Abstract
HLA-C remains the strongest susceptibility candidate gene in psoriasis. Evidence for interaction between HLA-C and endoplasmic reticulum aminopeptidase 1 (ERAP1) confined to individuals carrying the HLA-C risk allele was recently reported. Psoriasis displays wide variation, and genetic heterogeneity is likely to contribute to clinical diversity. Age at disease onset is a putative discriminator, and separating psoriasis into early- (<40 years) and late-onset disease has been useful. To sharpen the age-dependent phenotype, we compared genotypes for ERAP1 (rs26653, rs30187, and rs27524) and HLA-C*06:02 in healthy controls and cases stratified for onset of psoriasis at <10, 10–20, 20–40, and >40 years of age. This approach revealed that association with ERAP1 was confined to cases with onset between 10 and 20 years (odds ratio 1.59, 95% confidence interval: 1.28–1.98, P=0.00008) and no association was detected in cases with onset below 10 years, reflecting genetic heterogeneity within the childhood psoriasis population. In contrast to earlier findings, association with ERAP1 was neither dependent on nor interacting with HLA-C*06:02. ERAP1 single-nucleotide polymorphism rs26653, which, to our knowledge, has not previously been reported in psoriasis, is nonsynonymous, has suggestive functional consequences, and herein displays strong association with disease.
Introduction
Psoriasis is a highly heterogeneous disease with a complex genetic background, and the strategy to stratify into subgroups is likely to be fruitful for fully understanding the spectrum of the disease. The underlying genetics may affect the age at onset, and separating psoriasis into early- and late-onset disease using 40 years as the divider has been useful (Henseler and Christophers, 1985). Using this definition, early-onset psoriasis has been shown to have a higher degree of heritability and a higher prevalence of HLA-C*06:02 (Henseler and Christophers, 1985; Gudjonsson et al., 2006). However, onset before the age of 40 years has been reported in ∼75% of patients with psoriasis and thus this definition of early-onset comprises the majority of patients with psoriasis (Zhang et al., 2002). Nevertheless, most genetic studies use this classification and lack a more detailed stratification.
Other stratification attempts separate patients according to HLA-C status. Presence of the risk allele HLA-C*06:02 is claimed to be associated with earlier disease onset, more severe disease course, and a higher prevalence of the guttate phenotype (Mallon et al., 2000; Gudjonsson et al., 2006). Nail and joint manifestations are reported to be more frequent in HLA-C*06:02-negative patients (Gudjonsson et al., 2006).
Onset of psoriasis in adolescence and early adulthood is common, and a potential impact of puberty has been discussed (Braun-Falco et al., 1972; Farber and Nall, 1974; Swanbeck et al., 1995; Raychaudhuri and Gross, 2000). However, before puberty the onset of psoriasis is more unusual and is estimated to occur in only 10% of patients before the age of 10 and in 2% before the age of 2 years (Farber and Nall, 1974; Farber and Jacobs, 1977). Children with psoriasis may differ in their clinical presentation compared with adults and a correct diagnosis may be delayed, which is a potential confounder when determining disease onset in this group (Morris et al., 2001; de Jager et al., 2010).
HLA-C*06:02 is the candidate gene allele with the strongest association with psoriasis (Elder, 2006; Fan et al., 2008). Exactly how HLA-C*06:02 contributes to disease is, however, still unclear. A recent genome-wide association study reported evidence for a genetic interaction between the HLA-C and endoplasmic reticulum aminopeptidase 1 (ERAP1) genes (combined P=6.95 × 10−6), with the ERAP1-associated synonymous single-nucleotide polymorphism (SNP) (rs27524) affecting psoriasis susceptibility only in individuals carrying the HLA-C risk allele (Strange et al., 2010). ERAP1 has an important role in MHC class I peptide processing in the endoplasmic reticulum and has also been shown to be involved in shedding of proinflammatory cytokine receptors, altogether suggestive of a potential involvement in the pathogenesis of psoriasis (Cui et al., 2003a; Hearn et al., 2009; Haroon and Inman, 2010). Other nonsynonymous ERAP1 SNPs have been shown to be associated with ankylosing spondylitis (rs30187, rs26653) and multiple sclerosis (rs30187) (Burton et al., 2007; Reveille, 2011; Guerini et al., 2012). Altogether, these data suggest that ERAP1 may be essential in regulating immunity.
Herein we present a cohort of psoriasis patients stratified for phenotype (plaque and guttate) and age at onset (Table 1). The age groups were chosen in an attempt to reflect biologically significant transitions. In particular, we aimed to explore the impact of puberty. Setting the limit for puberty at 10 years represents a stringent approach (Parent et al., 2003). Herein we report a strong association of ERAP1 SNP rs26653 not restricted to HLA-C*06:02-positive individuals. Stratification for age at onset revealed that ERAP1 association was confined to cases with disease onset between 10 and 20 years (odds ratio (OR) 1.59, 95% confidence interval (CI): 1.28–1.98, P=0.00008), whereas prepubertal children with onset between 0 and 9 years lacked association (OR 1.24, 95% CI: 0.93–1.65, P=0.4). This work reveals striking genetic heterogeneity among patients with early-onset psoriasis and stresses the importance of detailed stratification in future genetic studies in psoriasis.
Table 1. Characteristics of participants.
|
Age at onset |
||||||
|---|---|---|---|---|---|---|
| 0–9 (N=119) | 10–20 (N=203) | 21–40 (N=283) | >40 (N=349) | All (N=954) | Controls (N=1,748) | |
| Women (%) | 58 (49) | 112 (55) | 137 (48) | 216 (62) | 523 (55) | 1,026 (59) |
| Mean age at onset | 5.4 | 14.9 | 31.0 | 56.8 | 33.9 | |
| Plaque phenotype (%) | 96 (81) | 157 (77) | 201 (71) | 309 (89) | 763 (80) | |
| Plaque phenotype HLA-Cw06 (%) | 44 (46) | 104 (66) | 84 (42) | 55 (17) | 287 (38) | |
| Guttate phenotype (%) | 23 (19) | 46 (23) | 82 (29) | 40 (11) | 191 (20) | |
| Guttate phenotype HLA-Cw06 (%) | 19 (83) | 38 (83) | 69 (84) | 25 (57) | 151 (79) | |
Abbreviation: N, number of subjects.
Results
The association of HLA-C*06:02 varies with the age at disease onset in psoriasis
The established and strong association of HLA-C*06:02 with psoriasis was confirmed in our cases (OR 4.51, 95% CI: 3.81–5.3, P=4.9 × 10−76) (Table 2). For analysis of a possible effect of age at onset, we separated the cases into four groups (Table 1). The strongest association for HLA-C*06:02 was detected in the age group 10–20 years (OR 8.46, 95% CI: 6.65–10.77, P=1.3 × 10−86), which was significantly stronger than that in the groups 0–9 (OR 5.71, 95% CI: 4.19–7.79, P=1.2 × 10−33) and 21–40 (OR 5.52, 95% CI: 4.40–6.92, P=8.9 × 10−58) (Table 3). In the oldest group with onset of disease above 40 years, the association with HLA-C*06:02 was significant, albeit much lower (OR 1.89, 95% CI: 1.45–2.46, P=9.1 × 10−6) than published data (Allen et al., 2005). The distribution of plaque and guttate phenotypes was similar in the three younger age cohorts but more skewed toward plaque phenotype in the oldest group (Table 1).
Table 2. Markers analyzed in ERAP1 and HLA-C*06 as well as allelic associations.
|
MAF |
All cases |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene | Marker | Region1 | Variation2 | Alleles3 | Controls | Cases | P4 | OR (95% CI) |
| 5 | ERAP1 | rs26653 | Exon 2 | Arg127Pro | C/G | 0.26 | 0.31 | 0.00006 | 1.31 (1.16–1.48) |
| 5 | ERAP1 | rs30187 | Exon 11 | Lys528Arg | T/C | 0.34 | 0.38 | 0.02 | 1.16 (1.03–1.30) |
| 5 | ERAP1 | rs27524 | 3′ UTR | None | A/G | 0.36 | 0.39 | 0.11 | 1.10 (0.98–1.23) |
| 6 | HLA-C | rs10484554 | None coding | None | T/C | 0.10 | 0.28 | 1.8 × 10−65 | 3.55 (3.06–4.13) |
| 6 | HLA-C | Cw06:025 | P/N | 0.07 | 0.24 | 4.9 × 10−76 | 4.51 (3.81–5.34) | ||
Abbreviations: CI, confidence interval; ERAP, endoplasmic reticulum aminopeptidase 1; MAF, minor allele frequency; OR, odds ratio; UTR, untranslated region.
Data from www.ensembl.org.
Data from www.ncbi.nlm.nih.gov.
Minor allele/major allele.
Adjusted P-value for five tests with Holm as implemented in PLINK adjust.
Table 3. Allelic associations for ERAP1 and HLA-C genes in psoriasis patients stratified for age at onset (case versus controls).
|
Age at onset |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Age 0–9 |
Age 10–20 |
Age 21–40 |
Age >40 |
|||||||||
| Marker | MAF ca | P1 | OR (95% CI) | MAF ca | P1 | OR (95% CI) | MAF ca | P1 | OR (95% CI) | MAF ca | P1 | OR (95% CI) |
| rs26653 | 0.30 | 0.4 | 1.24 (0.93–1.65) | 0.36 | 0.00008 | 1.59 (1.28–1.98) | 0.31 | 0.05 | 1.27 (1.05–1.54) | 0.30 | 0.1 | 1.21 (1.01–1.44) |
| rs30187 | 0.39 | 0.4 | 1.20 (0.91–1.57) | 0.43 | 0.0001 | 1.46 (1.18–1.80) | 0.35 | 1 | 1.03 (0.85–1.24) | 0.37 | 0.5 | 1.11 (0.93–1.31) |
| rs27524 | 0.39 | 0.4 | 1.12 (0.86–1.47) | 0.42 | 0.02 | 1.28 (1.04–1.58) | 0.36 | 1 | 0.98 (0.81–1.18) | 0.38 | 0.5 | 1.09 (0.92–1.29) |
| rs10484554 | 0.33 | 2.5 × 10−28 | 4.66 (3.48–6.24) | 0.42 | 2.9 × 10−72 | 6.65 (5.29–8.35) | 0.31 | 4.6 × 10−46 | 4.25 (3.44–5.24) | 0.15 | 0.0005 | 1.59 (1.25–2.02) |
| HLA-Cw06:02 | 0.29 | 1.2 × 10−33 | 5.71 (4.19–7.79) | 0.38 | 1.3 × 10−86 | 8.46 (6.65–10.77) | 0.28 | 8.9 × 10−58 | 5.52 (4.40–6.92) | 0.12 | 9.1 × 10−6 | 1.89 (1.45–2.46) |
Abbreviations: CI, confidence interval; ERAP, endoplasmic reticulum aminopeptidase 1; MAF ca, minor allele frequency cases; OR, odds ratio.
Adjusted P-value for five tests with Holm as implemented in PLINK adjust.
Replication of association for ERAP1 in psoriasis
The association of ERAP1 in psoriasis was reported recently (Strange et al., 2010; Bergboer et al., 2012). We investigated three SNPs in the ERAP1 gene (Table 2). Significant allelic association was found for rs26653 (OR 1.31, 95% CI: 1.16–1.48, P=0.00006) and rs30187 (OR 1.16, 95% CI: 1.03–1.30, P=0.02) in the total data set. No association was found for rs27524 (OR 1.10, 95% CI: 0.98–1.23, P=0.11) in the total data set (Table 2), although we did find association with rs27524 in the group with disease onset between 10 and 20 years (OR 1.28, 95% CI: 1.04–1.58, P=0.02) (Table 3).
Association with ERAP1 is not dependent on HLA-C*06:02 and is confined to cases with disease onset between 10 and 20 years
In contrast to previous findings, in which the association of ERAP1 rs27524 was confined to individuals carrying the HLA-C*06:02 allele, the association with rs26653 was independent of HLA-C*06:02 in the total data set (P=0.001, OR 1.39 95% CI: 1.14–1.70) (Table 4a). Association independent of HLA-C*06:02 with rs27524 and rs30187 was found only in the group with onset between 10 and 20 years (Table 4b). No interaction between ERAP1 (rs26653, rs30187, rs27524) and HLA-C*06:02 was detected, neither in the total data set (Supplementary Table S1 online) nor in the significantly associated group with onset between 10 and 20 years (data not shown).
Table 4a. Independent association of ERAP1 SNP rs26653 and HLA-C*06 in all subjects.
| rs26653 | HLA-C*06 | P-value | OR (95% CI) |
|---|---|---|---|
| GG | NN | 1 | 1 |
| GC/CC | NN | 0.001 | 1.39 (1.14–1.70) |
| GG | P | <2 × 10−16 | 6.21 (4.73–8.14) |
| GC/CC | P | <2 × 10−16 | 7.17 (5.47–9.39) |
Abbreviations: CI, confidence interval; ERAP, endoplasmic reticulum aminopeptidase 1; NN, negative for HLA-C*06; OR, odds ratio; P, positive for HLA-C*06; SNP, single-nucleotide polymorphism.
Generalized linear model as implemented in R software package. Individuals with the low risk genotype for rs26653 and HLA-C*06 NN were set as baseline.
Table 4b. Independent association of ERAP1 SNPs and HLA-C*06 in the age group 10–20.
| ERAP1 SNP | ERAP1 genotype | HLA-C*06 | P-value | OR (95% CI) |
|---|---|---|---|---|
| rs26653 | GG | NN | 1 | 1 |
| GC/CC | NN | 0.009 | 2.02 (1.19–3.43) | |
| GG | P | <2 × 10−16 | 19.43 (11.55–32.66) | |
| GC/CC | P | <2 × 10−16 | 27.11 (16.35–44.95) | |
| rs30187 | CC | NN | 1 | 1 |
| TC/TT | NN | 0.0005 | 3.10 (1.64–5.87) | |
| CC | P | <2 × 10−16 | 28.00 (14.26–54.96) | |
| TC/TT | P | <2 × 10−16 | 38.70 (20.63–72.58) | |
| rs27524 | GG | NN | 1 | 1 |
| GA/AA | NN | 0.002 | 2.68 (1.41–5.08) | |
| GG | P | <2 × 10−16 | 30.10 (15.43–58.70) | |
| GA/AA | P | <2 × 10−16 | 33.96 (18.09–63.76) |
Abbreviations: CI, confidence interval; ERAP, endoplasmic reticulum aminopeptidase 1; NN, negative for HLA-C*06; OR, odds ratio; P, positive for HLA-C*06; SNP, single-nucleotide polymorphism.
Generalized linear model as implemented in R software package. Individuals with risk alleles for respective ERAP1 SNPs and HLA-C*06.
Stratification for age at onset as described above showed association for all three nonsynonymous ERAP1 SNPs rs26653 (OR 1.59, 95% CI: 1.28–1.98, P=0.00008), rs30187 (OR 1.46, 95% CI: 1.18–1.80, P=0.0001), and rs27524 (OR 1.28, 95% CI: 1.04–1.58, P=0.02) in the group of patients with onset between 10 and 20 years. The association with ERAP1 in the 10–20 age group was significantly stronger compared with all other age groups (rs26653; OR 1.28 95% CI: 1.01–1.62, P=0.04, rs30187; OR 1.30, 95% CI: 1.03–1.63, P=0.03). In the group with age at onset between 21 and 40 years, there was borderline association for rs26653 (OR 1.27, 95% CI: 1.05–1.54, P=0.05).
No significant difference in ERAP1 association was found when we stratified for the clinical phenotypes, plaque or guttate (Supplementary Figure S1 online).
Discussion
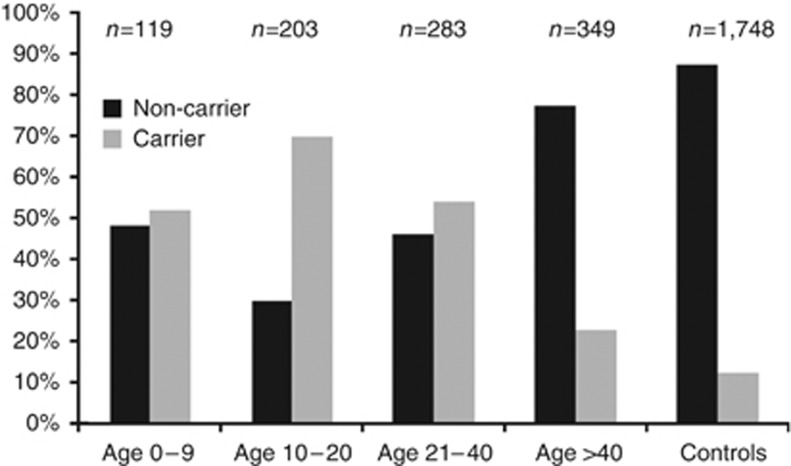
The present view within the field of psoriasis is that the earlier the onset of disease, the stronger the association with HLA-C*06:02 and the more severe the disease (Gudjonsson et al., 2006). The data presented herein reveal a more complex scenario where patients with onset of psoriasis during adolescence (here 10–20 years) emerge as a distinct group compared with those with onset before puberty (Figure 1). Children with disease onset below the age of 10 years have a prevalence of HLA-C*06:02 similar to that of patients with adult onset (21–40 years) and lack association with ERAP1, whereas cases with onset of disease between 10 and 20 years have a significantly higher prevalence of HLA-C*06:02 irrespective of phenotype and also show a significant association with ERAP1 (Table 3). These data indicate an age-dependent difference in the genetic background among patients with early onset of psoriasis. Keeping in mind the limited number of cases in each group, we believe that the stringent inclusion criteria and careful phenotyping by only a handful of dermatologists around the time of disease onset serve to reinforce the results. Whether these genetic differences will translate into clinical differences in terms of disease development, severity, and comorbidity profiles should be explored in future studies.
Figure 1.
HLA-C*06:02 carrier and noncarrier distribution in the whole sample set. Frequency of HLAC*06:02 allele in the different age-at-onset groups.
Obviously, any stratification involves simplification, but we believe that a cutoff at 10 years of age represents a valid approximation for puberty that constitutes a significant biological transition (Parent et al., 2003). In patients with juvenile psoriatic arthritis a similar pattern of two distinct populations of early-onset disease was presented, one with a peak of onset around the age of 2–3 years and the other group with a peak of onset around 10–12 years. Genetic and clinical differences were detected between these groups of juvenile psoriatic arthritis patients (Stoll et al., 2006; Stoll and Punaro, 2011). An additional study on juvenile arthritis showed subtype-specific association with ERAP1 and IL23R, in which the enthesitis-related arthritis subtype associated with ERAP1, whereas juvenile psoriatic arthritis showed association with IL23R (Hinks et al., 2011). A recent study in a small group of childhood psoriasis patients, separating early and late onset at 18 years of age, showed results in accordance with the data presented herein, with a stronger association with ERAP1 rs27524 in the group with onset of disease before 18 years (Bergboer et al., 2012).
A peak of disease onset at puberty has been shown in cases of psoriasis, which is slightly earlier in girls than in boys (Swanbeck et al., 1995). The mechanisms underlying this increase are not known today. A possible effect of increased levels of sex hormones at puberty, in particular estrogens, which are known to promote keratinocyte proliferation via receptor-mediated mechanisms and also to influence inflammatory pathways, has been discussed (Moverare et al., 2002; Kanda and Watanabe, 2005). Other potential triggers may be different infectious panorama in different age groups.
The distribution of phenotypes in the present study reveals a relatively high proportion of the guttate subtype. This may in part be explained by the recruitment strategy, which has been described in detail (Mallbris et al., 2005). In short, patients were recruited in association with onset of first psoriasis episode and the diagnosis verified by a trained dermatologist in a single center. In the majority of patients (77%) an associated streptococcal infection was confirmed. Earlier studies have shown a higher prevalence of HLA-C*06:02 in patients with guttate psoriasis (Mallon et al., 2000; Gudjonsson et al., 2006). The distribution between phenotypes (plaque versus guttate) in prepubertal and postpubertal children was similar in our study (Table 1). Thus, differences in HLA-C*06:02 prevalence between prepubertal children and postpubertal children cannot be attributed to a higher percentage of the guttate phenotype in the latter group but was rather contributed by a larger percentage of plaque cases carrying HLA-C*06:02 in the 10–20-year age group.
As expected, cases with late onset of disease (above 40 years) had significantly lower association with HLA-C*06:02 and also lacked association with ERAP1, confirming that 40 years represents a biologically valid approximation for a genetically distinct subgroup of psoriasis.
This work replicates the association of ERAP1 in psoriasis. We studied three SNPs in the ERAP1 gene (Table 2). The SNPs rs26653 (Arg127Pro) and rs30187 (Arg528Lys) have been shown to be associated with ankylosing spondylitis (rs26653 and rs30187) and multiple sclerosis (rs30187) and showed association with psoriasis in the present study (Harvey et al., 2009; Maksymowych et al., 2009; Reveille, 2011; Guerini et al., 2012). In contrast to the previously reported synonymous SNP rs27524, these latter SNPs are nonsynonymous and have been shown to be functional, affecting the catalytic activity of the enzyme (Goto et al., 2006; Mehta et al., 2009; Evnouchidou et al., 2011). The lack of association with the synonymous ERAP1 SNP rs27524 in our data set could be because of two reasons: (a) our associated group (age 10–20) comprises only 21% of our total cases, which may not be sufficient to reach significance. If we randomly reduce the number of cases in the other age groups so that the 10–20-year group would make up 36% of the total data set, comparable to data recently presented in Bergboer et al. (2012), the association with rs27524 would be significant in our study as well (Bergboer et al., 2012). (b) There are too few individuals carrying two minor alleles as rs27524 follows a recessive model, meaning that two minor alleles are needed for risk (Supplementary Table S2 online). The SNPs rs26653 and rs30187 on the other hand display an additive genotypic effect (Supplementary Table S2 online). The finding of significant association with rs30187 strengthens this conclusion, as rs30187 and rs27524 are in linkage disequilibrium.
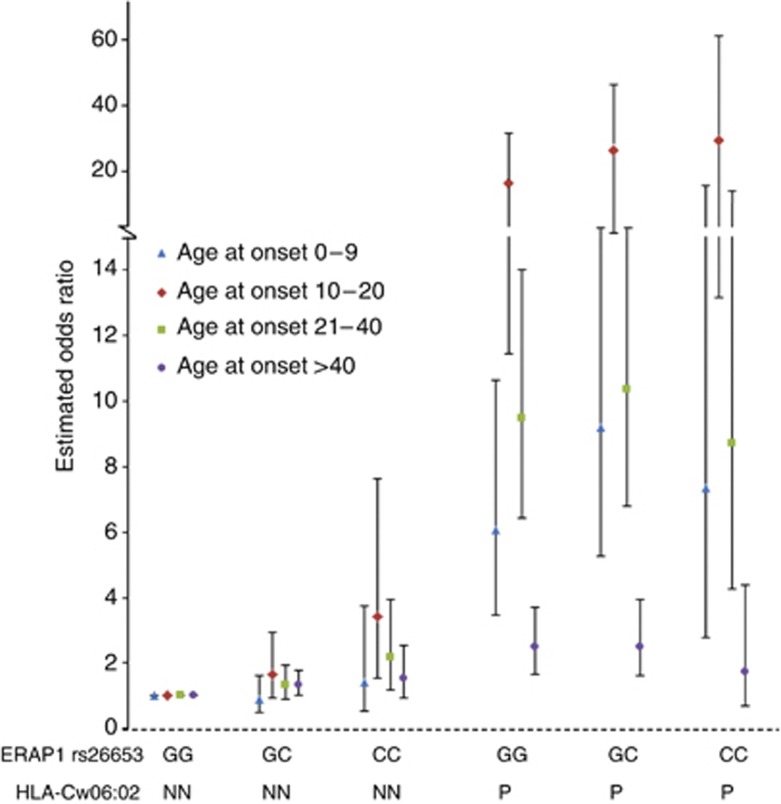
Thus, stratification for age at onset revealed a clear age-dependent association with ERAP1 (Table 3). Surprisingly, in our cases, association with ERAP1 was not restricted to individuals carrying HLA-C*06:02, and patients negative for HLA-C*06:02 with the rs26653 minor allele show an OR of 1.39 (95% CI: 1.14–1.70, P=0.001) (Table 4a). The additive effect of carrying the minor allele (C) for rs26653 in HLA-C*06:02-positive patients was seen between GG/P and GC/P genotype groups (Figure 2). Calculations for additive interaction and multiplicative interaction with stratification for different phenotypic and genetic data did not reveal any interactions. At present, we cannot explain the discrepancy between our findings and published data on ERAP1 association being dependent on HLA-C*06:02 (Strange et al., 2010). In the latter publication, the authors applied typing of rs10484554 to determine HLA-C status, which is different from the present method, which captures only HLA-C*06 genotypes (Nikamo and Stahle, 2012). However, using rs10484554 in our study did not alter the results (not shown).
Figure 2.
Association for ERAP1 SNP rs26653 stratified for HLA-Cw06 status and age. Estimates of genotype effects were calculated using logistic regression in the R software package. Individuals with the low-risk genotypes for rs26653 GG and HLA-C*06 NN were set as the baseline. The other genotype combinations were coded according to a series of dichotomous indicator variables. Odds ratios were derived by exponentiation of the relevant coefficient from the logistic regression.
At present, the functional consequences of ERAP1 polymorphisms in psoriasis are unknown. ERAP1 has several functions. It may act as a “molecular ruler” clipping peptides for optimal HLA class I binding and presentation (Saric et al., 2002; Chang et al., 2005). Second, in vitro studies suggest that ERAP1 cleaves cytokine receptors off the cell surface, including tumor necrosis factor receptor I (Cui et al., 2002), IL-1R2 (Cui et al., 2003b), and IL6Rα (Cui et al., 2003a). However, a recent study showed that the ERAP1 polymorphisms associated with ankylosing spondylitis did not influence the serum cytokine receptor levels in patients with ankylosing spondylitis (Haroon et al., 2010). The peptide-trimming function theoretically implies a biological interaction with HLA and has so far been thought to be causative for most of the effect in psoriasis. However, functional studies are lacking, and the importance of a “sheddase” function for ERAP1 in psoriasis and other inflammatory needs further exploration.
To investigate whether clinical subtype (plaque or guttate) influenced association with ERAP1, we stratified for phenotype. No significant difference was detected (Supplementary Figure S1 online).
In summary, we found that childhood psoriasis is a genetically heterogeneous group with significant differences between prepubertal and postpubertal disease onset. Prepubertal cases had a similar profile for HLA-C and ERAP1 as the group with onset between 21 and 40 years, whereas the group with onset between 10 and 20 years emerges as a distinct cluster carrying the strongest genetic HLA-C and ERAP1 association. In contrast to previous reports, the association with ERAP1 (rs26653) was neither dependent on nor interacting with HLA-C*06:02. Our work stresses the importance of detailed stratification for age at disease onset in further genetic and mechanistic studies in psoriasis. We do acknowledge the importance of replicating the present findings in a separate cohort of psoriasis patients. It will then be crucial to use a stringent clinical classification of the psoriasis diagnosis, which may be particularly difficult in children. In addition, reliable data on age at onset must be secured to avoid dilution of data.
Materials and Methods
Study population
All patients included in the study were examined and the diagnosis of psoriasis verified by dermatologists at the department of Dermatology, Karolinska University Hospital, Stockholm, Sweden. The study was approved by the Regional Committee of Ethics and conducted according to the Declaration of Helsinki Principles. Patients and controls gave their informed consent; for children younger than 18 years of age, consent was also obtained from their parents. For inclusion, patients needed to present with at least one typical psoriasis lesion at the time of examination. Scalp involvement and/or diaper rash alone was not regarded as diagnostic. Only patients categorized as having plaque or guttate psoriasis were enrolled in the study to minimize clinical heterogeneity (Table 1). Other more rare phenotypes were excluded. Disease severity ranged from mild to severe psoriasis, representing a cross-section of psoriasis patients. Healthy controls were recruited from the Dermatology (n=513) clinic at the Karolinska University Hospital and from the Epidemiological Investigation of Rheumatoid arthritis study (EIRA study) (n=1235) (Stolt et al., 2003). Only cases and controls of Caucasian origin based on ethnicity SNP genotyping were included (Giardina et al., 2008). Controls were matched for sex (59% female/41% male).
Genotyping
Peripheral blood samples were collected and genomic DNA extracted by standard procedures. SNPs were selected on the basis of previous publications (Haroon and Inman, 2010; Strange et al., 2010; Szczypiorska et al., 2011). All SNPs were genotyped on a 7900HT Fast Real-Time PCR System Instrument by using allele-specific Taqman MGB probes labeled with fluorescent dyes FAM and VIC (Applied Biosystems), according to the manufacturer's protocols. Allelic discrimination was made with the ABI PRISM 7900HT SDS and the SDS 2.2.1 program (Applied Biosystems). Ten percent of the samples were run as duplicates to check for genotyping errors. HLA-C*06:02 typing was carried out as described and referred to as positive when the HLA-C*06:02 allele was present and negative when not present (Nikamo and Stahle, 2012). We studied three SNPs in the ERAP1 gene, rs26653 (Arg127Pro), rs30187 (Lys528Arg), and rs27524 (UTR3′). rs27524 and rs30187 were in linkage disequilibrium (r2=0.75).
Statistical analysis
Case–control analysis was performed to test genetic markers for susceptibility to psoriasis. Case–case analysis was used to compare frequencies of genetic markers in plaque and guttate phenotypes and for different age groups. Both case–control and case–case analyses were tested for allele frequency differences for the three ERAP1 SNPs (rs26653 (Arg127Pro), rs30187 (Lys528Arg), and rs27524 (UTR3′)) and for the HLA-C genotype using logistic regression. Association between genetic markers and disease status was analyzed using PLINK v1.07 (Purcell, 2007, #85). Association for ERAP1 SNP rs26653 stratified for HLA-C*06:02 status and age, as well as independent association for ERAP1 using patients negative for ERAP1 and HLA-C*06:02 minor allele as the reference group, was calculated using R v2.15.0 with GLM (http://www.r-project.org/). We also used R v2.15.0 with GLM (http://www.r-project.org/) to test for interaction between ERAP1 and HLA-C*06:02 using both additive and multiplicative modes. Hardy–Weinberg equilibrium was evaluated for each SNP using the χ2-test. Significant P-values were corrected for multiple testing using the adjust mode as implemented in PLINK v1.07 (Purcell, 2007 #85).
Acknowledgments
This work was funded by the Medical Research Council, Swedish Psoriasis Association, Welander Finsen Foundation, Berth von Kantzow's Foundation, Karolinska Institutet, the Magnus Bergwall Foundation, and the Royal Physiographic Society in Lund. We express our gratitude to patients and controls who were part of this study and to those who contributed to this work. In addition to the authors (JL and MS), dermatologists who performed clinical phenotyping of patients were: Lotus Mallbris, Katarina Wolk, and Petra Kjellman. The research nurses involved were Annelie Gren, Susanne Bergqvist, Papeli Kassari, Helena Griehsel, and Maria Lundqvist. Technical assistance from Anna-Lena Kastman and Kerstin Bergh is gratefully acknowledged.
Glossary
- 95% CI
95% confidence interval
- ERAP1
endoplasmic reticulum aminopeptidase 1
- OR
odds ratio
- SNP
single-nucleotide polymorphism
The authors state no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Allen MH, Ameen H, Veal C, et al. The major psoriasis susceptibility locus PSORS1 is not a risk factor for late-onset psoriasis. J Invest Dermatol. 2005;124:103–106. doi: 10.1111/j.0022-202X.2004.23511.x. [DOI] [PubMed] [Google Scholar]
- Bergboer JG, Oostveen AM, de Jager ME, et al. 2012Paediatric onset psoriasis is associated with ERAP1 and IL23R loci, LCE3C_LCE3B deletion and HLA-C*06 Br J Dermatole-pub ahead of print 18 April 2012 [DOI] [PubMed]
- Braun-Falco O, Burg G, Farber EM. [Psoriasis. A questionnaire study of 536 patients] Munch Med Wochenschr. 1972;114:1105–1110. [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Momburg F, Bhutani N, et al. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci USA. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Hawari F, Alsaaty S, et al. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J Clin Invest. 2002;110:515–526. doi: 10.1172/JCI13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Rouhani FN, Hawari F, et al. An aminopeptidase, ARTS-1, is required for interleukin-6 receptor shedding. J Biol Chem. 2003a;278:28677–28685. doi: 10.1074/jbc.M300456200. [DOI] [PubMed] [Google Scholar]
- Cui X, Rouhani FN, Hawari F, et al. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J Immunol. 2003b;171:6814–6819. doi: 10.4049/jimmunol.171.12.6814. [DOI] [PubMed] [Google Scholar]
- de Jager ME, de Jong EM, Meeuwis KA, et al. No evidence found that childhood onset of psoriasis influences disease severity, future body mass index or type of treatments used. J Eur Acad Dermatol Venereol. 2010;24:1333–1339. doi: 10.1111/j.1468-3083.2010.03645.x. [DOI] [PubMed] [Google Scholar]
- Elder JT. PSORS1: linking genetics and immunology. J Invest Dermatol. 2006;126:1205–1206. doi: 10.1038/sj.jid.5700357. [DOI] [PubMed] [Google Scholar]
- Evnouchidou I, Kamal RP, Seregin SS, et al. Coding single-nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186:1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Yang S, Huang W, et al. Fine mapping of the psoriasis susceptibility locus PSORS1 supports HLA-C as the susceptibility gene in the Han Chinese population. PLoS Genet. 2008;4:e1000038. doi: 10.1371/journal.pgen.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber EM, Jacobs AH. Infantile psoriasis. Am J Dis Child. 1977;131:1266–1269. doi: 10.1001/archpedi.1977.02120240084019. [DOI] [PubMed] [Google Scholar]
- Farber EM, Nall ML. The natural history of psoriasis in 5,600 patients. Dermatologica. 1974;148:1–18. doi: 10.1159/000251595. [DOI] [PubMed] [Google Scholar]
- Giardina E, Pietrangeli I, Martinez-Labarga C, et al. Haplotypes in SLC24A5 gene as ancestry informative markers in different populations. Curr Genomics. 2008;9:110–114. doi: 10.2174/138920208784139528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Hattori A, Ishii Y, et al. Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 2006;580:1833–1838. doi: 10.1016/j.febslet.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Karason A, Runarsdottir EH, et al. Distinct clinical differences between HLA-Cw*0602 positive and negative psoriasis patients—an analysis of 1019 HLA-C- and HLA-B-typed patients. J Invest Dermatol. 2006;126:740–745. doi: 10.1038/sj.jid.5700118. [DOI] [PubMed] [Google Scholar]
- Guerini FR, Cagliani R, Forni D, et al. A functional variant in ERAP1 predisposes to multiple sclerosis. PLoS One. 2012;7:e29931. doi: 10.1371/journal.pone.0029931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon N, Inman RD. Endoplasmic reticulum aminopeptidases: biology and pathogenic potential. Nat Rev Rheumatol. 2010;6:461–467. doi: 10.1038/nrrheum.2010.85. [DOI] [PubMed] [Google Scholar]
- Haroon N, Tsui FW, Chiu B, et al. Serum cytokine receptors in ankylosing spondylitis: relationship to inflammatory markers and endoplasmic reticulum aminopeptidase polymorphisms. J Rheumatol. 2010;37:1907–1910. doi: 10.3899/jrheum.100019. [DOI] [PubMed] [Google Scholar]
- Harvey D, Pointon JJ, Evans DM, et al. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum Mol Genet. 2009;18:4204–4212. doi: 10.1093/hmg/ddp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn A, York IA, Rock KL. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J Immunol. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13:450–456. doi: 10.1016/s0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- Hinks A, Martin P, Flynn E, et al. Subtype specific genetic associations for juvenile idiopathic arthritis: ERAP1 with the enthesitis related arthritis subtype and IL23R with juvenile psoriatic arthritis. Arthritis Res Ther. 2011;13:R12. doi: 10.1186/ar3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda N, Watanabe S. Regulatory roles of sex hormones in cutaneous biology and immunology. J Dermatol Sci. 2005;38:1–7. doi: 10.1016/j.jdermsci.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Maksymowych WP, Inman RD, Gladman DD, et al. Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arthritis Rheum. 2009;60:1317–1323. doi: 10.1002/art.24467. [DOI] [PubMed] [Google Scholar]
- Mallbris L, Larsson P, Bergqvist S, et al. Psoriasis phenotype at disease onset: clinical characterization of 400 adult cases. J Invest Dermatol. 2005;124:499–504. doi: 10.1111/j.0022-202X.2004.23611.x. [DOI] [PubMed] [Google Scholar]
- Mallon E, Bunce M, Savoie H, et al. HLA-C and guttate psoriasis. Br J Dermatol. 2000;143:1177–1182. doi: 10.1046/j.1365-2133.2000.03885.x. [DOI] [PubMed] [Google Scholar]
- Mehta AM, Jordanova ES, Corver WE, et al. Single-nucleotide polymorphisms in antigen processing machinery component ERAP1 significantly associate with clinical outcome in cervical carcinoma. Genes Chromosomes Cancer. 2009;48:410–418. doi: 10.1002/gcc.20648. [DOI] [PubMed] [Google Scholar]
- Morris A, Rogers M, Fischer G, et al. Childhood psoriasis: a clinical review of 1262 cases. Pediatr Dermatol. 2001;18:188–198. doi: 10.1046/j.1525-1470.2001.018003188.x. [DOI] [PubMed] [Google Scholar]
- Moverare S, Lindberg MK, Faergemann J, et al. Estrogen receptor alpha, but not estrogen receptor beta, is involved in the regulation of the hair follicle cycling as well as the thickness of epidermis in male mice. J Invest Dermatol. 2002;119:1053–1058. doi: 10.1046/j.1523-1747.2002.00637.x. [DOI] [PubMed] [Google Scholar]
- Nikamo P, Stahle M. Cost-effective HLA-Cw06:02 typing in a Caucasian population. Exp Dermatol. 2012;21:221–223. doi: 10.1111/j.1600-0625.2011.01423.x. [DOI] [PubMed] [Google Scholar]
- Parent AS, Teilmann G, Juul A, et al. The timing of normal puberty and the age limits of sexual precocity: variations around the world, secular trends, and changes after migration. Endocr Rev. 2003;24:668–693. doi: 10.1210/er.2002-0019. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri SP, Gross J. A comparative study of pediatric onset psoriasis with adult onset psoriasis. Pediatr Dermatol. 2000;17:174–178. doi: 10.1046/j.1525-1470.2000.01746.x. [DOI] [PubMed] [Google Scholar]
- Reveille JD. The genetic basis of spondyloarthritis. Ann Rheum Dis. 2011;70 (Suppl 1:i44–i50. doi: 10.1136/ard.2010.140574. [DOI] [PubMed] [Google Scholar]
- Saric T, Chang SC, Hattori A, et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- Stoll ML, Punaro M. Psoriatic juvenile idiopathic arthritis: a tale of two subgroups. Curr Opin Rheumatol. 2011;23:437–443. doi: 10.1097/BOR.0b013e328348b278. [DOI] [PubMed] [Google Scholar]
- Stoll ML, Zurakowski D, Nigrovic LE, et al. Patients with juvenile psoriatic arthritis comprise two distinct populations. Arthritis Rheum. 2006;54:3564–3572. doi: 10.1002/art.22173. [DOI] [PubMed] [Google Scholar]
- Stolt P, Bengtsson C, Nordmark B, et al. Quantification of the influence of cigarette smoking on rheumatoid arthritis: results from a population based case-control study, using incident cases. Ann Rheum Dis. 2003;62:835–841. doi: 10.1136/ard.62.9.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanbeck G, Inerot A, Martinsson T, et al. Age at onset and different types of psoriasis. Br J Dermatol. 1995;133:768–773. doi: 10.1111/j.1365-2133.1995.tb02753.x. [DOI] [PubMed] [Google Scholar]
- Szczypiorska M, Sanchez A, Bartolome N, et al. ERAP1 polymorphisms and haplotypes are associated with ankylosing spondylitis susceptibility and functional severity in a Spanish population. Rheumatology. 2011;50:1969–1975. doi: 10.1093/rheumatology/ker229. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang H, Te-Shao H, et al. The genetic epidemiology of psoriasis vulgaris in Chinese Han. Int J Dermatol. 2002;41:663–669. doi: 10.1046/j.1365-4362.2002.01596.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.