SUMMARY
Here we identify and characterize a cytoskeletal myosin protein required for IRE1α oligomerization, activation, and signaling. Proteomic screening identified nonmuscle myosin heavy chain IIB (NMHCIIB), a subunit of nonmuscle myosin IIB (NMIIB), as an ER stress-dependent interacting protein specific to IRE1α. Loss of NMIIB compromises XBP1s and UPR target gene expression with no effect on the PERK pathway. Mechanistically, NMIIB is required for IRE1α aggregation and foci formation under ER stress. The NMIIB-mediated effect on IRE1α signaling is in part dependent on the phosphorylation of myosin regulatory light chain and the actomyosin contractility of NMIIB. Biologically, the function of NMIIB in ER stress response is conserved as both mammalian cells and C. elegans lacking NMIIB exhibit hypersensitivity to ER stress. Thus, optimal IRE1α activation and signaling require concerted coordination between the ER and cytoskeleton.
INTRODUCTION
Perturbations in endoplasmic reticulum (ER) homeostasis culminate in ER stress and activate an ER-to-nucleus signal transduction cascade termed the unfolded protein response (UPR) (Hetz et al., 2011; Walter and Ron, 2011). In mammals, the ER luminal environment is sensitively monitored by three transmembrane sensors: inositol-requiring enzyme 1α (IRE1α), PKR-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6), with IRE1α as the most conserved (Hetz et al., 2011; Walter and Ron, 2011). A current model of mammalian IRE1α activation speculates that upon alterations in ER homeostasis, the luminal domain of IRE1α undergoes a conformational change and subsequent homodimerization. Juxtaposition of IRE1α proteins promotes transautophosphorylation and may facilitate higher-order oligomerization of IRE1α dimers, followed by activation of its ribonuclease (RNase) domain to splice 26 nucleotides from the X-box binding protein 1 (Xbp1) mRNA (Walter and Ron, 2011). This processing event produces a transcription factor XBP1 spliced (XBP1s) required for UPR target gene induction (Acosta-Alvear et al., 2007; He et al., 2010). Given that activation of the IRE1α-XBP1 pathway is emerging as a central paradigm underlying human pathologies, such as cancer, diabetes, and conformational diseases (Hetz et al., 2011; Sha et al., 2011), a comprehensive and mechanistic understanding of IRE1α activation is of critical therapeutic value.
One model for IRE1α activation proposes that under nonstress conditions, IRE1α is bound in an inactive state by the ER chaperone, glucose-regulated protein 78 (GRP78/BiP) (Bertolotti et al., 2000; Kimata et al., 2003). Accumulation of misfolded proteins promotes BiP dissociation, allowing for IRE1α homodimerization and activation. Alternatively, misfolded proteins may activate IRE1α through direct binding (Credle et al., 2005; Gardner and Walter, 2011; Kimata et al., 2007) to promote its oligomerization and optimal activation (Korennykh et al., 2009; Li et al., 2010; Walter and Ron, 2011). These two models are not mutually exclusive and may have different implications for yeast and mammalian IRE1α proteins (Oikawa et al., 2009). Here we identify a cytosolic factor required for mammalian IRE1α aggregation and optimal activation of this pathway.
Nonmuscle myosin II (NMII), a member of the myosin II motor superfamily (Ma and Adelstein, 2012), is a hexameric molecule composed of a pair of heavy chains (NMHCs), a pair of regulatory light chains (RLCs), and a pair of essential light chains (Vicente-Manzanares et al., 2009). In mammals, the identity of the heavy chain, NMHCIIA, NMHCIIB, or NMHCIIC determines the overall NMII isoform (Vicente-Manzanares et al., 2009). NMII activation is largely mediated by phosphorylation of its RLC at serine residue 19 (Ser19) (Adelstein and Conti, 1975) and to a lesser extent by increased transcriptional activity (Vicente-Manzanares et al., 2009). NMII has been widely studied in distinct biological processes that require remodeling of the actin cytoskeleton, such as cell migration, polarity determination, cell adhesion, and clustering of signal transduction molecules (Ma and Adelstein, 2012; Vicente-Manzanares et al., 2009). Consequently, NMII-dependent processes span diverse physiological functions, including tumor-necrosis factor signaling, T cell antigen receptor clustering, and viral entry (Arii et al., 2010; Flynn and Helfman, 2010; Ilani et al., 2009). Here, we report an indispensable role of NMIIB in IRE1α signaling of UPR.
RESULTS
NMHCIIB Physically Interacts with IRE1α upon ER Stress
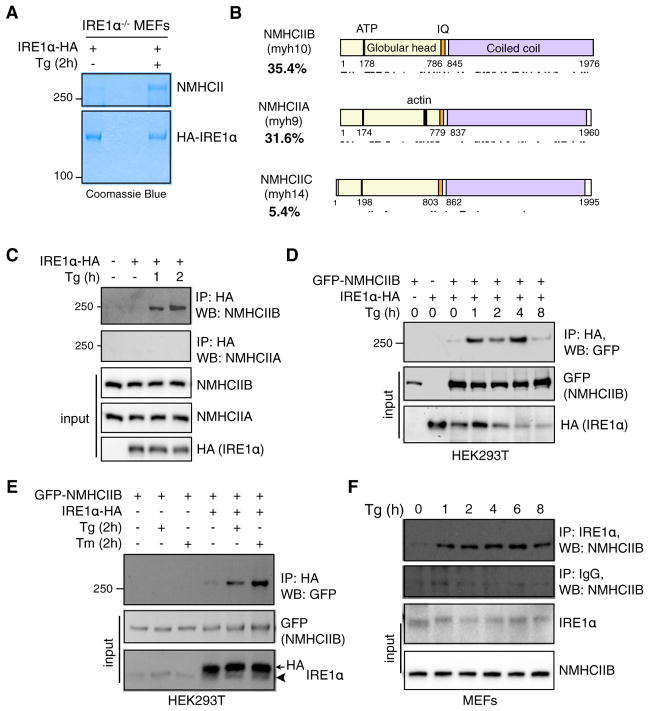
To investigate mechanisms regulating mammalian IRE1α signaling, we performed a proteomic screen using IRE1α−/− mouse embryonic fibroblasts (MEFs) stably expressing hemagglutinin (HA)-tagged IRE1α to identify potential IRE1α-interacting proteins. Coomassie blue staining revealed a band approximately 250 kDa in size that was responsive to thapsigargin (Tg)-induced ER stress (Figure 1A). Tandem mass spectrometry (MS/MS) identified this factor as the heavy chain of nonmuscle myosin II (NMHCII) with 35.4% identity to NMHCIIB, 31.6% to NMHCIIA, and 5.4% to NMHCIIC; peptide coverage spanned all functional domains (Figure 1B).
Figure 1. NMIIB Is an ER Stress-Induced IRE1α-Interacting Factor.
(A) Coomassie blue staining of immunoprecipitates (IPs) of IRE1α−/− MEFs stably expressing C-terminal hemagglutinin (HA)-tagged IRE1α untreated or treated with 300 nM Tg for 2 hr. Unknown Tg-specific band at 3250 kDa was excised and identified as NMHCII using tandem mass spectrometry (MS/MS) analysis.
(B) Schematic of the functional domains of the three mammalian isoforms of NMHCII with respective peptide coverage recovered (indicated by lines below) from MS/MS. Figure is drawn to scale using sequence annotation data from UniProt.
(C) Western blot showing recovery of endogenous NMHCIIB and NMHCIIA from immunoprecipitates of HA-tagged IRE1α prepared from transiently transfected HEK293T cells treated with 150 nM Tg for the indicated time.
(D and E) Western blot showing recovery of GFP-tagged NMHCIIB from IPs of HA-tagged IRE1α prepared from transfected HEK293T cells treated with 300 nM Tg for the indicated time (D) or 300 nM Tg or 5 μg/ml Tm for 2 hr (E). Arrowhead points to endogenous protein.
(F) Western blot showing recovery of endogenous NMHCIIB from IPs of endogenous IRE1α prepared from MEFs treated with 150 nM Tg for the indicated time.
For (C)–(F), similar results were observed in 2–3 independent experiments.
As endogenous NMHCIIB interacted with IRE1α much more strongly than NMHCIIA (Figure 1C), the remainder of our study focused on NMIIB. The ER stress-dependent interaction between NMHCIIB and IRE1α was verified in HEK293T cells overexpressing IRE1α-HA and green fluorescent protein (GFP)-NMHCIIB, or IRE1α-HA alone (Figures 1C and 1D). Treatment with another ER stress agent tunicamycin (Tm) also promoted the interaction (Figure 1E), demonstrating that this event is dependent on ER stress and independent of nonspecific Ca2+ effects. Lastly, immunoprecipitation in MEFs with anti-IREα antibody confirmed the ER stress-dependent interaction between endogenous proteins (Figure 1F).
NMIIB Is Required and Sufficient for Optimal IRE1α Activation
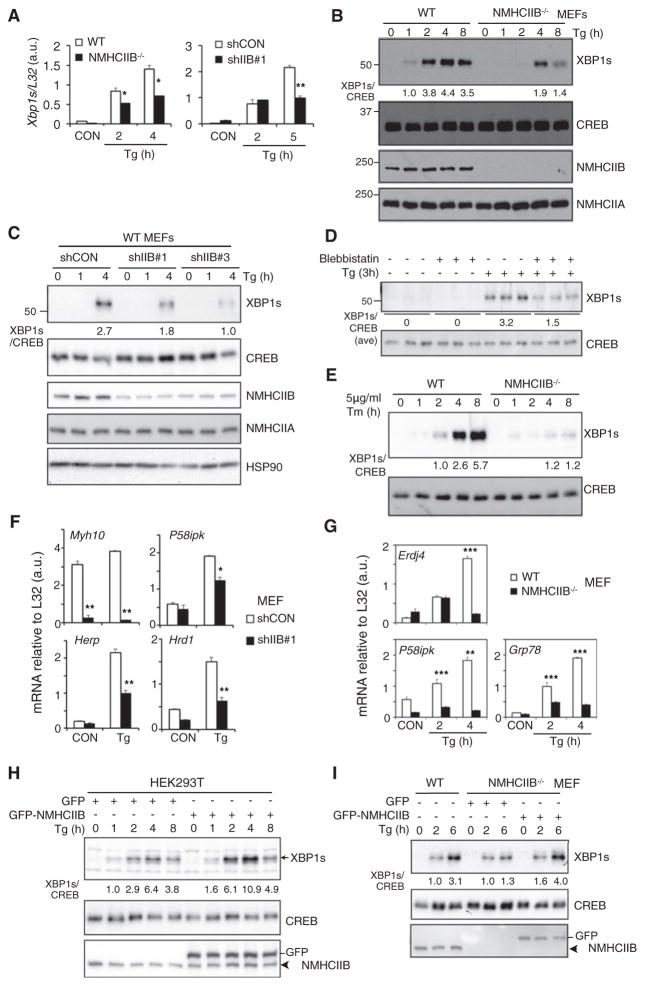
To study the functional role of NMIIB in IRE1α signaling, we used loss- and gain-of-function models. Induction of Xbp1s mRNA, a substrate of IRE1α, was blunted in NMHCIIB−/− and knockdown MEFs (Figure 2A). Concomitantly, nuclear XBP1s protein production was defective and delayed in NMHCIIB−/− MEFs (Figure 2B), NMHCIIB knockdown MEFs (Figure 2C) and cells treated with the NMII-specific inhibitor blebbistatin (Straight et al., 2003) (Figure 2D). A similar observation was made in NMHCIIB−/− MEFs treated with Tm (Figure 2E). Further pointing to defects in the IRE1α-XBP1 pathway, the expression of a subset of XBP1 target genes was attenuated in NMHCIIB knockdown (Figure 2F) and NMHCIIB−/− MEFs (Figure 2G).
Figure 2. NMIIB Is Required for Optimal IRE1α-XBP1 Signaling.
(A) Quantitative real-time PCR analysis of Xbp1s expression from NMHCIIB−/− or shIIB#1 MEFs treated with 150 nM Tg for the indicated time.
(B and C) Western blot of nuclear (XBP1s) and cytosolic extracts (NMHCIIA/B) from (B) WT and NMHCIIB−/− MEFs treated with 150 nM Tg for the indicated time or (C) WT MEFs stably expressing shRNA against control (shCON) or murine NMHCIIB (shIIB#1 and shIIB#3) treated with 150 nM Tg for the indicated time.
(D) Western blot of XBP1s in HEK293T cells untreated or pretreated with 50 μM blebbistatin followed by treatment with 150 nM Tg for 3 hr.
(E) Western blot of XBP1s in WT and NMHCIIB−/− MEFs treated with 5 μg/ml Tm for the indicated time.
(F and G) Quantitative real-time PCR analysis of UPR genes in (F) shCON and shIIB#1 MEFs treated with 150 nM Tg for 5 hr or (G) WT and NMHCIIB−/− MEFs treated with 150 nM Tg for the indicated time.
(H and I) Western blot of nuclear (XBP1s) and cytosolic extracts (NMHCIIA/B) from (H) HEK293T cells transiently overexpressing GFP vector or GFP-NMHCIIB treated with 150 nM Tg for the indicated time, or (I) WT or NMHCIIB−/− MEFs transiently overexpressing GFP vector or GFP-NMHCIIB treated with 150 nM Tg for the indicated time. Arrowhead points to endogenous protein. Quantitation of XBP1s protein levels shown below the gel after normalization to CREB. In western blots, CREB and HSP90 are loading controls. In quantitative real-time PCR analysis, data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. For all, similar results were observed in 2–3 independent experiments. See also Figure S1.
Conversely, overexpression of NMHCIIB in HEK293T cells increased XBP1s protein (Figure 2H) and overexpression in NMHCIIB−/− MEFs rescued the levels of XBP1s protein (Figure 2I). Notably, the gain-of-function effect was observed only under conditions of ER stress, further supporting the notion that NMIIB-mediated IRE1α signaling is ER stress dependent.
NMIIB Is Dispensable for the PERK Pathway
To determine the specificity of NMIIB for IRE1α, we studied its impact on the other two major UPR branches. Upon ER stress, PERK undergoes transautophosphorylation and subsequently phosphorylates Ser51 on eukaryotic initiation factor 2α (eIF2α), leading to attenuation of global protein synthesis. Paradoxically, this event selectively upregulates the translation of a subset of genes, including Chop (Walter and Ron, 2011). Upon ER stress, no interaction occurred between PERK and NMHCIIB (Figure S1A available online). Furthermore, PERK activation and signaling was unaffected in NMHCIIB knockdown MEFs as both PERK and eIF2α phosphorylation and Chop transcript levels were unchanged (Figures S1B–S1D). We were unable to study the relationship between NMIIB and ATF6 activation because of the lack of a good antibody against the endogenous active form of ATF6. Nonetheless, our data point to a specific role of NMHCIIB in IRE1α signaling and dispensable for the PERK pathway.
IRE1α Foci Formation and Oligomerization Require NMIIB
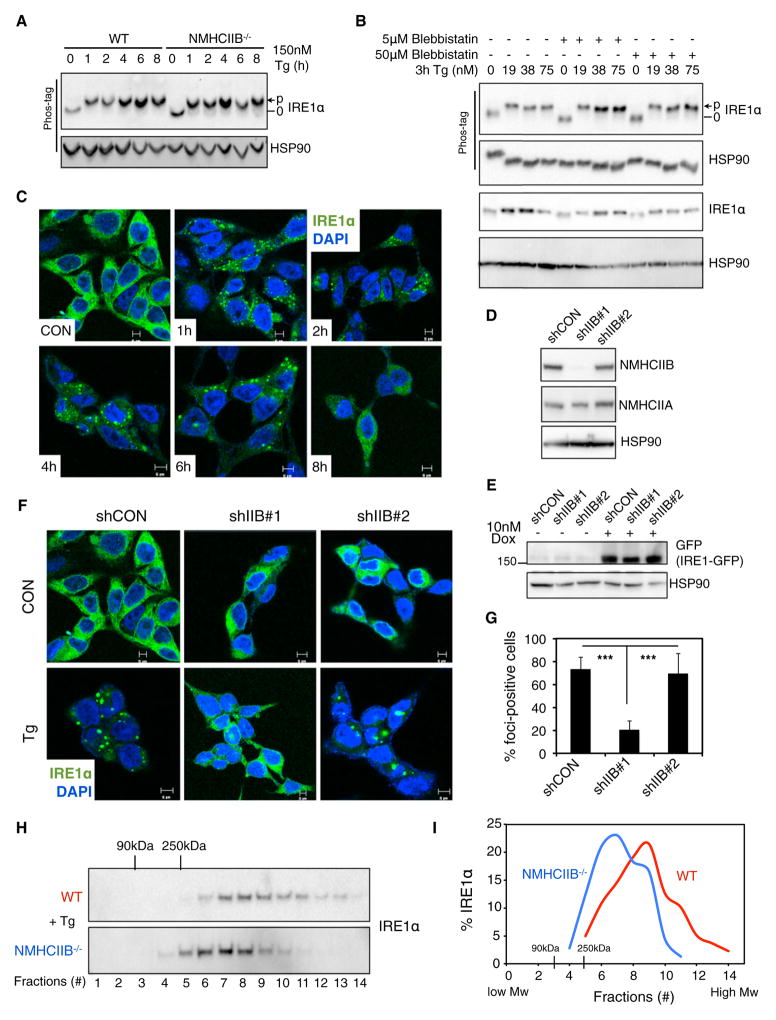
To understand mechanistically how NMIIB affects IRE1α signaling, we investigated two key steps in IRE1α activation, namely, dimerization/transautophosphorylation and oligomerization (Walter and Ron, 2011). Upon ER stress, IRE1α phosphorylation (Qi et al., 2011; Yang et al., 2010) was not affected in NMHCIIB−/− MEFs (Figure 3A), cells exposed to blebbistatin (Figure 3B), or cells overexpressing NMHCIIB (data not shown), suggesting that NMIIB is dispensable for IRE1α dimerization and transautophosphorylation.
Figure 3. NMIIB Promotes IRE1α Foci Formation upon ER Stress.
(A and B) Phos-tag western blot of IRE1α phosphorylation prepared from lysates of (A) WT or NMHCIIB−/− MEFs treated with 150 nM Tg for the indicated time, and (B) MEF cells pretreated with Blebbistatin prior to treatment with Tg at the indicated concentration for 3 hr.
(C) Confocal microscopic images of IRE1α-GFP in T-REx293 IRE1-3F6HGFP cells untreated (CON) or treated with 300 nM Tg for the indicated time.
(D) Western blot of NMHCIIB in T-REx293 IRE1-3F6HGFP cells stably expressing shRNA against control (shCON) or human NMHCIIB (shIIB#1 and shIIB#2). HSP90, loading control.
(E) Western blot of GFP in T-REx293 cells treated with 10 nM doxycycline for 24 hr.
(F and G) Confocal microscopic images of IRE1α-GFP in T-REx293 IRE1-3F6HGFP cells stably expressing shCON, shIIB#1, or shIIB#2 cells untreated (CON) or treated with 300 nM Tg for 4 hr. Quantitation of the number of foci-positive cells from at least 30 fields with 500–1,000 cells per group are shown in (G). Data are shown as the mean ± SEM. ***p < 0.001.
(H and I) Western blot showing the distribution of IRE1α (H) and quantitation (I) in fractions 1–14 prepared from WT and NMHCIIB−/− MEFs treated with 300 nM Tg for 3 hr and centrifuged using 20%–40% sucrose gradients.
For all, similar results were observed in 2–3 independent experiments. See also Figure S2.
As ER stress persists, mammalian IRE1α dimers oligomerize to form foci (Credle et al., 2005; Korennykh et al., 2009) that may correlate with RNase activation and Xbp1 splicing (Li et al., 2010). This event can be visualized using a T-REx293 cell system in which expression of a GFP-tagged IRE1α protein, IRE1-3F6HGFP, is driven by a doxycycline-inducible promoter (Li et al., 2010). In line with previous studies (Korennykh et al., 2009; Li et al., 2010), IRE1α distribution changed dramatically upon ER stress from diffuse to strong punctate foci beginning as early as 1 hr and peaking at 4 hr (Figure 3C). Knockdown of NMHCIIB (shIIB#1) drastically reduced foci formation from 73.3% ± 10.6% in control cells (shCON) to 20.2% ± 7.7% foci-positive cells in shIIB#1; cells with a shRNA-targeting sequence that failed to deplete NMHCIIB (shIIB#2) retained the ability to form foci with 70.0% ± 17.6% foci-positive cells (Figures 3D–3G). This effect was consistently observed at various stages of ER stress (Figure S2). Of note, IRE1α-GFP protein levels in all cell lines were comparable (Figure 3E), thus excluding the possibility that reduced protein levels accounted for diminished foci formation.
To confirm this observation under endogenous conditions, we examined oligomerization of endogenous IRE1α proteins using sucrose gradient fractionation. Loss of NMIIB reduced the formation of higher-order complexes of endogenous IRE1α under ER stress (Figures 3H and 3I). Overall, these results indicate that NMIIB regulates IRE1α aggregation and foci formation during ER stress, while having no effect on IRE1α dimerization and transautophosphorylation.
Effect of NMIIB on IRE1α Signaling Requires RLC Phosphorylation
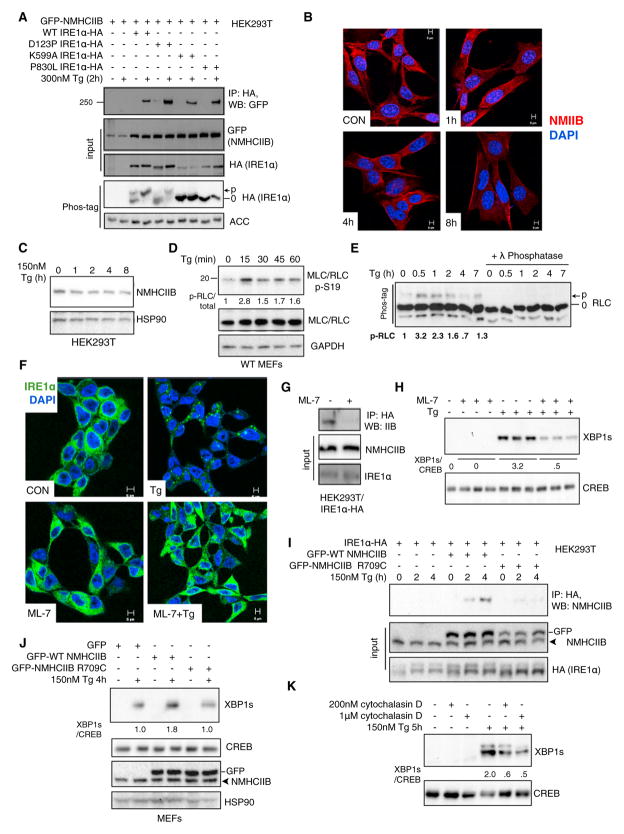
To understand the mechanistic basis of the ER stress-induced NMIIB-IRE1α complex, we first queried whether the interaction was contingent upon the activation status of IRE1α. We used to our advantage three IRE1α mutants: a dimerization-defective mutant D123P (Zhou et al., 2006), a kinase-dead mutant K599A (Tirasophon et al., 1998), and a loss-of-function mutant P830L lacking both kinase and RNase functions (Xue et al., 2011). Unexpectedly, the D123P IRE1α mutant that cannot undergo transautophosphorylation when stably expressed in IRE1α−/− MEFs (Xue et al., 2011) exhibited ER stress-induced phosphorylation in HEK293T cells (Figure 4A), presumably because of the presence of endogenous IRE1α. In contrast, both K599A and P830L IRE1α mutants were unable to undergo phosphorylation upon ER stress in HEK293T cells as shown previously (Xue et al., 2011). Surprisingly, both mutants interacted just as strongly with NMHCIIB as wild-type (WT) IRE1α (Figure 4A), suggesting that the interaction occurs independently of IRE1α kinase function and phosphorylation status.
Figure 4. RLC Phosphorylation and the Actomyosin Contractility of NMIIB Are Required for Optimal IRE1α Signaling.
(A) Western blot showing recovery of GFP-tagged NMHCIIB from immunoprecipitates of HA-tagged WT and mutant IRE1α (D123P, K599A, P830L) prepared from transfected HEK293T cells treated with 300 nM Tg for 2 hr. Bottom panel shows Phos-tag western blot analysis of IRE1α phosphorylation in HEK293T cells overexpressing WT or mutant IRE1α. ACC (acetyl-CoA carboxylase), loading control.
(B) Confocal microscopic analysis of endogenous NMHCIIB in MEF cells treated with 300 nM Tg for the indicated time. Images are representative of 25 fields. Scale bar, 5 μm.
(C) Western blot of NMHCIIB from HEK293T cells treated 150 nM Tg for the indicated time.
(D) Western blot of p-Ser19 and total RLC in whole-cell lysates prepared from MEFs treated with 300 nM Tg for indicated time.
(E) Phos-tag western blot analysis of total RLC phosphorylation in MEFs treated with 150 nM Tg for indicated time. Quantitation of p-RLC shown below.
(F) Confocal microscopic images of IRE1α-GFP in T-REx293 IRE1-3F6HGFP cells pretreated with 25 μM ML-7 for 30 min prior to treatment with 300 nM Tg for 4 hr. Data is representative of at least 30 fields with 500–1,000 cells per group. Scale bar, 5 μm.
(G) Western blot showing recovery of endogenous NMIIB from immunoprecipitates of HA-tagged IRE1α prepared from transiently transfected HEK293T cells untreated or pretreated with 25 μM ML-7 for 30 min prior to treatment with 300 nM Tg for 2 hr. Note that ML-7 pretreatment abolishes the interaction between IRE1α and NMIIB.
(H) Western blot analysis of nuclear XBP1s in HEK293T cells treated as in (F).
(I) Western blot showing recovery of GFP-tagged WT or the motor-defective R709C NMHCIIB mutant from immunoprecipitates of HA-tagged IRE1α prepared from transfected HEK293T cells treated with 150 nM Tg for the indicated time.
(J and K) Western blot of XBP1s in (J) MEFs transfected with GFP-tagged WT or R709C NMHCIIB treated with 150 nM Tg for 4 hr or (K) MEFs pretreated with an actin inhibitor, cytochalasin D, for 30 min prior to treatment with 150 nM Tg for 5 hr.
For all, similar results were observed in 2–3 independent experiments. CREB/GAPDH/HSP90, loading controls.
Next, we examined how ER stress may signal to NMIIB. Although ER stress had no noticeable effects on the intracellular distribution (Figure 4B) or protein levels (Figure 4C) of NMHCIIB, it induced phosphorylation of RLC at Ser19 (Figure 4D), a critical triggering event in NMII activation (Adelstein and Conti, 1975; Ma and Adelstein, 2012; Vicente-Manzanares et al., 2009). Additionally, an independent Phos-tag-based method examining total RLC phosphorylation (Figure 4E) demonstrated that RLC phosphorylation peaked within 60 min following ER stress (Figures 4D and 4E), preceding IRE1α foci formation ((Li et al., 2010) and Figure 3C).
Further demonstrating the significance of RLC phosphorylation, treatment with a myosin light chain kinase (MLCK)-specific inhibitor ML-7 (Saitoh et al., 1987) abolished ER stress-dependent IRE1α foci formation (Figure 4F), reduced the interaction between NMHCIIB and IRE1α (Figure 4G) and dramatically attenuated XBP1s protein levels by over 6-fold (Figure 4H). Thus, the physical interaction between NMIIB and IRE1α as well as the effect of NMIIB on IRE1α aggregation and signaling are largely dependent on MLCK-mediated RLC phosphorylation.
Motor Activity of NMIIB Is Indispensable for IRE1α Activation and Signaling
As NMIIB contains several functional domains (Vicente-Manzanares et al., 2009), we next queried whether the motor activity was required for optimal IRE1α signaling. Unlike wild-type NMHCIIB, a motor-defective mutant R709C with diminished ATPase activity (Kim et al., 2005; Takeda et al., 2003) did not exhibit ER stress-dependent association with IRE1α (Figure 4I), and overexpression of the mutant failed to enhance XBP1s protein expression upon ER stress (Figure 4J).
As myosin molecules propel along actin filaments to provide movement through the energy of ATP hydrolysis (Vicente-Manzanares et al., 2009), we next addressed the role of actin. Consistently, cells pretreated with an actin inhibitor cytochalasin D exhibited attenuated XBP1s levels (Figure 4K). A similar observation was obtained in cells treated with blebbistatin (Figure 2D), a drug specifically inhibiting the ATPase activity and motility of NMII proteins (Straight et al., 2003). Therefore, our data collectively suggest that the actomyosin contractility of NMIIB is required for IRE1α signaling.
NMIIB-Deficient Mammalian Cells Are Defective in ER Stress Response
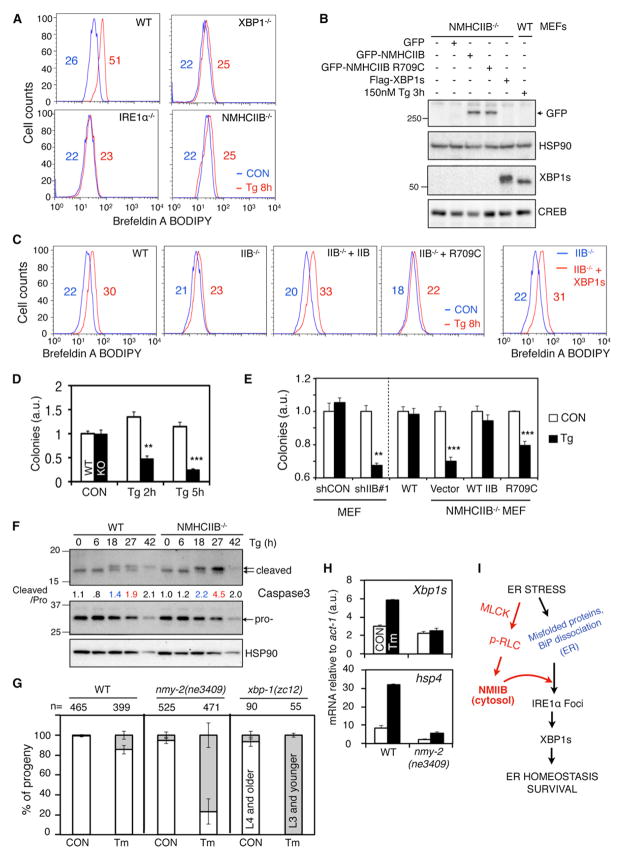
We studied the biological consequences of NMIIB on UPR at three levels: organelle, cellular, and organismal. At the organelle level, in line with the role of IRE1α-XBP1s in ER biogenesis (Hetz et al., 2006; Sriburi et al., 2004), loss of NMIIB, similar to the loss of IRE1α or XBP1, led to prominent defects in ER expansion in response to both Tg and Tm-induced ER stress (Figure 5A; Figure S3). This defect was rescued by overexpression of WT NMHCIIB but not by the R709C NMHCIIB mutant (Figures 5B and 5C), further supporting a functional requirement for the motor activity of NMIIB. Importantly, overexpression of XBP1s in NMIIB−/− MEFs rescued the ER expansion defect to a level similar to that of WT MEFs exposed to Tg (Figures 5B and 5C). Taken together, these data demonstrate that NMIIB and, specifically, its motor function is a critical component of ER stress response via IRE1α activation and XBP1s production.
Figure 5. NMIIB Deficiency Renders Cells and Worms Hypersensitive to ER Stress.
(A–C) Flow cytometric analysis of ER/Golgi mass of (A) WT and various mutant MEFs or (C) NMHCIIB−/− MEFs transfected with WT, R709C NMHCIIB, or XBP1s, followed by treatment with 300 nM Tg for 8 hr and stained with Brefeldin A-BODIPY. Numbers indicate mean channel fluorescence. Western blot analysis of protein levels in transfected cells shown in (B) with WT MEFs treated with 150 nM Tg for 3 hr in the far right lane. HSP90 and CREB, loading controls whole-cell extract and nuclear extract, respectively.
(D and E) Cell survival assays: Quantitative reading of crystal violet staining of WT and NMHCIIB−/− MEFs treated with 150 nM Tg for the indicated time, followed by recovery for 4–5 days. y axis indicates cell or colony numbers. In (E), MEFs expressing shCON or shIIB#1, or WT, NMHCIIB−/−, and NMHCIIB−/− MEFs rescued with WT or R709C NMHCIIB were used. Data are shown as mean ± SEM. **p < 0.01, ***p < 0.001.
(F) Western blot of active and procaspase 3 from WT and NMHCIIB−/− MEFs treated with 150 nM Tg for the indicated time. Quantitation of cleaved caspase 3 levels shown below the gel after normalization to procaspase 3.
(G and H) WT (N2) or nmy-2(ne3409) worms were shifted to 25°C after embryogenesis and grown on 6 μg/ml Tm to assay development at 72 hr. xbp-1(zc12) worms were included as controls. Data are shown as mean ± SD (p = 0.004 for nmy-2(ne3409) versus WT and p < 0.001 for xbp-1(zc12) versus WT). (H) Quantitative real-time PCR analysis of UPR genes in WT (N2) or nmy-2(ne3409) worms grown on 6 μg/ml Tm for 72 hr at 25°C. Data are shown as mean ± SEM.
(I) Model for the role of NMIIB in IRE1α aggregation and signaling. Our data suggest that optimal IRE1α activation and signaling require concerted coordination between the ER and cytoskeleton in the cytosol. Findings from this study are highlighted in red, whereas known activating signals from the ER are in blue. For all, similar results were observed in 2–3 independent experiments. See also Figure S3.
At the cellular level, NMHCIIB−/− MEFs were unable to recover after an ER stress challenge (Figure 5D), in line with a prosurvival role attributed to the IRE1α-XBP1s pathway (He et al., 2010). Consistent with the ER expansion phenotype, WT but not R709C NMHCIIB completely rescued cell survival defects (Figure 5E). This was further supported by an increase of caspase-3 cleavage, a marker for apoptosis, in NMHCIIB-deficient cells upon prolonged Tg treatment (Figure 5F).
NMIIB-Deficient Worms Are Hypersensitive to ER Stress
Lastly, we explored the physiological implications of NMIIB in ER stress response at the organismal level. As NMHCIIB−/− mouse embryos die before birth (Takeda et al., 2003), we turned to the nematode C. elegans, in which NMY-2, a protein required for polarity establishment during worm embryogenesis, is a homolog of NMHCIIB (Guo and Kemphues, 1996). We examined the ability of temperature-sensitive NMII mutant worms nmy-2(ne3409) (Liu et al., 2010) to reach L4 and adulthood in response to ER stress after shifting to the restrictive temperature following embryogenesis. In the absence of Tm, 99% ± 1% WT and 95% ± 3% nmy-2(ne3409) reached L4 or adulthood. However, in the presence of ER stress, a dramatic difference was consistently observed after 72 hr between the percent of WT (85% ± 4%) and nmy-2(ne3409) (23% ± 13%) animals that developed to L4 (Figure 5G), suggesting that nmy-2(ne3409) worms were hypersensitive to ER stress. This effect was similar to that of a Xbp1 deficiency as previously reported (Henis-Korenblit et al., 2010), though less severe (Figure 5G). At the molecular level, ER stress-dependent induction of Xbp1s and its target hsp-4, the worm homolog of the ER chaperone GRP78/BiP, was completely abrogated in nmy-2(ne3409) (Figure 5H). Together, these in vivo studies reveal a functional and conserved role for NMII specifically in IRE1α signaling.
DISCUSSION
Our study has identified and characterized NMIIB as a specific and essential component of the IRE1α-XBP1 signaling axis of UPR. We propose a model in which ER stress promotes an IRE1α-NMIIB interaction and subsequently, NMIIB facilitates the oligomerization, activation, and signaling of IRE1α (Figure 5I). This model is supported by our findings that NMIIB is required for IRE1α aggregation and foci formation, induction of XBP1s protein and downstream UPR targets, and cellular and organismal responses to ER stress in vivo. The actomyosin contractility of NMIIB is required as both the motor domain and involvement of the actin filament are indispensable.
Our data point to a concerted coordination between the ER and cytoskeleton that is essential for optimal IRE1α activation and cell fate determination in response to ER stress (Figure 5I). Upon ER stress, RLC phosphorylation is required for NMIIB activation and its regulation of IRE1α; these events are at least in part dependent on MLCK. How MLCK or other RLC kinases respond to ER stress remains unclear. An intriguing possibility is that IRE1α itself may serve as a kinase for MLCK directly or indirectly, and thus form a feedback regulatory loop. Although other scenarios, such as NMIIB-mediated Xbp1 mRNA trafficking or ER membrane reorganization, are exciting possibilities that remain to be explored, our data demonstrate that NMIIB directly impacts IRE1α activation and signaling by interacting with IRE1α and regulating its oligomerization and activation.
A number of studies have identified a repertoire of IRE1α regulatory cofactors that modulate its activity, including BAX/BAK, Bax inhibitor-1, RACK1, HSP90, and others (Hetz et al., 2006, 2011; Lisbona et al., 2009; Qiu et al., 2010). Collectively, these interacting proteins comprise the “UPRosome,” a scaffolding complex believed to dynamically regulate IRE1α in a cell or tissue-specific manner (Hetz and Glimcher, 2009; Hetz et al., 2011). Identification of these regulatory cofactors of IRE1α signaling has greatly advanced our understanding of how IRE1α activity and output can be regulated mechanistically. Unlike other IRE1α-interacting proteins that have been identified, NMIIB is unique in that it is an essential component involved in the oligomerization step of IRE1α activation. Speculatively, NMIIB may not only promote IRE1α aggregation but also recruit other regulatory components to the foci. The interplay between NMIIB and the “UPRosome” is an open but exciting question.
Findings from this study and others showed that IRE1α foci can be visualized as early as 1 hr, peaked around 4 hr and are resolved by 8 hr of ER stress (Figure 3C; Li et al., 2010). However, questions such as how foci dispersion and deoligomerization are regulated and whether NMIIB or other cytoskeletal proteins are involved in this process remain unaddressed. Interestingly, our data convey that NMIIB activation as indicated by RLC phosphorylation is extremely dynamic and transient, peaking within the first hour and resolved after prolonged ER stress (Figures 4D and 4E), suggesting an intimate relationship between NMIIB activation and the kinetics of IRE1α foci formation and dispersion. A recent study in yeast reported that the kinase activity of IRE1α is important for its deactivation and foci dispersion (Rubio et al., 2011). Whether and how NMIIB are linked to the activation and inactivation of IRE1α during ER stress remains to be elucidated, and future studies are warranted to tease out the mechanistic details and dynamics of IRE1α foci formation and dispersion.
IRE1α has been reported to cleave other nonspecific mRNAs upon ER stress in a process termed regulated IRE1-dependent decay (RIDD), presumably to decrease ER load (Han et al., 2009; Hollien et al., 2009; Hollien and Weissman, 2006; So et al., 2012). However, the molecular mechanism and signals by which IRE1α regulates RIDD remain largely unknown. For example, does IRE1α RNase activation depend on the substrate (i.e., Xbp1 versus RIDD targets), and is IRE1α aggregation a prerequisite for RIDD? As NMIIB regulates IRE1α oligomerization, it will be of great interest to determine whether and how NMIIB regulates IRE1α RIDD activity.
Although IRE1α dimers may possess RNase activity to splice Xbp1 mRNA, IRE1α oligomers are believed to have maximal splicing efficiency as the arrangement of dimers into oligomers brings multiple Xbp1 mRNA binding pockets into close proximity (Korennykh et al., 2009; Li et al., 2010; Walter and Ron, 2011). This is supported by our observation that a NMIIB deficiency does not completely abolish XBP1s production. Nonetheless, NMIIB-mediated IRE1α oligomerization is significant as shown by our in vivo data on the cellular and organismal response to ER stress in which NMIIB is essential for initiating and engaging an optimal IRE1α signaling and UPR.
One outstanding question is whether the involvement of NMIIB in IRE1α activation and signaling is relevant in vivo under physiological conditions, where ER stress can be much milder relative to the pharmacological insults used in cell culture to disrupt ER homeostasis (Pfaffenbach et al., 2010; Sha et al., 2011; Yang et al., 2010). Because of the lack of a good antibody for immunostaining, the question of whether endogenous IRE1α forms foci in vivo under physiological UPR remains unanswered. This is important as it will shed light on the activating mechanism of IRE1α under physiological and pathophysiological settings. Overall, our finding linking the IRE1α branch of the UPR and the cytoskeletal machinery of the cell enhances our comprehension of the cellular and molecular basis of mammalian ER stress response and may shed light on therapeutic targets for UPR-associated diseases.
EXPERIMENTAL PROCEDURES
Cell Lines and Drug Treatment
NMHCIIB−/− MEFs were generated as previously described (Meshel et al., 2005). XBP1−/− and IRE1α−/− MEFs were generously provided by Drs. L. Glimcher (Weill Cornell) and D. Ron (University of Cambridge), respectively. Matching WT MEFs were used in this study. T-REx293 IRE1-3F6HGFP was previously described (Li et al., 2010; Xue et al., 2011) and generously provided by Dr. P. Walter (UCSF). All cells were maintained in DMEM supplemented with 10% FBS (Hyclone, Logan, UT, USA) and 1% penicillin/streptomycin (Cellgro, Herndon, VA, USA). Tg and Tm (EMD Calbiochem, Darmstadt, Germany), Blebbistatin (Cayman Chemical, Ann Arbor, MI, USa), and ML-7 (Sigma-Aldrich, St. Louis, MO, USA) were dissolved in DMSO. Cytochalasin D (Enzo Life Sciences, Farmingdale, NY, USA) and doxycycline (EMD Millipore, Billerica, MA, USA) were dissolved in H2O. In most experiments, cells were treated with 150–300 nM Tg or 2.5–5 μg/ml Tm over a time course (up to 8 hr) as indicated in the figures and figure legends. A drug concentration titer was performed in one experiment to examine the dose-response curve. For experiments requiring pretreatment, cells were pretreated with the drug (e.g., blebbistatin, ML-7, cytochalasin D) for 30 min prior to treatment with Tg.
IRE1α Immunopurification and Mass Spectrometry
Whole-cell lysates from IRE1α−/− MEFs stably expressing human IRE1α-HA (with or without 300 nM Tg for 2 hr) were harvested from five 15 cm plates and immunoprecipitated with HA-agarose. Immunoprecipitates were eluted with SDS sample buffer and separated on a 6% polyacrylamide SDS-PAGE gel. Gel was incubated with Coomassie Brilliant Blue and destained. Protein bands were excised from Coomassie-stained gels and destained, and subjected to the mass spectrometric analysis (LC-MS/MS). Details are described in the Supplemental Experimental Procedures.
Immunoprecipitation
Whole-cell lysates were harvested in lysis buffer (150 mM NaCl, 1% Triton X-100, 1 mM EDTA, and 50 mM Tris HCl [pH 7.5]) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and 5 mM ATP (Sigma-Aldrich) and incubated on ice for 25 min. Samples were sonicated 10 s once with Branson Digital 250 Cell Disruptor at amplitude 10%. For endogenous IP, supernatant was precleared with Protein A-agarose (Invitrogen, Carlsbad, CA, USA) for 30 min and rocked with 2 μg primary antibody overnight at 4°C. Immune complexes were recovered with Protein A-agarose for 2 hr at 4°C with rocking. Beads were washed four times with wash buffer (20 mM Tris HCl [pH 7.5], 137 mM NaCl, 20 mM EDTA, 1% Triton X-100, and 10% glycerol) and eluted in boiling 2X SDS sample buffer, followed by SDS-PAGE and western blot. In the case of overexpressed tagged proteins, the only difference was that cells were incubated overnight with HA- or Flag-agarose at 4°C with rocking followed by washes as described above.
Western Blot and Image Quantitation
Preparation of cell lysates and western blot were performed as we previously described (Sha et al., 2009). Primary antibodies were diluted in 5% milk/TBST or 2% BSA/TBST and incubated with PVDF membrane overnight at 4°C, whereas secondary antibodies were incubated at room temperature for 45 min. Antibody information is available in the Supplemental Experimental Procedures. To study IRE1α phosphorylation, we used a Phos-tag-based western blot method that can sensitively monitor the phosphorylation status of UPR sensors (Qi et al., 2011; Sha et al., 2009; Yang et al., 2010). Membranes were routinely strip-reprobed for HSP90 as a positional control. In addition, a Phos-tag-based method was also used to visualize total RLC phosphorylation as previously described (Aguilar et al., 2011). Of note, to ensure sufficient signal for cleaved caspase 3, the membrane for caspase 3 was cut around the 25 kDa line and then probed separately with the caspase 3 antibody. Band density was quantitated using the Image Lab software on the ChemiDOC XRS+ system (Bio-Rad, Hercules, CA, USA) and presented as the mean ± SEM from several independent experiments or as representative data from at least two independent experiments.
siRNA Knockdown and Retroviral Transduction
Retroviral transduction and stable cell lines were carried out as previously described (Sha et al., 2009). Stable cell lines expressing siRNA were selected in hygromycin at 200 μg/ml or puromycin at 5 μg/ml. Stable cell lines were made and tested independently at least twice. siRNA sequences for mouse myh-10 (in MEFs): shIIB#1: 5′ GAGAAGAAACTGAAAGAAA 3′; shIIB#3: 5′ GGAACAAGGCTGAGAAACA 3′. siRNA sequences for human myh-10 (in HEK293T cells): shIIB#1: 5′ GAGAAGAAGCTGAAAGAAA 3′; shIIB#2: 5′ CCAAAGATGATGTGGGAAA 3′.
RNA Extraction and Quantitative Real-Time PCR
The experiments using mammalian cells were performed as previously described (Sha et al., 2009). For nematodes, WT (N2) or nmy-2(ne3409) worms were shifted to 25°C and grown on 6 μg/ml Tm. Seventy-two hours later, approximately 300 animals of each genotype and condition were washed off plates with M9 buffer, pelleted, and mixed with 10× volume of Trizol. Quantitative real-time PCR primer sequences are available in the Supplemental Experimental Procedures.
Immunofluorescence
MEFs were plated and grown on collagen-coated coverslips in 6-well plates overnight, followed by treatment with the indicated concentration and time of Tg. After a brief wash with PBS, cells were fixed in fresh 3.7% formaldehyde in PBS for 10 min and washed three times with PBS for 5 min each. Cells were lysed with 0.2% Triton X-100 in PBS for 5 min and washed three times with PBS for 5 min each. Cells were blocked in 5% BSA in PBS for 10 min and incubated 1 hr at room temperature with anti-myosin IIB at 1:50 dilution (Developmental Studies Hybridoma Bank, CMII23). Cells were washed three times with PBS for 5 min each, followed by 1 hr incubation with donkey anti-mouse Cy3 (Jackson ImmunoResearch, West Grove, PA, USA), with or without anti-rabbit FITC (Jackson ImmunoResearch), at 1:200 dilution in the dark. Cells were washed and mounted with Prolong Gold Antifade Reagent with DAPI (Invitrogen). Fluorescent microscopic images were taken with a Zeiss 710 Confocal microscope using a 63×/1.4 objective (Cornell Microscopy and Imaging Facility).
Foci Imaging
Analysis of IRE1α foci formation was performed as previously described (Xue et al., 2011). Briefly, T-REx293 IRE1-3F6HGFP cells were treated with 5 μg/ml doxycycline for 24 hr to induce IRE1α-GFP expression, followed by 300 nM Tg treatment for the indicated time. Fluorescent microscopic images were taken with a Zeiss 710 Confocal microscope using a 63x/1.4 objective (Cornell Microscopy and Imaging Facility). Foci-positive was calculated as the number of cells with one or more foci out of total number of cells.
Sucrose Gradient Sedimentation
Confluent WT and NMHCIIB−/− MEFs in 10 cm plates nontreated or treated with 300 nM Tg for 3 hr were harvested and lysed in 300 μl lysis buffer (described in Immunoprecipitation). Extracts were centrifuged through 20%–40% sucrose gradients prepared freshly by progressively layering higher to lower density sucrose fractions in 5% increments in polyallomer tubes of 11 × 60 mm (Beckman Coulter, Brea, CA, USA). Extracts were centrifuged at 60,000 rpm for 14.5 hr at 4°C using an SW 60 Ti rotor (Beckman Coulter). Each 4 ml gradient was divided evenly into 16 fractions (250 μl), and aliquots of fractions 3–16 (labeled 1–14 in Figures 3H and 3I) were subjected to SDS-PAGE analysis. IRE1α-containing complexes were detected using anti-IREα antibody. The density of IRE1α in each fraction was quantitated using the Image Lab software on the ChemiDOC XRS+ system (Bio-Rad), and the percent of IRE1α in each fraction was calculated as the amount of IRE1α in a fraction relative to the total IRE1α levels in all 14 fractions.
Cell Survival Assay
WT and NMHCIIB−/− or shCON and shIIB#1 MEFs were grown in 6-well plates and treated with 150–300 nM Tg for the indicated time. For rescue experiments, NMHCIIB−/− MEFs were transfected with WT or R709C NMHCIIB plasmids using Lipofectamine 2000, per the supplier’s protocol, for 24 hr prior to Tg treatment. Cells were counted by hemocytometer, and 1.5 × 105 cells were re-plated onto 10 cm plates. Four days later, the cells were briefly washed in PBS and fixed in freshly prepared 3.7% formaldehyde in PBS for 15 min, followed by 30 min incubation in 0.05% crystal violet in distilled water (filtered before use) with gentle rocking at room temperature. Cells were washed three times for 5 min each with ddH2O, permeabilized with methanol for 15 min, and sampled aliquots were read at OD 540 nm with Bio-Tek Synergy 2 plate reader (Bio-Tek Inc., Winooski, VT, USA).
Brefeldin-A BODIPY and Flow Cytometry
WT and various mutant MEFs were grown in 24-well plates overnight, followed by treatment with 150 nM Tg or 2.5 μg/ml Tm for the indicated time. For rescue experiments, NMHIIB−/− MEFs were transfected with WT, R709C NMHCIIB, or XBP1s plasmids with Lipofectamine 2000, per the supplier’s protocol, for 24 hr, followed by Tg treatment. Cells were incubated at 37°C for 30–45 min with 0.4 μg/ml Brefeldin A-BODIPY (Invitrogen) in culture media, followed by trypsinization and flow cytometric analysis using a BD FACSCalibur flow cytometer. Data were analyzed using the CellQuest and FlowJo software.
ER Stress Resistance in C. elegans
Nematodes were cultured using standard conditions (Brenner, 1974), with N2 (Bristol) as wild-type. The mutations used in this analysis include nmy-2(ne3409) (Liu et al., 2010) and xbp-1(zc12) (Calfon et al., 2002). For ER stress resistance assays, embryos were laid onto plates containing 0 or 6 μg/ml Tm. After 18 hr, the number of hatching larvae was noted and compared to the number of L4/adult stage worms after 72 hr. In experiments using nmy-2(ne3409), embryos were allowed to complete embryogenesis at the permissive temperature of 16°C and then shifted to the restrictive temperature of 25°C after hatching. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota) funded by the National Institutes of Health (NIH) National Center for Research Resources.
Statistical Analysis
Results are expressed as the mean ± SEM unless indicated otherwise. Comparisons between groups were made by unpaired two-tailed Student’s t test, where p < 0.05 was considered as statistically significant. All experiments were repeated at least two to three times, and representative data are shown.
Supplementary Material
Acknowledgments
We thank Drs. C. Hetz, D. Ron, P. Walter, M. Montminy, L. Glimcher, L. Kraus, F. Hu, A. Bretscher, and V. Vogt for reagents and equipment; A. Bretscher for suggestions; M. Montminy, J.T. Lis, and M.A. Conti for reading of the manuscript; D. Garbett and R. Dick for assistance with sucrose gradient; D. Morton for assistance with C. elegans experiments; S. Sun for the drawing of the graphical abstract; and members of the Qi laboratory for technical assistance and discussions. L.Q. is the recipient of the Junior Faculty and Career Development Awards from American Diabetes Association (ADA). This study is supported by the NIH (HL004228 to X.M. and R.S.A.; R01GM079112 to K.K.; P41 GM103533 to J.R.Y.; and R21AA020351 and R01DK082582 to L.Q.) and the ADA (1-12-CD-04 to L.Q.).
Footnotes
Supplemental information includes three figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2012.11.006.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Adelstein RS, Conti MA. Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature. 1975;256:597–598. doi: 10.1038/256597a0. [DOI] [PubMed] [Google Scholar]
- Aguilar HN, Tracey CN, Tsang SC, McGinnis JM, Mitchell BF. Phos-tag-based analysis of myosin regulatory light chain phosphorylation in human uterine myocytes. PLoS ONE. 2011;6:e20903. doi: 10.1371/journal.pone.0020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arii J, Goto H, Suenaga T, Oyama M, Kozuka-Hata H, Imai T, Minowa A, Akashi H, Arase H, Kawaoka Y, Kawaguchi Y. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature. 2010;467:859–862. doi: 10.1038/nature09420. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn PG, Helfman DM. Non-muscle myosin IIB helps mediate TNF cell death signaling independent of actomyosin contractility (AMC) J Cell Biochem. 2010;110:1365–1375. doi: 10.1002/jcb.22653. [DOI] [PubMed] [Google Scholar]
- Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Sun S, Sha H, Liu Z, Yang L, Xue Z, Chen H, Qi L. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 2010;15:13–25. doi: 10.3727/105221610x12819686555051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, Kenyon C. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat Immunol. 2009;10:531–539. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Kovács M, Kawamoto S, Sellers JR, Adelstein RS. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J Biol Chem. 2005;280:22769–22775. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell. 2003;14:2559–2569. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Maduzia LL, Shirayama M, Mello CC. NMY-2 maintains cellular asymmetry and cell boundaries, and promotes a SRC-dependent asymmetric cell division. Dev Biol. 2010;339:366–373. doi: 10.1016/j.ydbio.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Adelstein RS. In vivo studies on nonmuscle myosin II expression and function in heart development. Front Biosci. 2012;17:545–555. doi: 10.2741/3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshel AS, Wei Q, Adelstein RS, Sheetz MP. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat Cell Biol. 2005;7:157–164. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res. 2009;315:2496–2504. doi: 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Pfaffenbach KT, Nivala AM, Reese L, Ellis F, Wang D, Wei Y, Pagliassotti MJ. Rapamycin inhibits postprandial-mediated X-box-binding protein-1 splicing in rat liver. J Nutr. 2010;140:879–884. doi: 10.3945/jn.109.119883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Yang L, Chen H. Detecting and quantitating physiological endoplasmic reticulum stress. Methods Enzymol. 2011;490:137–146. doi: 10.1016/B978-0-12-385114-7.00008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Mao T, Zhang Y, Shao M, You J, Ding Q, Chen Y, Wu D, Xie D, Lin X, et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal. 2010;3:ra7. doi: 10.1126/scisignal.2000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C, Pincus D, Korennykh A, Schuck S, El-Samad H, Walter P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol. 2011;193:171–184. doi: 10.1083/jcb.201007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M, Ishikawa T, Matsushima S, Naka M, Hidaka H. Selective inhibition of catalytic activity of smooth muscle myosin light chain kinase. J Biol Chem. 1987;262:7796–7801. [PubMed] [Google Scholar]
- Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H, He Y, Yang L, Qi L. Stressed out about obesity: IRE1α-XBP1 in metabolic disorders. Trends Endocrinol Metab. 2011;22:374–381. doi: 10.1016/j.tem.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So JS, Hur KY, Tarrio M, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Lichtman AH, Iwawaki T, Glimcher LH, Lee AH. Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kishi H, Ma X, Yu ZX, Adelstein RS. Ablation and mutation of nonmuscle myosin heavy chain II-B results in a defect in cardiac myocyte cytokinesis. Circ Res. 2003;93:330–337. doi: 10.1161/01.RES.0000089256.00309.CB. [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Xue Z, He Y, Ye K, Gu Z, Mao Y, Qi L. A conserved structural determinant located at the interdomain region of mammalian inositol-requiring enzyme 1alpha. J Biol Chem. 2011;286:30859–30866. doi: 10.1074/jbc.M111.273714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xue Z, He Y, Sun S, Chen H, Qi L. A Phos-tag-based approach reveals the extent of physiological endoplasmic reticulum stress. PLoS ONE. 2010;5:e11621. doi: 10.1371/journal.pone.0011621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.