomb pathway, which controls the lifespan of human fibroblasts (Itahana et al., 2003). EEF2 (eukaryotic translation elongation factor 2) is also an RG, and has been reported to associate with age-related declines in protein synthesis in rats (Parado et al., 1999). Finally, a number of tumor-suppressor genes that are cell-cycle checkpoints and ensure genome integrity, such as ING4 tumor suppressor, DAXX, and MSH2, are also RGs. Thus, BBL treatment appears to be capable of restoring many molecular features of youthful skin to aged human skin, at least in the short term. Notably, we did not see gene expression changes associated with wounding or scarring.
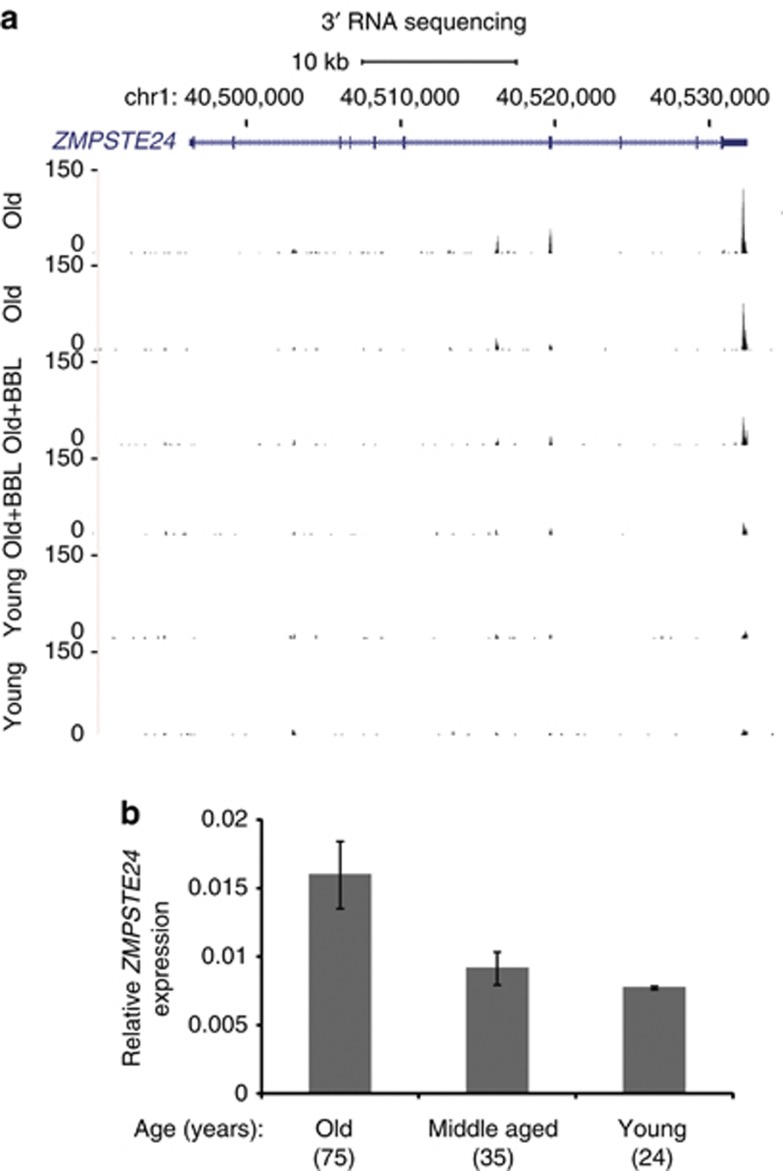
To confirm the findings on 3-seq, we performed quantitative reverse transcription–PCR (qRT–PCR) to confirm the levels of ZMPSTE24 on an independent group of untreated women across a spectrum of ages. The 3-seq had showed that untreated aged skin had the highest levels of ZMPSTE24 transcript expression level, treated aged skin had intermediate levels of ZMPSTE24 transcript level, and untreated young skin had the lowest levels (Figure 3a). By qRT–PCR, untreated aged arm skin (age 75 years) had the highest ZMPSTE24 transcript levels, untreated middle-aged arm skin (age 35 years) had intermediate levels, and untreated young arm skin (age 24 years) had the lowest levels (Figure 3b). This gradient has not been reported earlier in humans, but is an independent indicator suggesting that our findings are of biological relevance to physiological aging.
Figure 3.
ZMPSTE24 transcript levels increase after broadband light (BBL) treatment. (a) Schematic of ZMPSTE24 locus on chromosome 1, hg18. The 3′-seq (deep sequencing of RNA 3′ end) reads were plotted for two old individuals with and without BBL treatment and two untreated young samples. (b) ZMPSTE24 transcript expression is lower in untreated old skin compared with untreated young skin by quantitative reverse transcription–PCR (RT–qPCR). ZMPSTE24 transcript expression in untreated middle-aged skin is intermediate (n=1).
The enrichment in mRNAs encoding genes involved in RNA processing prompted us to evaluate the expression levels of additional RNA classes. The lncRNAs are a newly recognized class of genetic elements that are pervasively transcribed in the human genome (Wang et al., 2009; Wapinski and Chang, 2011). The roles of lncRNAs in aging and in skin have not been studied, as they have not been represented on microarray platforms in the past. However, the 3-seq technology can readily capture and quantify lncRNA expression. Of the 3,530 transcripts with altered levels between untreated young and untreated aged, 151 are lncRNAs. The chromosomal locations and most proximate genes of these lncRNAs are shown in Supplementary Table S3 online. Of the 1,293 RGs, 42 were lncRNAs. The chromosomal locations and most proximate genes of these lncRNAs are listed in Supplementary Table S4 online, with heat map in Supplementary Figure S1 online. These findings suggest that lncRNAs are potentially involved in the process of aging and rejuvenation, paralleling their roles in development and cellular reprogramming (Gupta et al., 2010; Loewer et al., 2010). Our data provide an initial set of lncRNAs associated with human aging that sets the groundwork for functional studies in the future.
GO analysis of the 151 lncRNAs with significant difference in expression between young untreated and aged untreated skin showed no significant enrichment for terms. Similarly, GO analysis of the 42 “rejuvenated” lncRNAs showed no significant enrichment for terms. However, the importance of lncRNAs in “rejuvenation” is not necessarily diminished. For instance, our 42 lncRNAs are a small number and future studies with greater sample size may identify more lncRNAs, enabling identification of significant GO terms. In addition, lncRNAs are a new class of RNA, and our GO analysis relied on the proximity of lncRNA sequences to known genes; it is possible that lncRNAs are important for regulating genes that are not necessarily proximal to the lncRNA (Gupta et al., 2010).
The effect of BBL treatment on the immune response includes altering the immune profile in a way that resembles untreated young skin. Figure 2a shows that although genes related to immune response are “up” after treatment, this “up” profile more closely resembles untreated young samples. This suggests that at least a portion of the immune response that is “up” after treatment is part of the “rejuvenated” profile and not specific to being treated with BBL.
The NF-κB pathway had been shown to be important in skin aging and rejuvenation (Adler et al., 2007), and we found that the RGs are indeed highly enriched for genes bound by NF-κB as measured by chromatin immunoprecipitation sequencing experiments. In all, 827 of the 1,293 RGs are bound by NF-κB (P=1.2 × 10−75, hypergeometric test). Interestingly, NF-κB itself was not one of the identified RGs.
RNA 3′ termination appears unaffected by aging or BBL
The 3-seq captures the 3′ polyadenylated (polyA) RNA fragments for deep sequencing, and thus has the potential to detect alterations in the location of 3′ transcript termination. The 3-seq method samples RNA sequences immediately upstream of the polyA tails. If there were changes in the use of the polyA site within the last exon such that the last exon is lengthened or truncated, this would be detected in the sequencing reads. This method does not evaluate the length of the polyA tail.
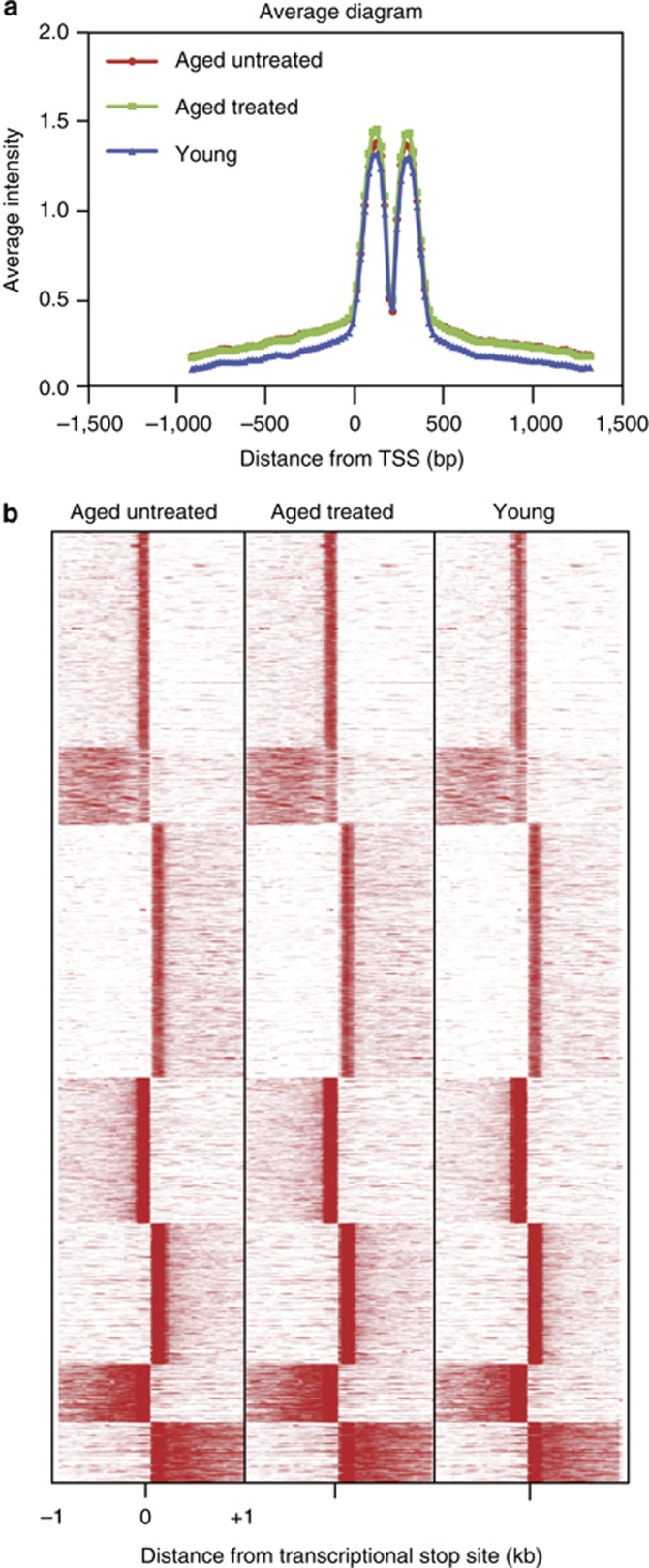
Alternative 3′-end usage is an important regulatory mechanism (Mayr and Bartel, 2009), and can alter gene expression output by changing the content of the 3′ untranslated region, which may then alter the repertoire of microRNA targets or RNA-binding proteins (such as those known to occur in cancer; Shapiro et al., 2011). Thus, in addition to quantifying changes in transcript abundance, we also searched for changes in transcript termination in association with aging or BBL treatment. Systematic comparison of all 3-seq reads showed that, as anticipated, the majority of reads fell into the annotated last exon, i.e., <1,000 bp from the transcriptional stop site (Figure 3a), and there were no consistent changes of 3′-end usage associated with aging or BBL treatment (Figure 3b). For instance, if the distribution of distances from transcriptional stop site for the RNAs from young untreated and aged treated samples were different, then aging may be associated with systemic changes in mRNA 3′ terminations (Figure 4).
Figure 4.
Broadband light (BBL) treatment and aging show no systematic changes of 3′-end usage. (a) Systematic comparison of all 3-seq (deep sequencing of RNA 3′ end) reads showing that the majority of reads fell into the annotated last exon (based on distance of within 1,000 bp from transcriptional start site (TSS)) for untreated aged, treated aged, and untreated young groups. The y-axis shows the average intensity of the 3-seq signal. (b) There were no systematic changes of 3′-end usage associated with aging or BBL treatment, as the reads showed similar length distributions between the untreated aged, treated aged, and untreated young groups.
Treatment-specific effects of BBL
In addition to affecting the age-associated gene expression program, we also considered the possibility that BBL treatment may induce unique treatment-specific effects that are distinct from aging. For instance, BBL treatment could induce wound healing or scarring response in addition to rejuvenation effects. We identified consistent changes in the expression of 1,112 genes that occur only in BBL-treated samples but not in either untreated young or untreated aged samples. Among these treatment-specific genes, the top five GO term categories significantly associated with increased expression after treatment were as follows: immune response (P=3.8 × 10−12), positive regulation of immune system process (P=2.0 × 10−8), cell activation (P=5.7 × 10−8), T-cell activation (6.0 × 10−7), and defense response (P=1.4 × 10−7). These categories are suggestive of an immune response to BBL separate from the immune response genes that are also increased in untreated young samples (as mentioned in the above section). The top five GO term categories significantly associated with decreased expression after treatment were as follows: regulation of transcription (P=2.0 × 10−6), transcription (P=1.7 × 10−5), response to organic substance (P=1.1 × 10−4), response to hormone stimulus (P=4.4 × 10−4), and negative regulation of transcription (P=4.7 × 10−4). These genes are distinct from a previously described “wound signature” that characterizes response to skin wounding (Chang et al., 2005); however, it is difficult to directly compare signatures with those in our study, as there are no published data at the equivalent time point after wounding as used in this study (4 weeks).
Finally, the top 10 genes that are most highly upregulated and downregulated in the treated aged samples compared with the untreated aged samples are listed in Table 1.
Table 1. Top 10 most significantly changed gene expression levels overall between BBL-treated aged samples and untreated aged samples.
| Gene symbol | Fold change: treated aged versus untreated aged | Directionality of change |
|---|---|---|
| HEPHL1 | 3.19 | Down |
| ZNF660 | 3.09 | Down |
| LY6G6D | 2.53 | Down |
| COCH | 2.38 | Down |
| CCL18 | 2.36 | Up |
| CEP78 | 2.36 | Down |
| ANGPTL7 | 2.34 | Down |
| SLN | 2.17 | Down |
| CPXM1 | 2.10 | Up |
| SAMD5 | 2.08 | Down |
Abbreviation: BBL, broadband light.
Discussion
Our results suggest that regulators of organismal aging can be altered in human skin using commonly available BBL technology. How such plasticity in aging may be modulated for healthful benefits such as prevention or treatment of age-associated skin conditions remains to be seen. Although BBL technology has been harnessed for its ability to produce a more clinically “youthful” appearance, our study suggests that “rejuvenation” at a molecular level has also occurred, with a number of genes linked to the aging process being altered in expression after treatment to more closely resemble young skin. Hence, it is possible that the clinical phenotype represents a functional rejuvenation (at least in the short term), rather than just a cosmetic mimic of youthful appearance.
As the BBL technology has been in existence for <20 years, the long-term effects of BBL remain to be determined. Although this study assessed the skin 4 weeks after treatment, it is unclear how durable the clinical and molecular response is. Also unknown is whether there is a decrease of age-associated skin changes such as seborrheic keratosis or actinic keratosis with time. It may be informative to follow these current participants in the long term (e.g., >5 years) with photographs and skin biopsies to determine the duration of clinical, histologic, and molecular effect of BBL treatment.
The precise mechanisms by which BBL (noncoherent wavelengths of light) alters gene expression are currently not well understood. For instance, it is known that BBL is absorbed by different targets including melanin and hemoglobin, leading to decreased erythema and pigmentation. It is thought that the decrease in fine wrinkling is partly due to the production of new collagen (Fisher et al., 2008). However, the genes identified in this study were not collagen specific. It may be possible that if the posttreatment skin biopsies were performed earlier than 4 weeks, some of the gene expression changes related to collagen production might have been captured.
Gene expression programs associated with human aging appear to differ between organ types. For instance, the aging human kidney and human muscle seem to have distinct gene expression signatures (Rodwell et al., 2004; Zahn et al., 2006). The aging gene expression profiles of human skin generated in this study do not appear to be the same as other reported organ types; however, future direct comparison studies may shed more light on this issue.
NF-κB is an important regulator of gene expression in many contexts. In this case, the most relevant role of NF-κB is likely in controlling cell senescence (Bernard et al., 2004; Adler et al., 2007) and immune response. Our finding that RGs are highly enriched for NF-κB-bound genes suggests that BBL may influence pathways controlled by NF-κB. The precise mechanisms by which this occurs remain to be investigated. Nevertheless, our results are consistent with a prior study showing that inducible blockade of NF-κB in aged murine skin restores the gene expression program and phenotypes of young skin (Adler et al., 2007).
It is difficult to directly compare the results of our study with the gene expression profiles in humans reported currently in the literature for two reasons: (1) the time point of biopsies may not be exactly the same, and (2) the nature of the disease entity or treatment is not the same as BBL. For instance, gene expression patterns in human postburn hypertrophic scars at 6–15 months in two pediatric and two adult patients identified six genes as significantly increased (Paddock et al., 2003), none of which were significantly changed in our BBL study. In another example, an in vitro human keratinocyte model using scratch wounding has shown increased activation of NF-κB in cells between 1 and 14 days (Adams et al., 2007). Our study captured the 1-month time point when the effects of wound healing might be decreasing, and we are more likely to detect rejuvenation effects. At our 1-month time point, NF-κB levels were not significantly increased, but genes known to interact with NF-κB were significantly increased, which is a distinct and, to our knowledge, previously unreported finding.
Two RGs, RING1 and MOV10, are in the Polycomb pathway, with the potential to contribute to both rejuvenation effects and wound repair. In mice and cell culture, the Polycomb pathway controls the lifespan of human fibroblasts (Itahana et al., 2003) and associates with the upregulation of wound repair genes (Shaw and Martin, 2009).
The ligands for Toll-like receptors 2, 3, and 5 have been reported to affect the transcript and protein levels of matrix metalloproteinases 1 and 9 and induce the nuclear translocation of NF-κB after 24–48 hours in human keratinocyte culture (Lee et al., 2009). We did not detect significant increases in Toll-like receptors 2, 3, and 5, or NF-κB, but our study was in vivo and skin samples were obtained at the 1-month time point.
In addition, although our data show that coherent biological themes such as “translation” or “RNA processing” are altered after BBL treatment, our study does not identify the population of cells within the skin that undergo these changes. Future studies that get at this question may better explain how BBL treatment might lead to histological or structural changes such as resorption of elastosis or collagen deposition.
It would be interesting to compare whether other modalities known to reduce clinical skin aging parameters such as topical tretinoin result in gene expression changes that are in common with BBL-induced changes.
In addition, comparison of non-sun-exposed older skin before and after treatment may identify gene expression changes that are specific to intrinsic skin aging.
This is an exploratory study, and we will consider including treated young skin in future studies. In this study, we did not treat younger skin (defined as age <30 years for this study) because there was no clinical indication; these subjects did not have detectable photoaging or intrinsic aging on the arm skin. As it is unlikely that BBL would be used in practice on young skin without photoaging (except possibly for hair removal), we did not include this group in the study.
The current literature on the ability of BBL to induce collagen neogenesis is contradictory. Although some reports on histologic changes induced by BBL include collagen neogenesis (Negishi et al., 2001), there are other studies showing no change (Prieto et al., 2002). This latter study also reported no change in elastin content after treatment. In our study, there were no marked changes in collagen content after treatment on periodic acid–Schiff staining. There were decreases in the amount of elastin on von Giesen staining. We did not detect any significant changes in collagen or elastin gene expression levels after treatment. One possibility is that the histology was taken at a single time point, and may not have captured the time when collagen or elastin expression levels were more markedly changed. Future studies will indeed biopsy-treated skin longitudinally to reveal the kinetics of activation/suppression of target genes. In addition, it is precisely the goal of this study to extend beyond the conventional histologic analysis of skin and explore molecular changes of skin aging and BBL treatment. We observed numerous gene expression changes related to pathways beyond connective tissue organization that can be modulated by BBL.
Finally, future studies with larger sample size may enable us to identify additional significant genes (both coding and noncoding) whose expression is altered in untreated young versus untreated aged, as well as untreated aged and treated aged human skin samples. Larger sample size might also enable us to correlate the degree of clinical response with more “rejuvenated” gene expression changes.
Materials and Methods
Human subjects and sample acquisition
This study was conducted in accordance with the Declaration of Helsinki Principles. After Institutional Review Board approval and written informed consent was obtained, five female participants over the age of 50 years underwent BBL treatments to the left forearm. Inclusion criteria included Fitzpatrick skin type II or III, and a global assessment of forearm skin aging consistent with moderate or severe forearm skin aging (modified validated instrument from McKenzie et al., 2010) for treated participants. Treatments were performed on the Sciton Joule Platform using the BBL module. The same investigator performed the treatments at 4-week intervals for a total of three treatments using a 515-nm or a 560-nm cutoff filter at a single long pulse of 10–20 ms duration, with fluences of 8–14 J cm−2. At each treatment session, two or more passes were performed. At 4 weeks after the third BBL treatment, 4-mm skin biopsies were performed by the Keys punch technique from the treated and adjacent untreated skin. Punch biopsies (4 mm) were taken from non-sun-exposed arm skin of five participants <30 years old. These specimens were bisected and placed into either RNAlater (Ambion, cat. no. AM7022, Grand Island, NY) or formalin solution for staining with hematoxylin and eosin, von Giesen, or periodic acid–Schiff.
The 3-seq and bioinformatics
Total RNA was extracted using the RNeasy Fibrous Tissue Mini Kit (Qiagen, Germantown, MD). The 3-seq was performed as described in Beck et al. (2010). In brief, oligo-dT-directed reverse transcription generated complementary DNAs corresponding to 3′ ends of polyA transcripts; the complementary DNAs were cloned and subjected to deep sequencing on the Illumina GAIIx (San Diego, CA) platform with raw read length of 36 bp. Raw reads were aligned to human genome (hg18) using bowtie (Langmead et al., 2009); each sample generated 6.5–12.4 million uniquely mappable reads. The 3′ sequencing of skin transcripts was performed to assess length distributions.
Reads per kilobase of exon per million mappable reads (RPKM, a direct measure of transcript abundance) and the number of raw reads falling on to each gene were calculated using a self-developed script by Kun Qu. The Reference Sequence (RefSeq; www.ncbi.nlm.nih.gov/RefSeq) and Ensembl (http://www.ensembl.org) annotated noncoding genes were included. Significant genes were called using the DESeq package (http://www.bioconductor.org) comparing aged treated with aged untreated samples (genes changed because of treatment), and aged untreated with young untreated samples (genes changed because of aging). Unsupervised hierarchical clustering of significantly different expressed genes was performed using Cluster. The GO terms were generated using DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/). Genes close to lncGenes were identified using GREAT database (http://great.stanford.edu). These data have been deposited into the Gene Expression Omnibus.
To determine the overlap between RGs and NF-κB binding, we downloaded the NF-κB-bound genes identified by the ENCODE project (ENCODE Consortium, 2011) by chromatin immunoprecipitation sequencing experiments. A total of 9,650 genes bound NF-κB in one or more cell types, and these were compared with the RG gene list.
RT–qPCR
Total RNA was extracted with TRIzol (Invitrogen, Grand Island, NY) followed by RNeasy column purification (Qiagen) and DNAse Turbo Treatment (Ambion). RT–qPCR was performed using total RNA (10 ng), Taqman One Step RT–PCR master mix, and one of the following Taqman assays: GAPDH (Hs99999905_m1) and ZMPSTE24 (Hs00956778_m1; Applied Biosystems, Carlsbad, CA). Reactions were in triplicate for each sample and were performed a minimum of two times. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels.
Acknowledgments
This study was funded by a research grant from Sciton. We are indebted to Paul Khavari and Jean Tang for prereview of the manuscript. We thank Olena Mykhaylichenko and Sarah Jacobs for administrative support.
Glossary
- BBL
broadband light
- GO
Gene Ontology
- lncRNA
long noncoding RNA
- polyA
polyadenylated
- qRT–PCR
quantitative reverse transcription–PCR
- RG
rejuvenated gene
- 3-seq
3′-end sequencing for expression quantification
PB has given lectures on broadband light technology. The other authors state no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
This study was accepted as a poster presentation at the 2012 Society of Investigative Dermatology Annual Meeting
Supplementary Material
References
- Adams S, Pankow S, Werner S, et al. Regulation of NFKB activity and keratinocytic differentiation by the RIP4 protein: implications for cutaneous wound repair. J Invest Dermatol. 2007;127:538–544. doi: 10.1038/sj.jid.5700588. [DOI] [PubMed] [Google Scholar]
- Adler AS, Sinha S, Kawahara TLA, et al. Motif module map reveals enforcement of aging by continual NFKB activity. Genes Dev. 2007;21:3244–3257. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AH, Weng Z, Witten DM, et al. 20103′-end sequencing for expression quantification (3SEQ) from archival tumor samples PLoS One 195e8768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Gosselin K, Monte D, et al. Involvement of Rel/ NFkB transcription factors in keratinocyte senescence. Cancer Res. 2004;64:472–481. doi: 10.1158/0008-5472.can-03-0005. [DOI] [PubMed] [Google Scholar]
- Bitter Jr PH. Noninvasive rejuvenation of photodamaged skin using serial, full-face intense pulsed light treatments. Dermatol Surg. 2000;26:835–843. doi: 10.1046/j.1524-4725.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- Caslini C, Connelly JA, Serna A, et al. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol. 2009;29:4519–4526. doi: 10.1128/MCB.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, et al. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:p.ra71. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Consortium The user's guide to the encyclopedia of DNA elements. PLoS Biol. 2011;9:1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GJ, Varani J, Voorhees JJ. Looking older: fibroblast collapse and therapeutic implications. Arch Dermatol. 2008;144:666–672. doi: 10.1001/archderm.144.5.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, et al. Members of the histone H3 lysine 4 trimethylation complex regulate lifespan in germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C-C, Kuro-o M, Rosenblatt KP, et al. The ASK1-Signalosome regulates p38 MAPK activity in response to levels of endogenous oxidative stress in the Klotho mouse models of aging. Aging. 2010;2:597–611. doi: 10.18632/aging.100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JS, Hwang JS, Chang I, et al. Age-associated decrease in proteasome content and activities in human dermal fibroblasts: restoration of normal level of proteasome subunits reduces aging markers in fibroblasts from elderly persons. J Gerontol A Biol Sci Med Sci. 2007;62:490–499. doi: 10.1093/gerona/62.5.490. [DOI] [PubMed] [Google Scholar]
- Itahana K, Zou Y, Itahana Y, et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23:389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby K, Hu J, Hilliker AJ, et al. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc Natl Acad Sci USA. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim H, Kim S, et al. Activation of toll-like receptors 2, 3, or 5 induces matrix metalloproteinase-1 and -9 expression with the involvement of MAPKs and NFkB in human epidermal keratinocytes. Exp Dermatol. 2009;19:e44–e49. doi: 10.1111/j.1600-0625.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- Liang R, Khanna A, Muthusamy S, et al. Post-transcriptional regulation of IGF1R by key microRNAs in long-lived mutant mice. Aging cell. 2011;10:1080–1088. doi: 10.1111/j.1474-9726.2011.00751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;103:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, et al. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Wuttke D, Wood SH, et al. Genome-environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol Rev. 2012;64:88–101. doi: 10.1124/pr.110.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie NE, Saboda K, Duckett LD, et al. Development of a photographic scale for consistency and guidance in dermatologic assessment of forearm sun damage. Arch Dermatol. 2010;147:31–36. doi: 10.1001/archdermatol.2010.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TJ, Behfar A, Yamada S, et al. Stem cell platforms for regenerative medicine. Clin Transl Sci. 2009;2:222–227. doi: 10.1111/j.1752-8062.2009.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi K, Tezuka Y, Kushikata N, et al. Photorejuvenation for Asian skin by intense pulsed light. Dermatol Surg. 2001;27:627–631. doi: 10.1046/j.1524-4725.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- Paddock HN, Schultz GS, Baker HV, et al. Analysis of gene expression patterns in human postburn hypertrophic scars. J Burn Care Rehabil. 2003;24:371–377. doi: 10.1097/01.BCR.0000095508.96754.E0. [DOI] [PubMed] [Google Scholar]
- Parado J, Bougria M, Ayala A, et al. Effects of aging on the various steps of protein synthesis: fragmentation of elongation factor 2. Free Radic Biol Med. 1999;26:362–370. doi: 10.1016/s0891-5849(98)00202-0. [DOI] [PubMed] [Google Scholar]
- Partridge L. The new biology of ageing. Phil Trans R Soc B 12. 2010;365:147–154. doi: 10.1098/rstb.2009.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto VG, Sadick NS, Lloreta J, et al. Effects of intense pulsed light on sun-damaged human skin, routine, and ultra-structural analysis. Lasers Surg Med. 2002;30:82–85. doi: 10.1002/lsm.10042. [DOI] [PubMed] [Google Scholar]
- Rodwell G, Sonu R, Zahn JM, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M. Cancer: final act of senescence. Nature. 2011;479:481–482. doi: 10.1038/479481a. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Kim HJ, Jejelowo O, et al. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucl Acids Res. 2011;39:e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C, Huang J, Barzilai N, Suh Y. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging cell. 2011;10:551–554. doi: 10.1111/j.1474-9726.2011.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gersten M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, et al. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.