Abstract
Background:
Anopheles fluviatilis, one of the major malaria vectors in Iran, is assumed to be a complex of sibling species. The aim of this study was to evaluate Cytochrome oxidase I (COI) gene alongside 28S-D3 as a diagnostic tool for identification of An. fluviatilis sibling species in Iran.
Methods:
DNA sample belonging to 24 An. fluviatilis mosquitoes from different geographical areas in south and southeastern Iran were used for amplification of COI gene followed by sequencing. The 474–475 bp COI sequences obtained in this study were aligned with 59 similar sequences of An. fluviatilis and a sequence of Anopheles minimus, as out group, from GenBank database. The distances between group and individual sequences were calculated and phylogenetic tree for obtained sequences was generated by using Kimura two parameter (K2P) model of neighbor-joining method.
Results:
Phylogenetic analysis using COI gene grouped members of Fars Province (central Iran) in two distinct clades separate from other Iranian members representing Hormozgan, Kerman, and Sistan va Baluchestan Provinces. The mean distance between Iranian and Indian individuals was 1.66%, whereas the value between Fars Province individuals and the group comprising individuals from other areas of Iran was 2.06%.
Conclusion:
Presence of 2.06% mean distance between individuals from Fars Province and those from other areas of Iran is indicative of at least two sibling species in An. fluviatilis mosquitoes of Iran. This finding confirms earlier results based on RAPD-PCR and 28S-D3 analysis.
Keywords: Anopheles fluviatilis, Sibling species, Cytochrome oxidase I, Taxonomy
Introduction
Despite a considerable progress in malaria control in Iran over the past few years that led to significant reduction of cases, the disease still remains a major health problem in south and southeastern parts of the country. Inherent problems of drug resistance (Zakeri et al. 2008, Afsharpad et al. 2012) and insecticide resistance of Anopheles vectors (Enayati et al. 2003, Vatandoost et al. 2005, Hanafi-Bojd et al. 2012) are aggravated by continuous influx of imported cases, mostly with Plasmodium falciparum, from neighboring countries of Afghanistan and Pakistan (Zakeri et al. 2010), making control of disease much more difficult in these areas.
One of the key elements in fighting malaria is accurate identification of malaria vectors. The latest checklists of Iranian mosquitoes include 28 Anopheles species, identified mostly on the basis of morphological features, and a few by DNA-based approaches (Azari-Hamidian 2007). Seven species namely An. stephensi, An. culicifacies, An. dthali, An. fluviatilis, An. superpictus, An. sacharovi, and An. maculipennis are known to be responsible for transmission of malaria in the country (Manouchehri et al. 1992, Sedaghat and Harbach 2005). Some important malaria vectors in Iran are assumed to be members of species complexes or species groups, which are often difficult to distinguish morphologically.
Application of DNA-based approaches has resolved some cryptic species in Iranian complex species including An. culicifacies, An. maculipennis, An. superpictus, and An. fluviatilis, however, the identity of some members are still doubtful or were refuted later (Azari-Hamidian 2007, Naddaf et al. 2010). Molecular taxonomy of An. fluviatilis in Iran has received great attention over the past decade. This, to great extent, is due to introduction of molecular markers such as ITS2 and 28S-D3 genes for discriminating the members of this complex species in India (Manonmani et al. 2001, Singh et al. 2004). Biology, variation in behaviors, and role of this species in malaria transmission in different geographical areas of Iran has been extensively reviewed by others (Eshghi et al. 1976, Manouchehri et al. 1976, Edalat 1997–1998, Hanafi-Bojd et al. 2012). In an early study comparison of ITS2 sequence of Iranian specimens from various localities in south and southeastern Iran revealed only species Y, which is presumably species T (Naddaf et al. 2003), however, RAPD-PCR analysis of same specimens revealed two distinct patterns, separating representatives of Fars Province from other areas (Naddaf et al. 2002, Naddaf et al. 2003). Analysis of 28S-D3 gene from same populations corroborated RAPD results, Fars Province specimens showed to be identical to species U in India, while individuals from other areas exhibited heterozygocity at the only base pair position that identifies species U and T (Naddaf et al. 2010). In addition, in a separate study based on 28S-D3 analysis, species T and species U were reported from Jiroft of Fars Province and Chabahar of Sistan va Baluchestan Province, respectively (Mehravaran et al. 2011).
The aim of this study was to evaluate Cytochrome oxidase I (COI) gene alongside 28S-D3 as a diagnostic tool for identification of An. fluviatilis sibling species in Iran. COI gene sequences have been extensively used for population studies and resolving evolutionary relationship among closely related species groups of insects (Lunt et al. 1996) and Anopheline mosquitoes (Krzywinski and Besansky 2003). Variations in this fragment have been exploited as DNA barcodes for identifications of Culicidae mosquitoes including An. fluviatilis (Cywinska et al. 2006, Kumar et al. 2007).
Materials and Methods
Mosquitoes DNA
The DNA samples used in this study were obtained from An. fluviatilis mosquitoes originated from different localities in south and southeastern areas of Iran including Fars, Hormozgan, Kerman, and Sistan va Baluchestan Provinces. The extraction method and identity of some mosquitoes based on ITS2 and/or 28-D3 genes were described previously (Naddaf et al. 2002, Naddaf et al. 2003, Naddaf et al. 2010). The details for DNA samples used in this study are shown in Table 1.
Table 1.
Details for DNA samples used in this study
| NO. | Province | Specimen ID | Collection area | Identification on | |
|---|---|---|---|---|---|
| ITS2/Ref. | 28S-D3/Ref. | ||||
| 1 | Hormozgan | 117 | Siahoo/Koveh | NP | NP |
| 2 | Hormozgan | 164 | Siahoo/Koveh | T (14) | H (10) |
| 3 | Hormozgan | 169 | Siahoo/Koveh | NP | NP |
| 4 | Hormozgan | 170 | Siahoo/Koveh | NP | NP |
| 5 | Hormozgan | 172 | Siahoo/Koveh | NP | H (10) |
| 6 | Hormozgan | 173 | Siahoo/Koveh | T (14) | H (10) |
| 7 | Hormozgan | 186 | Siahoo/Koveh | NP | NP |
| 8 | Hormozgan | 120 | Minab/Tombe Basat | NP | NP |
| 9 | Hormozgan | 121 | Minab/Tombe Basat | NP | NP |
| 10 | Kerman | 815 | Kahnouj/ Condor Garmaei | NP | H (10) |
| 11 | Kerman | 834 | Kahnouj/Manoujan | NP | H (10) |
| 12 | Kerman | 836 | Kahnouj/Manoujan | NP | H (10) |
| 13 | Kerman | 839 | Kahnouj/Manoujan | NP | H (10) |
| 14 | Sistan va Baluchestan | 392 | Daman/Abchekan | NP | NP |
| 15 | Sistan va Baluchestan | 393 | Daman/Abchekan | NP | NP |
| 16 | Sistan va Baluchestan | 398 | Daman/Abchekan | T(14) | H (10) |
| 17 | Fars | 87 | kazeroun/Pirzabs | NP | NP |
| 18 | Fars | 91 | kazeroun/Pirzabs | NP | NP |
| 19 | Fars | 92 | kazeroun/Pirzabs | T (14) | U (10) |
| 20 | Fars | 97 | kazeroun/Pirzabs | NP | NP |
| 21 | Fars | 655 | Kazeroun/Islamabad | NP | NP |
| 22 | Fars | 926 | Khesht/Chiti | T (14) | U (10) |
| 23 | Fars | 928 | Khesht/Chiti | T (14) | U (10) |
| 24 | Fars | 930 | Khesht/Chiti | NP | NP |
NP= Not performed, Ref. = Reference, H= heterozygosity at the nucleotide position of 28S-D3 gene that identify species T and U.
PCR and sequencing of DNA
All the DNA samples were initially subjected to allele specific (AS)-PCR based on 28S-D3 gene as described by Singh et al. (2004). The COI gene was amplified using universal primers, UBC6 (5′- GGA GGA TTT GGA AAT TGA TTA GTT CC -3′) and UBC9 (5′-CCC GGT AAA ATT AAA ATA TAA ACT TC-3′), designed by Simon et al. (1994) and later used by Sedaghat (2003). The PCR reaction conditions were as outlined by Singh et al. (2004) with minor modifications. Each 25μl reaction contained 20 pmol of each primer, 2mM Mg Cl2, 10mM Tris-HCl, 50mM KCl, 150μM of dNTPs, 1U of Taq, and 2μl of DNA. PCR products were purified using a gel band purification kit (Pharmacia, Piscataway, NJ, USA) according to manufacturer’s recommendations and later sequenced using the same primers as used for amplification at SeqLAb laboratory in Germany. The sequences were manually edited and corrected using BioEdit software, version 7.1.3.0 (Hall 1999) and fragments of 474 bp length were selected for analysis. The COI sequences of our specimens were aligned with 59 similar sequences of An. fluviatilis, and one sequence of An. minimus as outgroup from GenBank database using Clustal X software (Thompson et al. 1997). The distances between groups and between individual sequences were calculated, and phylogenetic tree for Iranian sequences was generated using the Kimura two parameter (K2P) model of neighbor-joining method in a complete deletion procedure using MEGA 4 software (Tamura et al. 2007). The robustness of the topologies was estimated through 1000 bootstrap replications.
The sequence data for the COI gene sequences were submitted to GenBank with the accession numbers JX020706-JX020729.
Results
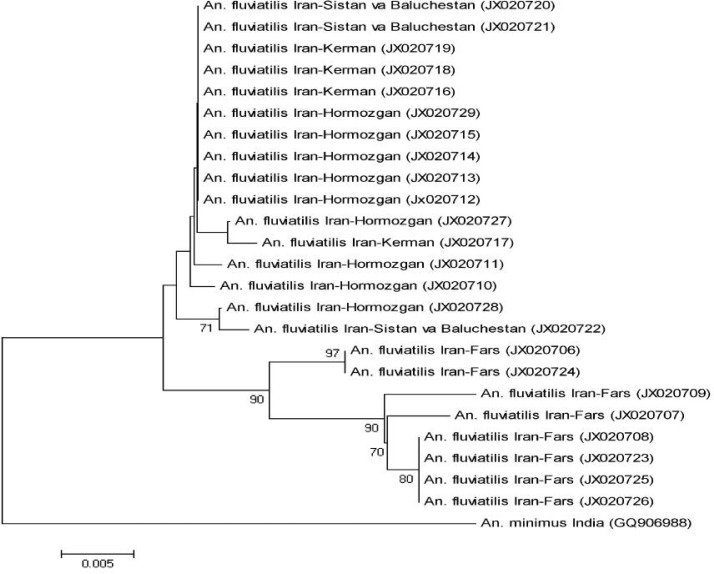
All the DNA specimens from Fars Province yield only a product of approximately 375 bp length indicative of species U, whereas specimens from Hormozgan, Kerman and Sistan va Baluchestan provinces amplified two bands of 375 bp and 128 bp length. Phylogenetic analysis using COI gene grouped individuals from Fars Province in two distinct clades separate from other Iranian individuals representing populations of Hormozgan, Kerman, and Sistan va Baluchestan (Fig. 1). Within group mean distances for Iranian and Indian individuals were 1% and 1.09%, respectively. The mean distance between Iranian and Indian groups was 1.66%, while the value between Fars group and the group comprising other Iranian members was 2.06%. The Indian group exhibited the same distance (2.06%) with Fars group. The highest distance (3.09%) among Iranian individuals was between specimens 930 from Fars Province and 398 from Sistan va Baluchestan Province. Six individuals belonging to two clades (87, 97, 655, and 928) and (92) from Fars Province showed 100% identity. In addition, ten individuals from other geographical areas including four from Koveh (170, 172, 173, and 186) and one from Minab (121) in Hormozgan Province, two from Abchekan (392–393) in Sistan va Baluchestan Province, and three from Kahnouj in Kerman Province (815, 836, and 839) were 100% identical.
Fig. 1.
Phylogenetic tree based on COI sequences. The scale bar corresponds to a 0.005 distance and the accession numbers of gene sequences are shown in parentheses.
Discussion
Accurate identification of malaria vectors is not only one of the most basic requisite for success of malaria control programs, but also has become an intriguing issue for understanding speciation process and evolution of Anopheles mosquitoes. In absence of cytotaxonomic evidence, RAPD-PCR methodology and variation in 28S-D3 gene have resolved two potential sibling species in An. fluviatilis mosquitoes of Iran. Here we report further evidence for the occurrence of these two sibling species by COI analysis of mitochondrial DNA from the same specimens.
Anopheles fluviatilis James is a complex of cryptic species; cytotaxonomic studies of polythene chromosomes has revealed three reproductively isolated species in India known as S, T, and U (Subbarao et al. 1994). Application of the first DNA-based method using ITS2 gene identified two putative species of X and Y that are presumably equivalent to species S and T, respectively (Manonmani et al. 2001). Later, a complete AS-PCR assay based on variations of 28S-D3 gene against chromosomally examined specimens identified all the members of the complex (Singh et al. 2004). Analysis of ITS2 gene revealed a single species in An. fluvialitis mosquitoes of Iran. However, the same specimens displayed variations in 28S-D3 gene; the individuals from Fars exhibited similarity with species U in India whereas individuals from others areas showed heterozygocity at the single nucleotide position that identifies species U and T. The identity of An. fluviatilis complex in Iran became complicated as the heterozygocity in 28-D3 was not reflected in ITS2 fragment and individuals from Fars Province exhibited dual identity of T and U based on ITS2 and 28S genes, respectively (Chen et al. 2006, Naddaf et al. 2010). Kumar et al. (2007) identified 62 mosquitoes species, including An. fluviatilis s.l. among members of the family Culicidae from India by COI analysis. The variation between An. fluviatilis sibling species were not addressed in their study, however, K2P genetic distances between different species of Culicidae were reported to be >2%. In our study, the mean distance between Iranian and Indian populations was 1.66%, whereas the value between Fars group and the group comprising other Iranian individuals was 2.06%. The Indian group exhibited the same distance (2.06%) with Fars group. The distance between most individuals from Fars Province and individuals from other areas was >2%. Two individuals (91–92) from Fars Province exhibited almost equivalent distance with all other members including those (930, 926, 928, 655, 87, and 97) from the same province. No variation was seen over 154 Amino acids shared by all 24 specimens. There was a sequence in GenBank from Bandar Abbas (accession no. JF966741, unpublished) that showed a high divergence from other sequences and appeared as an out group beyond An. minimus in early phylogenetic trees and hence was excluded. The results of present study were almost concordant with earlier results obtained by RAPD-PCR methodology and 28S-D3 analysis (Naddaf et al. 2002, Naddaf et al. 2010).
Conclusion
This study shows that COI gene can be used as a useful tool along other DNA markers like 28-D3 gene for dissolving closely related taxa of An. fluviatilis complex species. Analysis of more identified specimens of An. fluviatilis mosquitoes (by ITS2 and 28S-D3 genes) from India, Iran, and other geographical areas by this genetic marker can bring more insight into taxonomy of this sibling species.
Acknowledgments
We gratefully acknowledge technical assistance of our colleague Dr N Piazak from the Department of Parasitology, Pasteur Institute of Iran. This study was supported by a grant from the Pasteur Institute of Iran (No 292). The authors declare that there is no conflict of interest.
References
- Afsharpad M, Zakeri S, Pirahmadi S, Djadid ND. Molecular monitoring of Plasmodium falciparum resistance to antimalarial drugs after adoption of sulfadoxine-pyrimethamine plus artesunate as the first line treatment in Iran. Acta Trop. 2012;121(1):13–18. doi: 10.1016/j.actatropica.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Azari-Hamidian S. Checklist of Iranian mosquitoes (Diptera: Culicidae) J Vector Ecol. 2007;32(2):235–242. doi: 10.3376/1081-1710(2007)32[235:coimdc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Chen B, Butlin RK, Pedro PM, Wang XZ, Harbach RE. Molecular variation, systematics and distribution of the Anopheles fluviatilis complex in southern Asia. Med Vet Entomol. 2006;20(1):33–43. doi: 10.1111/j.1365-2915.2006.00604.x. [DOI] [PubMed] [Google Scholar]
- Cywinska A, Hunter FF, Hebert PD. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20(4):413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- Edalat H. Ecology of Anopheles fluviatilis, the main malaria vector, and its role in epedimiology of malaria. 1997–1998. [PhD Dissertation]. Tehran University of Medical Sciences, Iran.
- Enayati AA, Vatandoost H, Ladonni H, Townson H, Hemingway J. Molecular evidence for a kdr-like pyrethroid resistance mechanism in the malaria vector mosquito Anopheles stephensi. Med Vet Entomol. 2003;17(2):138–144. doi: 10.1046/j.1365-2915.2003.00418.x. [DOI] [PubMed] [Google Scholar]
- Eshghi N, Motabar M, Javadian E, Manoutcheri AV. Biological features of Anopheles fluviatilis and its role in the transmission of malaria in Iran. Trop Geogr Med. 1976;28(1):41–44. [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp. 1999;(41):95–98. [Google Scholar]
- Hanafi-Bojd AA, Vatandoost H, Oshaghi MA, Haghdoost AA, Shahi M, Sedaghat MM, Abedi F, Yeryan M, Pakari A. Entomological and epidemio logical attributes for malaria transmission and implementation of vector control in southern Iran. Acta Trop. 2012;121(2):85–92. doi: 10.1016/j.actatropica.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol. 2003;48:111–139. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44(1):1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Zhang DX, Szymura JM, Hewitt GM. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 1996;5(3):153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Manonmani A, Townson H, Adeniran T, Jambulingam P, Sahu S, Vijayakumar T. rDNA-ITS2 polymerase chain reaction assay for the sibling species of Anopheles fluviatilis. Acta Trop. 2001;78(1):3–9. doi: 10.1016/s0001-706x(00)00154-6. [DOI] [PubMed] [Google Scholar]
- Manouchehri AV, Djanbakhsh B, Eshghi N. The biting cycle of Anopheles dthali. A. fluviatilis and A. stephensi in southern Iran. Trop Geogr Med. 1976;28(3):224–227. [PubMed] [Google Scholar]
- Manouchehri AV, Zaim M, Emadi AM. A review of Malaria in Iran, 1975–1990. J Am Mosq Control Assoc. 1992;8:381–385. [PubMed] [Google Scholar]
- Mehravaran A, Oshaghi MA, Vatandoost H, Abai MR, Ebrahimzadeh A, Roodi AM, Grouhi A. First report on Anopheles fluviatilis U in southeastern Iran. Acta Trop. 2011;117(2):76–81. doi: 10.1016/j.actatropica.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Naddaf SR, Oshaghi MA, Vatandoost H, Assmar M. Molecular characterization of Anopheles fluviatilis species complex in the Islamic Republic of Iran. East Mediterr Health J. 2003;9(3):257–265. [PubMed] [Google Scholar]
- Naddaf SR, Oshaghi MA, Vatandoost H, Djavadian E, Telmadarei Z, Assmar M. Use of random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) and ITS2 PCR assays for differentiation of populations and putative sibling species of Anopheles fluviatilis (Diptera: Culicidae) in Iran. Iranian J Publ Health. 2002;32:133–137. [Google Scholar]
- Naddaf SR, Razavi MR, Bahramali G. Molecular variation and distribution of Anopheles fluviatilis (Diptera: Culicidae) complex in Iran. Korean J Parasitol. 2010;48(3):231–236. doi: 10.3347/kjp.2010.48.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghat MM. Molecular characterization and species distribution of Anopheles maculipennis complex in Iran. 2003. [PhD Dissertation]. Tehran University of Medical Sciences, Iran.
- Sedaghat MM, Harbach RE. An annotated checklist of the Anopheles mosquitoes (Diptera: Culicidae) in Iran. J Vector Ecol. 2005;30(2):272–276. [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook PK. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- Singh OP, Chandra D, Raghavendra K, SuNanda N, Sunil S, Sharma SK, Dua VK, Subbarao SK. Differentiation of members of the Anopheles fluviatilis species complex by an allele-specific polymerase chain reaction based on 28S ribosomal DNA sequences. Am J Trop Med Hyg. 2004;70(1):27–32. [PubMed] [Google Scholar]
- Subbarao SK, Nanda N, Vasantha K, Dua VK, Malhotra MS, Yadav RS. Cytogenetic evidence for three sibling species in Anopheles fluviatilis (Diptera: Culicidae) Ann Entomol Soc Am. 1994;87:116–121. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatandoost H, Mashayekhi M, Abaie MR, Aflatoonian MR, Hanafi-Bojd AA, Sharifi I. Monitoring of insecticides resistance in main malaria vectors in a malarious area of Kahnooj district, Kerman province, southeastern Iran. J Vector Borne Dis. 2005;42(3):100–108. [PubMed] [Google Scholar]
- Zakeri S, Afsharpad M, Kazemzadeh T, Mehdizadeh K, Shabani A, Djadid ND. Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2008;78(4):633–640. [PubMed] [Google Scholar]
- Zakeri S, Kakar Q, Ghasemi F, Raeisi A, Butt W, Safi N, Afsharpad M, Memon MS, Gholizadeh S, Salehi M, Atta H, Zamani G, Djadid ND. Detection of mixed Plasmodium falciparum and P. vivax infections by nested-PCR in Pakistan, Iran and Afghanistan. Indian J Med Res. 2010;132:31–35. [PubMed] [Google Scholar]