Abstract
The importance of the trifluoromethyl group in the polypharmacological profile of nilotinib was investigated. Molecular editing of nilotinib led to the design, synthesis and biological evaluation of analogues where the trifluoromethyl group was replaced by a proton, fluorine and a methyl group. While these analogues were less active than nilotinib toward Abl, their activity toward Kit was comparable, with the monofluorinated analogue being the most active. Docking of nilotinib and of analogues 2a–c to the binding pocket of Abl and of Kit showed that the lack of shape complementarity in Kit is compensated by the stabilizing effect from its juxtamembrane region.
Keywords: CML, Kit, Abl, kinase, molecular editing
The receptor tyrosine kinase Kit (also known as c-Kit, or CD117) is involved in a host of cellular processes, including cell differentiation, proliferation and survival.1 Kit is found on the surface of hematopoietic stem cells (HSC), common myeloid progenitors, common lymphoid progenitors, myeloblasts, megakaryocytes, and mast cells. Upon binding of its ligand, the stem cell factor (SCF), the receptor dimerizes, leading to the activation of the cytoplasmic kinase domain. The phosphorylated tyrosine residues of Kit are then involved in a series of signal transduction events including the PI3K pathway, the Jak-Stat pathway, Src-family kinases, and the Ras-Erk pathway.2 Gain-of-function mutations of Kit are associated with a number of cancers such as acute myeloid leukemia (AML), chronic myeloid leukemia (CML), mast cell leukemia, and gastrointestinal stromal tumors (GIST). Another cause of CML is the oncogenic breakpoint cluster region protein-abelson kinase (Bcr-Abl) fusion protein, caused by the Philadelphia chromosome.3 Bcr-Abl becomes constitutively active, and unregulated phosphorylation of proteins found in HSC leads to uncontrolled growth and survival.
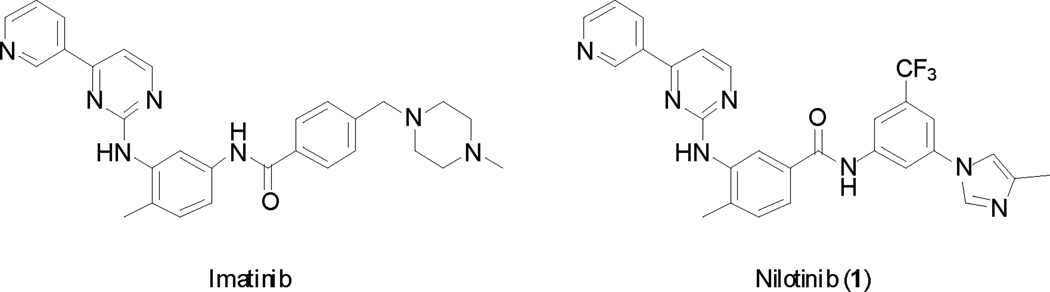
Imatinib (Gleevec, Novartis) (Fig. 1) is an inhibitor of the Bcr-Abl fusion protein, Kit and of the receptor tyrosine kinases platelet-derived growth factor receptor subtype alpha (PDGFRα). It was approved in 2001 for the treatment of CML, and in 2003 for the treatment of GIST. Despite its success, resistance to the drug was observed in patients because of mutations of the Abl protein. The second generation kinase inhibitor nilotinib (Tasigna, Novartis) (Fig. 1) also targets Abl and Kit and was approved in 2007 for the treatment of CML. The drug was then advanced to phase III clinical for the treatment of GIST resistant to imatinib, but was discontinued in 2011 because of a lack of superior efficacy compared to the current standard of care. Understanding the polypharmacology of nilotinib (and other kinase inhibitors) is paramount to understanding their ultimate utility, and it is likely that the capacity for nilotinib to target both Abl and Kit, along with other kinases such as PDFRα, accounts for the clinical utility of the drug. Pathway redundancy via Kit signaling, in particular, may abrogate Bcr-Abl inhibition requiring that Kit must be inhibited in addition to Bcr-Abl for efficacy to be realized.4
Figure 1.
Structure of imatinib and of nilotinib
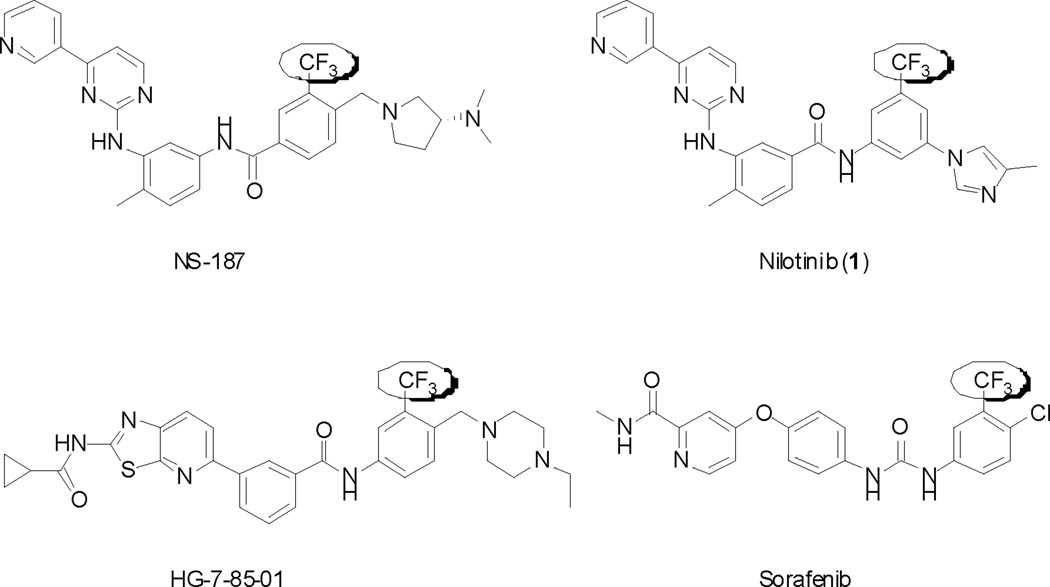
Molecular design of nilotinib (1) was based upon the crystal structure of the imatinib-Abl complex. Binding to the inactive form of Abl was maintained through the [4-(3-pyridinyl)-2-pyrimidinyl]aniline portion of imatinib and of nilotinib. However, nilotinib relies on fewer hydrogen-bonding interactions with amino acid residues of the hinge region of Abl, and on a larger number of hydrophobic interactions. Such van der Waals interactions are found by the 4-methyl imidazole moiety and especially by the trifluoromethyl group. In fact, structural studies have shown that one of the CF3 fluorines interacts with the carbonyl of the Asp381 backbone in Abl.5 Olsen showed that protein backbone fragments H-Cα-C=O provide a favorable “fluorophilic” environment for the C-F bond, including multipolar interactions between the intrinsically polar C-F and C=O bonds.6 A significant number of molecular probes and drug candidates targeting kinases feature a trifluoromethyl group (Fig. 2). Examples include NS1877 (a Bcr-Abl and Lyn kinase inhibitor), HG-7-85-018 (a Bcr-Abl, Kit, PDGFR, and Src family kinase inhibitor), and sorafenib9 (Nevaxar, Bayer/Onyx)(a c-Raf, Kit, isoform specific VEGFR inhibitor). Since protein kinases display a high degree of homology at their active sites, ATP-competitive small molecule kinase inhibitors often possess activity against several targets. In the particular case of nilotinib, the activity profile is well established.10 In this study, we endeavored to define the influence of the trifluoromethyl group present in nilotinib with respect to its polypharmacology.
Figure 2.
Structures of kinase inhibitors bearing a trifluoromethyl group
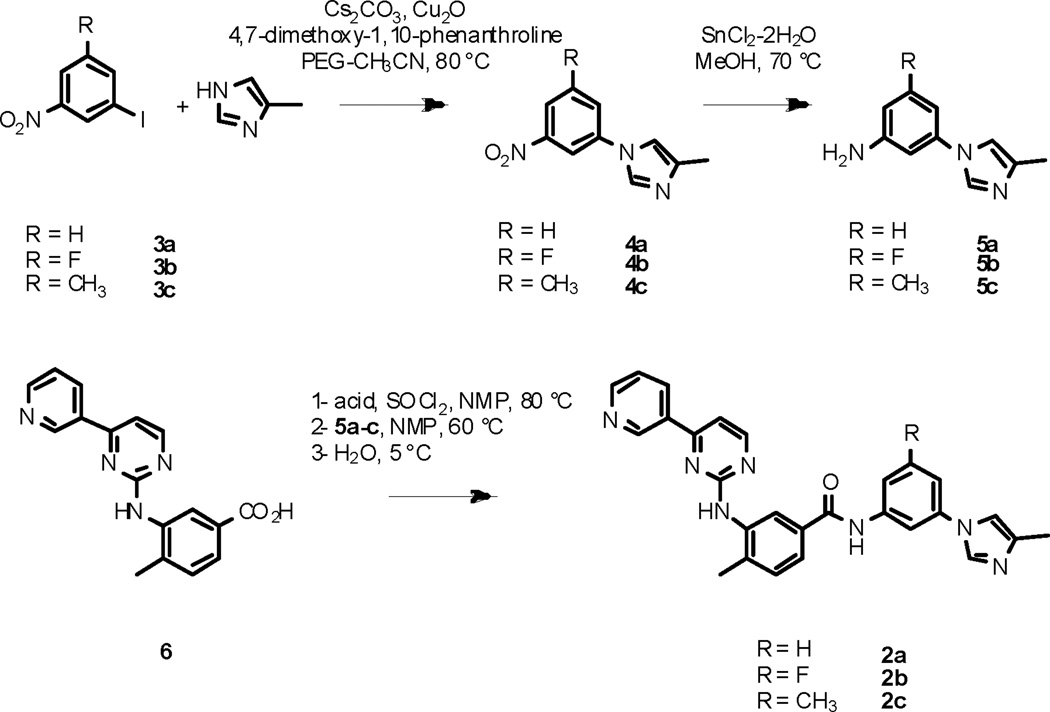
A modular synthetic sequence was developed to access nilotinib analogues (scheme 1). The syntheses of analogues 2a–c commenced with the Ullman-type coupling between aryl iodide 3a–c and 4-methyl-1H-imidazole, using a procedure developed by Buchwald et al.11 Three challenges associated with this reaction were (1) the poor solubility of the reagents in the reaction mixture, (2) the lack of reactivity of the starting aryl iodide (especially 3b), and (3) the possible isomerization of 4-methyl-1H-imidazole during the reaction. Using the more soluble cesium carbonate as base and polyethylene glycol as a co-solvent to acetonitrile improved the solubility of the reagents, affording a clear reaction solution when stirred at reflux. Activation of the copper catalyst with 4,7-dimethoxy-1,10-phenanthroline12 allowed not only the reaction to proceed in higher yields than when using copper(I) iodide and ethylene diamine were used, but also led to the formation of N-aryl imidazoles 4a–c as single isomers. The nitro group was reduced using tin(II) chloride-dihydrate in methanol. Formation of the amide bond between benzoic acid 6 and anilines 5a–c was more difficult than expected. Common procedures such as EDCI or DCC coupling in DCM, DCE, or acetonitrile failed; HATU coupling in DMF was not successful either; deprotonation of the aniline using potassium tert-butoxide in THF, followed by treatment of the resulting anilide13 with the methyl ester of 6 also failed. Eventually, conversion of 6 to the corresponding acyl chloride, using thionyl chloride in the polar aprotic solvent NMP, followed by addition of anilines 5a–c provided nilotinib analogues 2a–c.14 The low solubility of 5a–c and of 6 in solvents such as DCM, DCE or THF, along with the poor reactivity of anilines 5a–c may account for the difficulty to form the amide bond. The nature of the substitution on the aromatic ring of aryl iodide 3 had a great influence on its reactivity, as well as the reactivity of its subsequent synthetic intermediates. Thus, fluorinated aryl imidazole 4b was consistently obtained in lower yield, as compared to 4a and 4c. Similarly, aniline 5b was less reactive toward the acyl chloride of 6 in the final coupling step leading to analogue 2b. Dilution of the reaction mixture with ice-cold water led to the precipitation of the crude products which could then be collected by filtration. The solids thus obtained were finally purified by reverse-phase HPLC.
Scheme 1.
Syntheses of analogues 2a–c
Utilizing two commercial organizations (Reaction Biology Corp. www.reactionbiology.com and DiscoveRx http://www.kinomescan.com/), we generated IC50 and Kd values for nilotinib (1) and analogues 2a–c for a panel of kinases (table 1). The activity profile of nilotinib versus a broad panel of kinases had been reported by Davis et al.,15 and those results were used as a starting point in our study to focus on the kinases involved in the signaling pathways primarily targeted by nilotinib.
Table 1.
Inhibitory activities of compounds 1 and 2a–c
| Compound IC50 (nM)a | ||||
|---|---|---|---|---|
| Kinase | 1 | 2a | 2b | 2c |
| Abl | <2.54 | 2953 | 749.8 | 135.8 |
| Kit | 14.70 | 32.50 | 9.96 | 54.94 |
| DDR2 | <2.54 | 653.3 | 178.6 | 17.22 |
| FLT3 | 9548 | 973.9 | 117.8 | 882.0 |
| LCK | 108.2 | 44620 | 11050 | 1448 |
| LYN | 1281 | 45420 | 15150 | 3534 |
| PDGFRα | <2.54 | 83.87 | 25.96 | 8.20 |
| PDGFRβ | 60.11 | 35400 | >5000 | 768.4 |
Assay details are provided by Reaction Biology Corp. via the internet at www.reactionbiology.com
The results obtained in both enzymatic assays were in good agreement with those previously reported for nilotinib.16 When comparing the four compounds activities within the functional assay (table 1), nilotinib (1) was the most active toward Abl by a hundred fold (IC50 < 2.5 nM). The second most active analogue was methyl analogue 2c (IC50 = 135.8 nM), followed by 2b (IC50 = 750 nM) and 2a (IC50 = 2953 nM). These results underscore the importance of a methyl or of a trifluoromethyl group for the binding of the compound to Abl, as reported by Manley et al.5 The influence of the trifluoromethyl group, and, to a lesser extent, of the methyl group was also observed for the inhibition of the discoin domain receptor subtype 2 (DDR2), the lymphocyte-specific tyrosine kinase (LCK) and for the inhibition of LYN: once again, nilotinib was the most active compound, followed by methyl analogue 2c. Regarding the platelet-derived growth factor receptor (PDGFR), nilotinib (1) and methyl analogue 2c were the most active (IC50 < 2.54 nM and 8.20 nM respectively), compared to 2a and 2b (IC50 < 83.87 nM and 25.96 nM respectively). The comparative activity between methylated and non-methylated analogues was particularly noticeable with PDGFRβ (IC50 < 60.11 nM for 1 and 768.4 nM for 2c versus 35400 nM for 2a and >50000 nM for 2b). On the other hand, Nilotinib and analogues 2a–c displayed good inhibitory activity toward Kit, suggesting that unlike Abl, the presence of a methyl or of a trifluoromethyl group was not required. The same structure activity relationship was observed for the four compounds versus the Fms-like tyrosine kinase 3 (FLT3), with fluorinated analogue 2c being the most active compound (IC50 = 117.8 nM). While 2a and 2c displayed comparable activites (IC50 = 973.9 nM, and 882.0 nM respectively), no correlation was observed between 2c and 1 (IC50 = 9548 nM).
The binding affinity data (table 2) showed that nilotinib (1) and methylated analogue 2c displayed stronger binding affinities toward Abl (Kd = 3.6 nM for 1 and Kd = 3.4 nM for 2c), although the Kd values for 2a and 2b were close. Fluorinated analogue 2b had a stronger binding affinity than unsubstituted analogue 2a (Kd = 9.4 nM for 2b and Kd = 29 nM for 2a). This result may imply the presence of van der Waals interactions between the fluorine and a carbonyl of the backbone in the hydrophobic pocket. In addition, compounds 2a and 2b showed the strongest affinity for Kit (4.4 nM and 3.5 nM respectively). A good correlation between the measurement of the inhibition of kinase activity (IC50) and that of the thermodynamic binding affinity (Kd) was observed for Abl, Kit, DDR2, and FLT3. While the level of selectivity between compounds 1 and 2a–c was not high in the panel of kinases tested (the Kd values were in the same order of magnitude), a remarkable difference in affinity of the fluorinated analogue toward FLT3, whereby fluorinated analogue 2b showed a tighter binding by a factor twenty compared to the other compounds tested. This result supports the selective inhibitory activity of 2b as compared to the other analogues (table 1).
Table 2.
Dissociation constants of compounds 1 and 2a–c
| Compound Kd (nM)a | ||||
|---|---|---|---|---|
| Kinase | 1 | 2a | 2b | 2c |
| Abl | 3.6 | 29 | 9.4 | 3.4 |
| Kit | 17 | 4.4 | 3.5 | 14 |
| DDR2 | 6.2 | 320 | 14 | 5.6 |
| FLT3 | 40000 | 1400 | 58 | 1200 |
| PDGFRα | 74 | 14 | 24 | 65 |
| PDGFRβ | 16 | 7.2 | 8.5 | 12 |
Assay details are provided by Reaction Biology Corp. via the internet at http://www.kinomescan.com/.
Taken together, those results show that the four compounds tested fell into two categories: ‘methylated’ analogues (1 and 2c) versus ‘non methylated’ analogues (2a and 2b), each category possessing its own selectivity profile. In addition the influence of a fluorine further tuned the activity profile of the four analogues. For instance, nilotinib (1) is selective toward Abl and PGDFRα, while 2b is more selective toward Kit and FLT3.
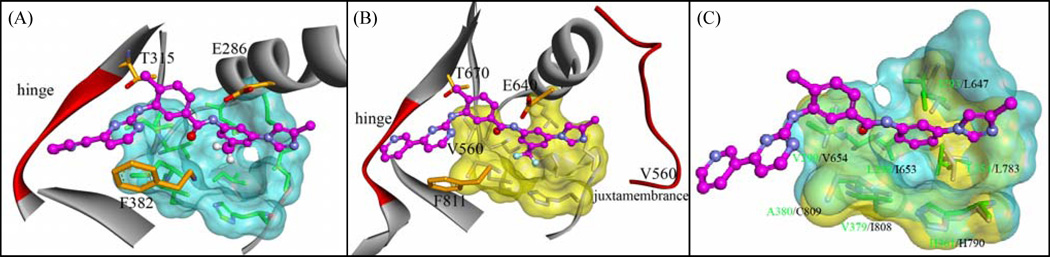
The binding modes of these nilotinib analogues to Abl and Kit were generated by docking them into the binding site followed by energy minimization refinement. As shown in figure 3, the small molecule inhibitor occupied in the ATP binding pocket of Kit17,18 in a similar manner as observed in the complex of Abl/nilotinib. Key interactions such as binding at the hinge region, H-bonding with the gatekeeper Thr-670 (Kit), and aromatic stacking interaction with Phe-811 of the DFG motif are typically retained in these kinases. Another remarkable binding feature associated with these compounds is that the edited group was orientated into a hydrophobic pocket at the bottom of the cleft, which is formed by a number of hydrophobic residues and highly conserved in Abl and Kit (Fig. 3). The trifluoromethyl group of nilotinib appears to fit more favorably into the pocket of Abl as compared to the proton and the fluoride substituent of compounds 2a and 2b that had a significantly decreased activity. Compared to Abl, the hydrophobic pocket in Kit is relatively small (estimated cavity volume is 755.4 Å3 and 822.5 Å3 for Kit and Abl, respectively), suggesting that it may have contributions to the differential activities found herein.
Figure 3.
Predicted binding mode from AutoDock: (A) compound 2c bound in the active site of Abl, (B) nilotinib bound in Kit and (C) structural overlay of the hydrophobic pocket between Kit and Abl. Compound 2a is illustrated in the binding complex. Residues of Abl are labeled in green and residues of Kit are labeled in black. Surface of the binding pocket of Abl is colored in cyan and the surface of Kit is colored in yellow.
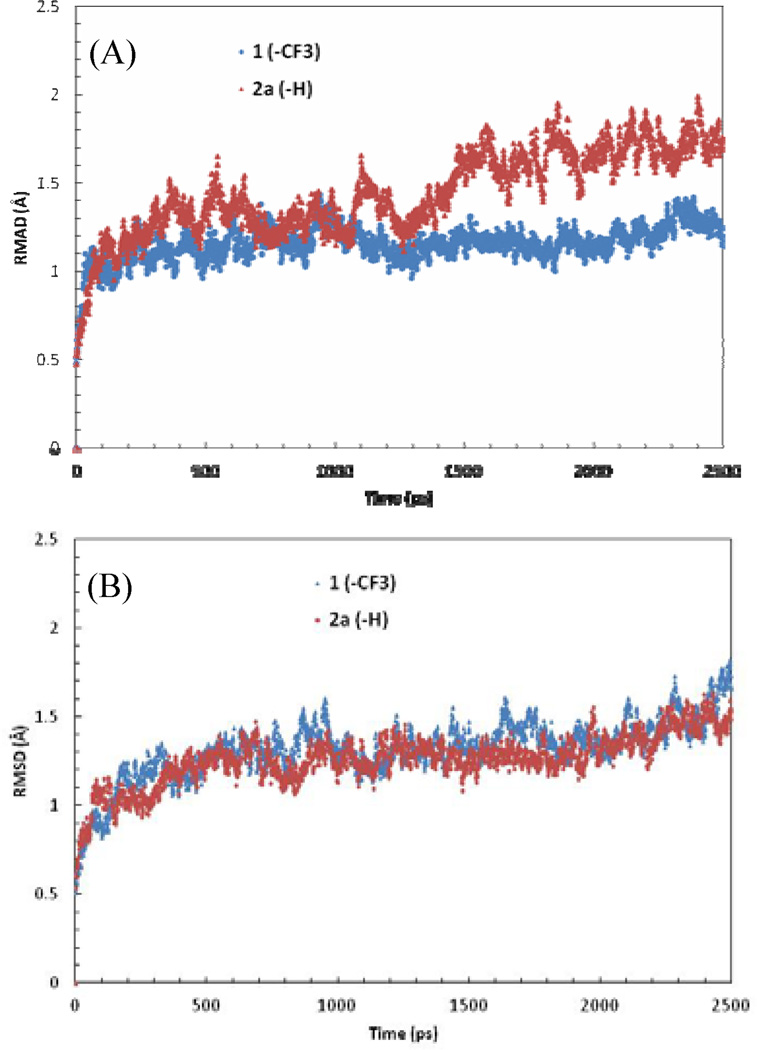
The binding and functional activities of nilotinib (1) and 2a–c versus Kit suggest that this kinase is much more tolerant of structural deviance at this specific molecular site. The protonated compound 2a, while retained the same activity as nilotinib to Kit, exhibited ~100 fold higher selectivity over Abl. This raised an interesting issue, as the binding interactions of 2a in Kit and Abl are highly conserved. Further analysis indicated that Kit possesses a unique juxtamembrane region at the N-terminal end followed by the kinase domain, which extends to the back door of the solvent exposed region of the substrate binding pocket. It is likely that the juxtamembrane loop participates in the binding interaction by stabilizing the inhibitor in the pocket. To test this hypothesis, MD simulations were performed to investigate the dynamics of inhibitor binding. As shown in figure 4, the complex of 2a bound to Abl exhibited high flexibility, but was significantly stabilized in the Kit binding complex. In contrast, the complexes of nilotinib in Abl and Kit were comparable and remained stable during the simulations. Therefore, a plausible explanation for the higher inhibitory activity and selectivity of these analogues to Kit is that, while the trifluromethyl group plays a dominant role in nilotinib binding to Abl, lack of such van der Waals interaction and shape complementarity in Kit is largely compensated by the stabilizing effect from the juxtamembrane region.
Figure 4.
RMSD plot of the inhibitor binding complexes with (A) Abl and (B) Kit from MD simulations.
To gain further insight into this possible explanation we examined nilotinib (1) and 2a–c within a series of functional assays at selected, well characterized Kit mutants (D816V, D816H, T670I, V560G, V654A) (Reaction Biology Corp. www.reactionbiology.com). Residue Asp-816 lies in front of the DFG motif in the activation loop. Though it is not directly involved in the binding interactions with inhibitors, mutation of this residue with a His or Val is expected to affect the conformation of the activation loop and, consequently the binding affinity of small molecules. This was, in fact, found with each agent possessing > 1 M activity versus D816V and > 200 nM versus D816H. The gatekeeper residue Thr-670 forms a hydrogen bond and plays a vital role in inhibitor binding and each agent in this study lost activity (> 1 M) versus the T670I mutant. It is worth noting that mutant T670I, while disrupting the key H-bonding interaction, also likely poses a steric hindrance to inhibitor in the pocket. Val-654 is found at the bottom of the binding pocket which contributes to the hydrophobic interaction. The decreased activity of these compounds (1, 2a and 2c > 1 M; 2b > 400 nM) against the mutant V654A corroborated the importance of the hydrophobic interaction in the pocket. More interestingly, these analogues showed increased inhibitory activities to mutant V560G (nilotinib (1) and 2a–c < 10 nM). Val560 lies in the juxtamembrane region and a replacement of Val to Gly may possibly make the juxtamembrane loop more flexible allowing more facile coverage of the solvent-exposed region and further stabilize inhibitor binding.
In conclusion, the influence of the trifluoromethyl group in the activity profile of nilotnib was investigated, by synthesizing analogues of nilotinib bearing a fluorine (2b), a methyl group (2c), or no substitutent (2a) in that position. Presence of a trifluoromethyl group in nilotinib (1), and of a methyl group in 2c, leads to tight fitting in Abl, and probably in DDR2 as well. Furthermore, the trifluoromethyl group allows van der Waals interactions between one of the fluorine and the one of the carbonyl backbone located in the hydrophobic pocket adjacent to the hinge region in Abl, thus further enhancing the selectivity of nilotnib toward this kinase. On the other hand, the corresponding hydrophobic pocket found in Kit is larger in size, accommodating each one of the four compounds studied. However, the role juxtamembrane region in Kit may account for the higher binding affinity of 2a and 2b toward Kit. Fluorinated analogue 2b possesses a good inhibitory activity toward Kit and FLT3, and binds tightly to those two kinases. Since kinases possess a high degree of homology, taking advantage of the subtle difference between a methyl and a trifluoromethyl group, or between a proton and fluorine may allow tuning of the pharmacological profile of kinase inhibitors.
Acknowledgements
We thank Jim Bougie, Thomas Daniel and William Leister for compound purification, as well as Paul Shinn, Danielle VanLeer and Christopher LeClair for assistance with compound management. This research was supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research grant U54HG005033 and the Intramural Research Program of the National Human Genome Research Institute at the National Institutes of Health. S. Shukla and S.V. Ambudkar were supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Lennartsson J, Jelacic T, Linnekin D, Shivakrupa R. Stem Cells. 2005;23:16. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R. Biochem. Biophys. Res. Commun. 2005;337:1. doi: 10.1016/j.bbrc.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 3.Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M. Ann. Intern. Med. 2003;138:819. doi: 10.7326/0003-4819-138-10-200305200-00010. [DOI] [PubMed] [Google Scholar]
- 4.Belloc F, Airiau K, Jeanneteau M, Garcia M, Guerin E, Lippert E, Moreau-Gaudry F, Mahon FX. Leukemia. 2009;23:679. doi: 10.1038/leu.2008.364. [DOI] [PubMed] [Google Scholar]
- 5.Manley PW, Cowan-Jacob SW, Fendrich G, Mestan J. Blood. 2005;106:940A. [Google Scholar]
- 6.Olsen JA, Banner DW, Seiler P, Wagner B, Tschopp T, Obst-Sander U, Kansy M, Müller K, Diederich F. ChemBioChem. 2004;5:666. doi: 10.1002/cbic.200300907. [DOI] [PubMed] [Google Scholar]
- 7.Kimura S, Naito H, Segawa H, Kuroda J, Yuasa T, Sato K, Yokota A, Kamitsuji Y, Kawata E, Ashihara E, Nakaya Y, Naruoka H, Wakayama T, Nasu K, Asaki T, Niwa T, Hirabayashi K, Maekawa T. Blood. 2005;106:3948. doi: 10.1182/blood-2005-06-2209. [DOI] [PubMed] [Google Scholar]
- 8.Weisberg E, Choi HG, Ray A, Barrett R, Zhang J, Sim T, Zhou W, Seeliger M, Cameron M, Azam M, Fletcher JA, Debiec-Rychter M, Mayeda M, Moreno D, Kung AL, Janne PA, Khosravi-Far R, Melo JV, Manley PW, Adamia S, Wu C, Gray N, Griffin JD. Blood. 2010;115:4206. doi: 10.1182/blood-2009-11-251751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, Cao Y, Shujath J, Gawlak S, Eveleigh D, Rowley B, Liu L, Adnane L, Lynch M, Auclair D, Taylor I, Gedrich R, Voznesensky A, Riedl B, Post LE, Bollag G, Trail PA. Cancer Res. 2004;64:7099. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 10.Weisberg E, Manley P, Mestan J, Cowan-Jacob S, Ray A, Griffin JD. Br. J. Cancer. 2006;94:1765. doi: 10.1038/sj.bjc.6603170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altman RA, Koval ED, Buchwald SL. J. Org. Chem. 2007;72:6190. doi: 10.1021/jo070807a. [DOI] [PubMed] [Google Scholar]
- 12.Altman RA, Buchwald SL. Org. Lett. 2006;8:2779. doi: 10.1021/ol0608505. [DOI] [PubMed] [Google Scholar]
- 13.Ueda S, Su MJ, Buchwald SL. J. Am. Chem. Soc. 2012;134:700. doi: 10.1021/ja2102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Analogue 2a 1H NMR (400 MHz, DMSO-d6) δ 10.54 (s, 1H), 9.53 (s, 1H), 9.28 (s, 1H), 9.17 (s, 1H), 8.70 (d, J = 4.8 Hz, 1H), 8.55 (d, J = 5.1 Hz, 1H), 8.48 (d, J = 8.0 Hz, 1H), 8.32 (t, J = 2.1 Hz, 1H), 8.29 (d, J = 1.9 Hz, 1H), 7.95 (s, 1H), 7.81 (d, J = 8.4 Hz, 1H), 7.75 (d, J = 7.9 Hz, 1H), 7.61 (t, J = 8.1 Hz, 1H), 7.58 – 7.52 (m, 1H), 7.49 (d, J = 5.1 Hz, 1H), 7.47 (m, 1H), 7.44 (d, J = 8.4 Hz, 1H), 2.36 (s, 3H), 2.36 (s, 3H).Analogue 2b 1H NMR (400 MHz, DMSO-d6) δ ppm 10.69 (s, 1 H), 9.52 (d, J=1.8 Hz, 1 H), 9.30 (d, J=1.6 Hz, 1 H), 9.19 (s, 1 H), 8.72 (dd, J=4.8, 1.5 Hz, 1 H), 8.56 (d, J=5.1 Hz, 1 H), 8.51 (ddd, J = 8.2, 2.0, 1.8 Hz, 1 H), 8.30 (d, J = 1.8 Hz, 1 H), 8.12 (m, 1 H), 7.95(m, 1 H), 7.79 (m, 1 H), 7.75 (dd, J = 8.0, 2.0 Hz, 1 H), 7.59 (dd, J = 8.1, 4.8 Hz, 1 H), 7.50 (d, J = 5.3 Hz, 1H), 7.49 (dt, J = 9.4, 2.2 Hz, 1 H), 7.45 (d, J = 8.2 Hz, 1 H), 2.36 (s, 3 H), 2.35 (s, 3 H).Analogue 2c 1H NMR (400 MHz, DMSO-d6) δ 10.45 (s, 1H), 9.52 (s, 1H), 9.28 (s, 1H), 9.16 (s, 1H), 8.71 (m, 1H), 8.55 (d, J = 5.1 Hz, 1H), 8.48 (d, J = 7.9 Hz, 1H), 8.28 (d, J = 1.9 Hz, 1H), 8.12 (m, 1H), 7.94 (m, 1H), 7.74 (m, 1H), 7.65 (s, 1H), 7.55 (m, 1H), 7.49 (d, J = 5.2 Hz, 1H), 7.44 (d, J = 7.6 Hz, 1H), 7.32 (s, 1H), 2.40 (s, 3H), 2.35 (s, 6H).
- 15.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Nat. Biotech. 2011;29:1046. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 16.Manley PW, Cowan-Jacob SW, Fendrich G, Jahnke W, Fabbro D. Blood. 2008;118:727. [Google Scholar]
- 17.Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, Snell GP, Zou H, Sang B-C, Wilson KP. J. Biol. Chem. 2004;279:31655. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 18.Mol CD, Lim KB, Sridhar V, Zou H, Chien EYT, Sang B-C, Nowakowski J, Kassel DB, Cronin CN, McRee DE. J. Biol. Chem. 2003;278:31461. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]