Abstract
Integration of the viral DNA in the cellular genome has been suggested to be critical in carcinogenic progression of HPV-associated cervical neoplasia. This event can be accompanied by disruption of the open reading frame (ORF) encoding the E2 repressor, thus leading to transcriptional up-regulation of the E6 and E7 viral oncogenes. At this stage, it is unclear whether disruption of the E2 ORF is mandatory for carcinogenic progression. We measured E2 RNA and protein expression in clinical samples of various grades of HPV16-associated cervical neoplasia and compared it with the status of the viral genome. RNA extracted from paraffin embedded tissues was hybridized to specific probes and quantified by the NanoString technology. Protein expression was appreciated by immunohistochemistry and the status of viral DNA was determined by in situ hybridization, all performed on serial sections of the same samples. E2 protein was found highly expressed in CIN1, CIN2 lesions where the HPV DNA was highly replicative, while it was decreased in more advanced grade lesions where replication is decreased or lost (CIN3 and SCC). In contrast, E2 transcripts could be elevated even in conditions of no or low expression of the protein, as found in the Caski cell line. Our data demonstrate that integration of the viral DNA in the cellular genome does not always lead to disruption of the E2 ORF and drastic reduction of E2 transcripts, while in contrast, expression of the E2 protein is always drastically reduced.
Keywords: HPV, transcription, IHC staining, E2, E6, E7, cyclin B1, viral genome status.
INTRODUCTION
Human papillomavirus (HPV) infection is the necessary cause of cervical cancer and HPV DNA is found in the vast majority of these cancers. Only the high risk HPV types are found associated with cancer, among which HPV16 is prominent as it is found in about 60% of cervical cancers worldwide. HPV are small DNA viruses containing an 8kb circular genome which replicates as an episome in the nuclei of infected cells. The viral DNA replication is however totally dependent on epithelial differentiation since the viral vegetative DNA replication, giving rise to thousands of new viral genomes, only occurs in terminally differentiated cells of the upper layers of the infected epithelium, reviewed in [1]. It has also been shown that viral replication requires the cellular machinery together with two viral proteins, the E1 helicase and the E2 transcription factor. Both viral proteins cooperatively bind to the origin of replication in the viral genome, and then, E1 forms the hexameric DNA helicase allowing assembly of the replication initiation complex containing the required cellular elements to start episomal replication [2]. The viral E2 protein is therefore required to initiate viral DNA replication and this occurs through binding of E2 to specific binding sites in the origin of replication. Interestingly, it has been shown that binding of E2 to these same sites leads to repression of the transcription initiated at the overlapping promoter by both steric hindrance and recruitment of transcriptional repressors, reviewed in [3, 4]. HPV16 early genes transcription including the two main viral E6 and E7 oncogenes, initiates at this promoter referred to as P97 in HPV16 [5, 6].
The viral E2 transcription factor is a negative regulator of the viral oncogenes and is therefore expected to be down-modulated during neoplastic progression. Interestingly, in cervical carcinoma cell lines, the HPV genome is integrated in the cellular genome, and this often occurs by disruption of the E1 and/or E2 open reading frames, thereby preventing E2 expression [7-9]. However, while this pattern is clearly established for some cell lines associated with HPV16 or HPV18, it is less so regarding clinical samples and more particularly HPV16-associated squamous cell carcinomas, where the viral DNA can be found in both integrated and episomal forms. In these circumstances it is not known whether the E2 protein is expressed and whether it could still exert its repressive function on E6/E7 transcription. Until recently however, expression of the E2 protein in clinical samples could not be measured due to lack of suitable reagents. Using a recently described polyclonal antibody raised against the HPV16 E2 protein, we demonstrated previously that E2 is highly expressed in the upper layers of low grade CIN1, CIN2 lesions and that this expression tends to decrease in higher grade CIN3 and SCC lesions [10].
E2 is an unstable protein which cannot accumulate in actively cycling cells since it is degraded in a cell cycle-dependent manner [11, 12]. In contrast it is stabilized in cells that are arrested in the cell cycle such as the differentiated cells composing the intermediate and upper layers of the epithelium as observed in CIN1 and CIN2 samples. In all, these data provide a coherent view of the E2 protein expression in vivo: it is highly expressed in differentiating cells where it promotes episomal vegetative replication of the viral genome and represses E6/E7 transcription. It is repressed in advanced lesions, thereby allowing efficient transcription of the E6/E7 oncogenes and cellular transformation.
Recent data have however challenged this model. There is at least one example of expression of E2 in the HPV16-associated cell line W12, coexisting with E6 and E7 to maintain the HPV16 genome in an episomal form [13]. This situation has also been described in some HPV-associated lesions where both episomal and integrated DNA has been found probably correlating with simultaneous transcription of E2 together with E6 and E7 [9]. We decided to clarify this point by studying expression of E2, together with the status of the viral DNA, in HPV16-associated clinical samples at different stages of progression. E2 protein expression was studied by immunohistochemistry (IHC) and the status of the viral DNA by in situ hybridization (ISH) in serial sections of the same samples. We also correlated expression of the E2 protein with its transcription by quantifying the E2 transcripts extracted from serial sections of the same samples using the NanoString technology which enabled us to capture and count specific transcripts in an RNA mixture. These experiments indicated that levels of E2 protein and transcripts were not always correlated and moreover that E2 transcription could be found in SCC samples with integrated viral DNA. However disappearance of the E2 protein and decreased relative transcription of the E2 gene expression compared to the E6E7 oncogenes were indicators of progression in HPV16-associated neoplasia of the cervix.
MATERIALS AND METHODS
Tissue Specimens
A total of 13 formalin-fixed and paraffin-embedded (FFPE) cervical specimens were selected from the archives of the department of Pathology, National University Hospital affiliated to National University of Singapore. The specimens included three normal cervices, three CIN2, three CIN3 and four SCC. The pathologic diagnosis was validated by 2 pathologists. Normal cervical specimens came from hysterectomy due to benign uterine diseases. CIN2 and CIN3 lesions were from loop electrosurgical excision procedure (LEEP) and SCC from hysterectomy samples. FFPE blocks were set to cut 15 serial sections for immunohistochemistry staining, DNA in situ hybridization (5μm) as well as for microdissection (10μm) in order to perform RNA extraction preparing for NanoString. Additional 3 sections were used for DNA extraction to perform HPV genotyping.
The study was approved by the Institutional Review Board of the National University of Singapore (continuing review of NUS-IRB 09-218)
Gene Expression Quantification by NanoString Technology
Total RNA was extracted from the FFPE samples using Qiagen’s RNeasy FFPE kit. Gene expression was profiled by the NanoString nCounter Analysis System according to manufacturer’s instructions. The color-coded probe sets specific to HPV 16 E6-E7, E2-E4, E2N and human cyclin B1 were designed and synthesized by NanoString Technology. Each probe pair consists of a 50-base capture probe links to biotin and an adjacent 50-base reporter probe which carries a color-coded molecular tag that provides the detection signal. The linear order of these differently colored probes, annealed to the specific transcripts in the mixture, creates a unique code for each gene of interest. All probes are mixed together with total extracted RNA from the clinical samples in a single step hybridization in solution. The sequences of the probes are as follows: HPV 16 E6-E7: ATGTCTTGTTGCAGATCATCAAGAACACGTAGAGAAACCCAGCTGTAATCATGCATGGAGATACACCTACATTGCATGAATATATGTTAGATTTGCAACC; HPV 16 E2-E4: TTGTTGCACAGAGACTCAGTGGACAGTGCTCCA ATCCTCACTGCATTTAACAGCTCACACAAAGGACGG ATTAACTGTAATAGTAACACTACACCCATAG; HPV 16 E2N: GAAACACATGCGCCTAGAATGTGCTATTTA TTACAAGGCCAGAGAAATGGGATTTAAACATATTAACCACCAGGTGGTGCCAACACTGGCTGTATCAAAG; human cyclin B1: AACTTGAGGAAGAGCAAGCAGTCA GACCAAAATACCTACTGGGTCGGGAAGTCACTGGAAACATGAGAGCCATCCTAATTGACTGGCTAGTACAGGTTCA.
For sample preparation, 100 ng total RNA was hybridized overnight with the probe sets at 65°C in a thermocycler. Hybridization mixtures were then loaded into the nCounter Prep Station where excess probes and non-target transcripts were washed away and the purified target/probe complexes were oriented in one direction by an electric field and then immobilized into the sample cartridge. Color-coded barcodes on the reporter probes were read and quantified by the nCounter Digital Analyzer. The level of expression is measured by counting the number of codes for each RNA and values are then standardized with internal controls and 3 housekeeping genes (RPL27, RPS13 and ACTB).
Cell Culture, Infection, Western blot and Immunofluorescence
Caski, SiHa, IC3 [14] and C33a cells were grown in DMEM medium supplemented with 10% FBS. Caski cells were also infected with recombinant adenoviruses expressing GFP or GFP-HPV16 E2 at a multiplicity of infection (m.o.i) of 50. The cells were harvested after 24 h for further western blots and immunofluorescent staining.
Proteins were extracted in a buffer containing (300 mM NaCl, 50 mM Tris-HCl [pH 8], 0.5% Nonidet P-40 [NP-40], 1 mM EDTA, protease and phosphatase inhibitors) for 30 minutes at 4°C followed by centrifugation. Equal amounts of total proteins were denatured, boiled and separated in 10% SDS polyacrylamide gels. After transfer, nitrocellulose membranes were saturated in PBS-Tween buffer plus 5% milk, incubated with anti-16E2 (as described in [10]) and anti-Actin as first antibody for 1 hour, secondary antibody conjugated with Horseradish Peroxidase was used for another hour. Membrane was revealed by ECL-plus kit and detected by STORM (GE Healthcare).
Caski cells grown on coverslips in DMEM supplemented with 10% FBS were rinsed with phosphate buffered saline (PBS) 24 h after infection with recombinant adenoviruses and were then fixed in 4% formaldehyde. Cells were permeabilized with 0.1% Triton and incubated with anti-16E2 serum followed by an anti-rabbit antibody coupled to Texas red. Coverslips were counterstained with DAPI.
DNA Extraction for HPV Genotyping
DNA extraction was carried out by using a column-based extraction protocol with QIAamp DNA FFPE Tissue Kit (QIAGEN). 100 ng (1-5 μl) extracted total DNA was used for HPV genotyping. HPV genotyping was performed by using the commercial HPV GenoArray test kit (Hybribio Limited, Hong Kong). The methods used both DNA amplification and HybriBio’s proprietary flow through hybridization technique to simultaneously identify 21 HPV genotypes, including 5 low-risk types (types 6, 11, 42, 43, and 44), 14 high-risk types (types16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68), and 2 undetermined-type risk types (types CP8304 and 53). The test employs a macro-array format with a nylon membrane onto which HPV genotype-specific oligonucleotides probes have been immobilized. The final results were detected by colorimetric change on the membrane under direct visualization.
Immunohistochemistry
Consecutive sections were processed with 16E2 (1:100), p16INK4 (1:100) and cyclin B1 (1:100) immunohisto-chemistry staining. Sections were immersed in xylene to remove paraffin and rehydrated through grade ethanol. Endogenous peroxidase activity was blocked by 1% hydrogen. Epitope exposure was performed by heat using 10 mM citrate buffer (pH 6.0). After blocking of nonspecific binding by preincubation with 10% goat serum for 20 minutes, primary antibodies were added for 1 hour at room temperature, after wash, Dake EnVision+, Peroxidase system, with goat anti-rabbit or mouse immunoglobulins (Ig) conjugated to peroxidase labelled-dextran polymer in Tris-HCl buffer was used as secondary antibody for 30 minutes. 3,3’-diaminobenzidine (DAB+) substrate-chromogen (Dako) was added for color development which results in a brown colored precipitate at the antigen site. The sections were counterstained with hematoxylin, dehydrated, cleared, and mounted in DPX mount medium (Leica). Anti-16E2 purified antibody was prepared as previous described [10]. p16 (clone JC8, Santa Cruz) and anti-cyclin B1 (clone Y106, Epitomics) were commercially available.
In Situ hybridization
In situ hybridizations were performed according to the manufacture’s guidelines. DakoCytomation wide spectrum HPV biotinylated DNA probe set was used to detect replicated HPV DNA, which can detect 11 types of anogenital HPV including types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, and 52. DakoCytomation GenPoint HPV biotinylated DNA probe was used to detect integrated HPV DNA which detects types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 (Dako). Replicated HPV DNA shows pan-nuclear diffused blue or blue-purple color staining, whereas integrated HPV DNA signals are dot-like in the nucleus.
Statistical Analyses
NanoString data were first normalized to the NanoString spike-in positive controls followed by normalization to 3 housekeeping controls (RPL27, RPS13, ACTB). Correlations were assessed by Pearson’s correlation test; differences in categories were analyzed by Student’s t-test. Prism (GraphPad Inc) was used for calculations and graphing.
RESULTS
Absence of HPV16 E2 Protein Expression Without Disruption of the E2 ORF in Caski Cells
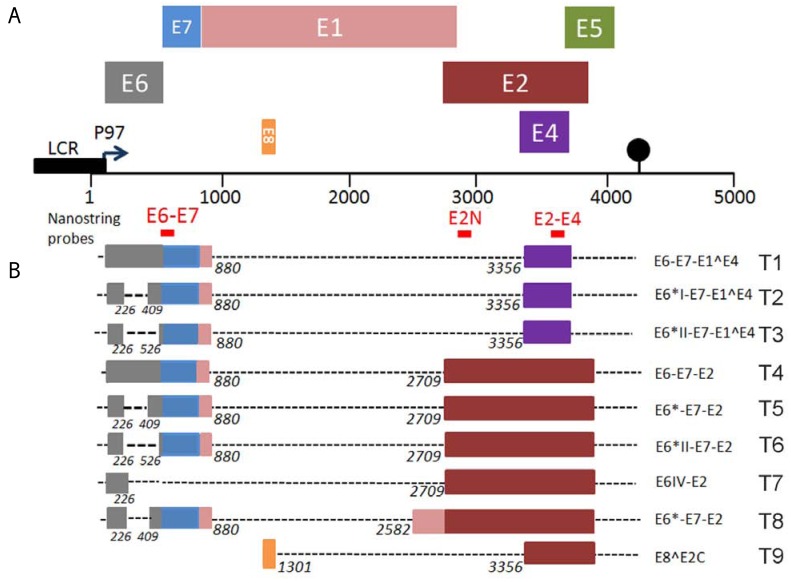
To analyze the viral transcriptome in Caski cells, total RNA was extracted and digitally quantified using the NanoString technology [15] with three HPV16-specific probes as depicted in Fig. (1A). The E6-E7 probe (nt 534-634) detects all E6-E7 transcripts (T1 – T6 and T8) (Fig. 1B). The E2N probe hybridizes to the 5’ part of the E2 RNA (nt 2855-2955), within the region encoding the N-terminal transactivation domain, and detect T4 to T8, while the E2-E4 (nt 3521-3621) detects the transcripts containing the 3’ part of the E2 including those spliced between 880 and 3356 (Fig. 1A). The latter probe can therefore detect all 9 early transcripts depicted in Fig. (1A) (T1 to T9), while the E6 and E2N probes are more selective, as they detect only 7 or 5 out of 9 of the total transcripts as summarized (Fig. 1B).
Fig. (1).
Schematic representation of the HPV16 early transcripts according to previously published map [5]. A) The P97 early promoter is shown as well as the structure of the HPV16 genome with the organization of the ORF of early genes on the DNA. The main early transcripts and the ORF that they contain are also shown. Localization of the NanoString probes relative to the viral ORFs is indicated as well as the early Poly A signal by a black dot. B) Recognition of the 9 different transcripts, as numbered in A, by the three NanoString probes.
The Caski cell line is derived from an intestinal metastasis of a cervical carcinoma associated with HPV16. It has been shown to contain about 400 copies of the HPV16 genome integrated in the cellular genome as both truncated and full-length integrates with intact E2 ORF [16]. This cell line is widely used as an in vitro model for cervical carcinoma and viral DNA integration. However, and as shown recently by Schmitt et al. [17], the E2 ORF is highly transcribed in these cells together with high transcription of the viral oncogenes. We therefore wanted to understand this situation by studying both transcription and protein expression of the E2 gene in these cells. A strong transcriptional signal, encompassing the entire E2 ORF was detected using both the E2N and the E2-E4 probes in our NanoString experiments in agreement with recently published data [17]. No significant E2 transcripts were detected in C33, a cervical carcinoma cell line not associated with HPV or in N-TERT cells, an immortalized human keratinocyte cell line (Table 1). The viral transcriptome was also analyzed in two other cervical carcinoma cell lines associated with integrated HPV16 genome: SiHa, containing a single integrated HPV16 genome with disruption of the E2 ORF [16], and IC3 containing 219 integrated copies [14], where strong signals were also obtained using the E2N probe. However, and in contrast with Caski, background levels were obtained with the E2-E4 probe confirming that, in SiHa and IC3, the E2 ORF has been disrupted by HPV16 genome integration (Table 1). In Caski however, genome integration did not disrupt the E2 ORF which is actively transcribed, as shown by the strong signals obtained with both E2N and E2-E4 probes (Table 1). Existence of these transcripts however does not necessarily infer that the specific E2 mRNA is being made since the HPV16 transcriptional map involves a large use of alternative splicing and polycistronic RNAs as depicted in Fig. (1) and in [5] and as previously discussed in [18].
Table 1.
Expression of Viral Genes and Cellular Targets was Analyzed in 6 Cell Lines, 3 Normal Cervix and 10 Clinical Samples in Relation to HPV DNA Status
| HPV16-Samples | E2-E4 | E2N | E6-E7 | Ratio E2N/E6-E7 | Ratio E2E4/E6-E7 | CyclinB1 | E2 IHC | P16INK4 | HPV DNA |
|---|---|---|---|---|---|---|---|---|---|
| Caski | 11000 | 2500 | 8500 | 0.3 | 1.3 | 4600 | - | ND | Int |
| Caski+E2 | 340 | 200 | 300 | 0.6 | 1.1 | 390 | - | ND | Int |
| SiHa | 4 | 360 | 1930 | 0.18 | - | 3900 | - | ND | Int |
| IC3 | 5 | 1960 | 4900 | 0.4 | - | 2300 | - | ND | Int |
| C33 | 4 | 5 | 2 | - | - | 3300 | - | ND | No |
| N-TERT | 5 | 7 | 4 | - | - | 2700 | - | ND | No |
| 2Ncervix | 5 | 9 | 2 | - | - | 130 | - | - | No |
| 4Ncervix | 5 | 10 | 6 | - | - | 110 | - | - | No |
| 8Ncervix | 4 | 4 | 3 | - | - | 60 | - | - | No |
| 2CIN2 | 15800 | 1520 | 570 | 2.6 | 27 | 290 | +++ | ++ | Rep ++ |
| 3CIN2 | 21500 | 4000 | 3100 | 1.3 | 7 | 1060 | +++ | ++ | Rep ++ |
| 4CIN2 | 33000 | 3900 | 2300 | 1.7 | 14 | 870 | +++ | + | Rep +++ |
| 2CIN3 | 3600 | 520 | 1130 | 0.5 | 3 | 460 | - | +++ | Rep+/- Int? |
| 12CIN3 | 136 | 29 | 70 | 0.4 | 2 | 200 | +/- | + | Rep+/- Int? |
| 18CIN3 | 81 | 38 | 38 | 1 | 2 | 190 | - | +++ | Rep+/- Int? |
| 2SCC | 3090 | 860 | 1680 | 0.5 | 1.8 | 600 | + | ++ | Int + rep? |
| 6SCC | 73600 | 37000 | 31500 | 1.17 | 2.3 | 3200 | - | +++ | Int |
| 7SCC | 11600 | 1500 | 5330 | 0.28 | 2.2 | 2000 | +/- | ++ | Int+ rep? |
| 8SCC | 15200 | 2200 | 7800 | 0.28 | 1.9 | 1600 | - | +++ | Int |
Transcription of the viral and cellular genes was measured by digital quantification using the NanoString technology. For each probe, the table shows the actual number of RNA molecules to which the probe has been bound, after standardization with 3 housekeeping genes: RPL27, RPS13 and ACTB. Coefficients of variation are typically < 7% for values above 10. Protein expression of E2 and p16INK4 was determined by immunohistochemistry (IHC) and HPV DNA status was assessed by in situ hybridization (ISH). RNA extraction, ISH and IHC were performed on serial sections of the same samples. Staining was summarized as +++ when at least half, ++ one third and + less than 1/3 of the cells of the samples were stained. For ISH, the 2 types of signals are indicated as replication (Rep) for uniform staining of the nuclei or integrated (Int) for homogenous punctate staining as shown in Fig. (3B). ND: Not Determined.
To understand the origin of E2 transcripts in Caski, we infected these cells with a recombinant adenovirus expressing the heterologous HPV18 E2. The E2 transcription factor, whether from HPV16 or HPV18, is a known repressor of the early promoter P97 (reviewed in [3]). As shown in Table 1, expression of HPV18 E2 in Caski cells induced a 32-fold decrease of all endogenous viral transcripts, as measured by the E2-E4 probe. Strong decreases were also observed with the more selective E6 (28-fold) and E2N (12-fold) probes (Table 1). Note that the probes used in this study are specific for HPV16 and do not detect ectopic HPV18 transcripts. This high sensitivity of all transcripts to E2 repression indicates that they initiate at the early P97 promoter [6] (Fig. 1A).
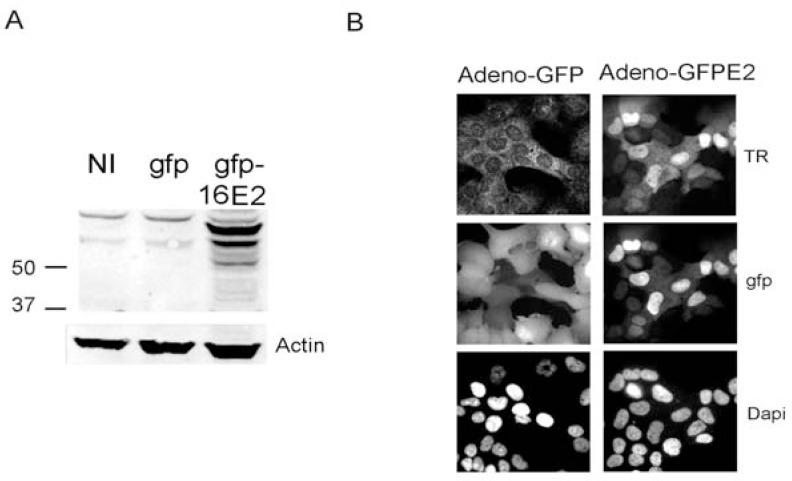
High levels of endogenous E2 transcripts in Caski cells prompted us to verify whether the protein was expressed. No endogenous E2 protein could be detected using either western blots or immunofluorescence using the HPV16 E2 antibody previously described [10] (Fig. 2). In contrast a strong signal was obtained in Caski cells infected with a recombinant ADV encoding a HPV16 GFP-E2 fusion protein (Fig. 2) or in CIN lesions, as previously demonstrated [10]. As E2 protein is known to be degraded by the proteasome in a cell cycle dependent manner [12], we also treated Caski cells with the proteasome inhibitor MG132. Even under these experimental conditions, no endogenous HPV16 E2 protein was detected (data not shown) suggesting that absence of E2 protein in Caski cells was due to the absence of proper mRNA or inefficient RNA translation rather than protein instability.
Fig. (2).
Caski cells do not express the HPV16 E2 protein. A) Caski cells were either not infected (NI) or infected with the GFP recombinant adenoviral vector (gfp) or the adenovirus expressing gfp-HPV16 E2 and were processed for immunofluorescence with the rabbit antibody raised against the HPV16 E2 protein previously described [10]. The expected MW for the untagged E2 protein is around 40Kd while the GFP tagged protein migrates at 66Kd [29]. B) Caski cells infected with the 2 recombinant adenoviruses, as indicated at the top of the pictures, were processed for immunofluorescence with the HPV16 E2 primary antibody and were revealed by secondary rabbit antibody coupled with Texas red (TR).
In summary we demonstrated here that integration of HPV16 DNA in Caski cells does not disrupt the E2 ORF, contrary to SiHa and IC3 cells. Moreover, in Caski cells the E2 ORF is actively transcribed with no detectable translation of the corresponding E2 protein.
Abundant E2 Transcripts and Absence of E2 ORF Disruption in SCC Clinical Samples
When viral transcripts were quantified using the same approach in ten HPV16 clinical samples including three CIN2, three CIN3 and four SCC, we noticed a significant decrease in total viral gene expression in CIN3 compared to CIN2 and cancer (Table 1).
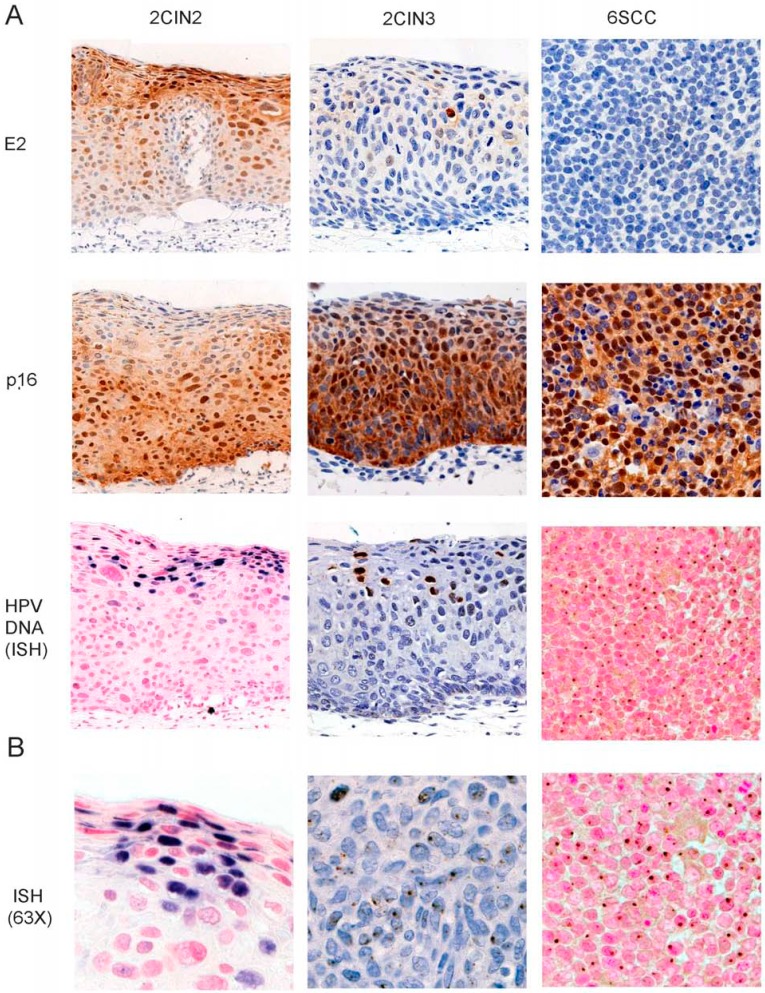
The viral DNA status was determined by ISH in serial sections of the same samples. Viral genome was episomal in the three CIN2 samples and purely integrated in two SCC samples. In two additional SCC samples, both episomal and integrated patterns were observed (Table 1 and Fig. 3A). Viral DNA was also detectable at low levels in CIN3 and when observed at high magnification, staining for HPV DNA was found both homogenous in some nuclei (episomal replication as in CIN2) or in punctate signals (which might indicate integration as in SCC) in others (Fig. 3B).
Fig. (3).
Strong E2 staining coincides with HPV DNA replication. A) Serial adjacent sections of paraffin embedded biopsies of CIN2, CIN3 and SCC were stained by IHC with the E2 and p16 antibodies as previously described [10] and as indicated. Viral DNA was revealed by in situ hybridization (ISH) with no amplification for CIN2 while the signal was amplified for the CIN3 and SCC samples in order to detect low copy number of HPV DNA (40X magnification). B) Higher magnification (63X) of cells exhibiting uniform strong dark blue nuclei in replicating cells of CIN2 while the punctate signal is homogeneous in SCC and more heterogeneous in CIN3.
In all SCC samples, despite integration of the viral genome, strong transcriptional signals were observed with both E2N and E2-E4 probes, indicating that integration of the viral genome did not disrupt the E2 ORF, a situation analogous to that found in Caski cells. As expected, strong signals were also obtained with both probes in CIN2 samples where HPV16 genomes replicate as episomes. We further compared carcinoma cell lines, CIN and SCC by computing the ratio of E2-E4/E2N transcripts therefore measuring the relative transcription of the 3’ and the 5’ moieties of the E2 ORF (Table 1). As expected, this ratio was close to 0 in cell lines with disrupted E2 ORF such as SiHa and IC3. It was between 5 and 10 in CIN2 (mean=8.1±1.5), where viral genome undergoes episomal replication. In CIN3 and SCC, lower values were observed (mean=4.7±1.4 and 5.1±1.4, respectively). The difference between CIN2 and CIN3/SCC was significant (t-test p-value=0.04), suggesting that, even if the integration event did not disrupt the E2 ORF, a different profile of transcripts might be associated with progression. In order to determine whether presence of E2 transcripts correlated with expression of the viral protein, we studied expression of E2 protein in serial sections of the same samples [10]. We also studied p16INK4 expression as a surrogate marker of E7 [19]. All 10 clinical samples exhibited an elevated expression of p16INK4. In CIN2, p16INK4 was restricted to the lower and intermediate layers, while it extended to the complete epithelium in higher grades CIN3 and SCC (Fig. 3A). E2 staining exhibited an inverted pattern, with high protein expression in the intermediate and upper layers of CIN2 and little if any protein expression in CIN3 and SCC confirming our previous findings [10] (Fig. 3A). Absence or low E2 staining was found in all SCC samples (n>50) that we have studied so far (data not shown). Therefore, absence of E2 protein, despite high transcription of E2 ORF seen in the four samples studied herein, is reminiscent of the findings described for the Caski cell line.
Transcriptional Assessment of Neoplastic Progression
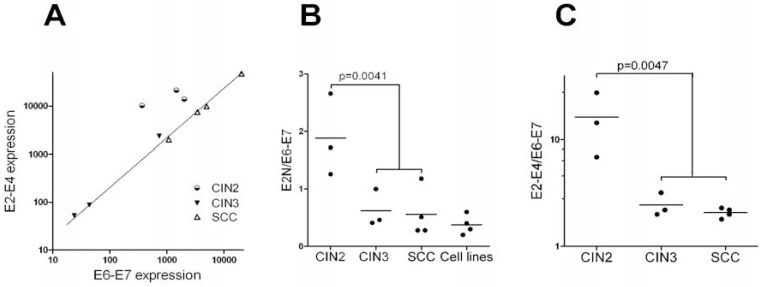
Since E2 is actively transcribed in HPV16 samples including SCC, the absolute transcription of E2N cannot be used to predict neoplastic progression (Table 1). We wondered how the relative transcription of E2 compared to E6 and/or E7 varies with the grade of the lesion. There are many alternative spliced products of the E6 and E7 ORF and our approach could not measure individual transcription of the two oncogenes as depicted in Fig. (1). Therefore we named the transcripts detected by our probe the E6-E7 transcripts. As discussed previously for the E2 transcripts, it does not mean that we detect only the E6 and/or E7 messenger RNAs. RT-PCR were done in SiHa and Caski cells with probes in the 5 and 3’ parts of the E2 ORF and a probe in E7 that confirmed the NanoString data given in Table 1 (data not shown). When E2-E4 transcripts level was plotted against E6-E7 transcripts, an almost perfect linear correlation was observed for CIN3 and SCC samples (Pearson r=0.99; 2-tail p-value<0.0001), while CIN2 samples were clearly excluded (Fig. 4A). We also calculated the E2N over E6-E7 ratio (Table 1 and Fig. 4B). In CIN2, this ratio was always above 1 (mean=1.89±0.41); in CIN3 and SCC it was usually less than 1 (mean=0.59±0.13). HPV16-associated cell lines behaved like SCC or CIN3, including the Caski cells expressing exogenous E2 (Fig. 4B). The difference between HPV lesions that can replicate the viral DNA (CIN2) and lesions where replication is low or inexistent (CIN3 and SCC) was significant (t-test p-value = 0.0038) (Table 1 and Fig. 4B). We also compared E2-E4 and E6-E7 transcription. Again a significant difference in the ratios of E2-E4 over E6-E7 transcripts was observed between CIN2 (mean=16.3±6.1) and high grade lesion CIN3 and SCC (mean=2.23±0.17; t-test p-value=0.0047) (Table 1 and Fig. 4C). Taken together, these comparisons show that even if the presence of E2 transcripts is not a significant predictor of neoplastic progression, in contrast, the relative amounts of E2 (either E2N or E2-E4) compared to E6-E7 transcripts is significantly correlated with progression between CIN2 and CIN3. In pathological diagnosis CIN2 belong to HSIL (High grade Squamous Intraepithelial Lesion) together with CIN3 while we show here that viral replication and gene expression are totally different between CIN2 and CIN3.
Fig. (4).
Relative transcription of viral genes: A) E2 transcription (measured with the E2-E4 probe) was plotted as a function of E6-E7 transcription. Note that CIN3 and SCC samples are perfectly aligned while the three CIN2 samples are not. B) The ratio of E2 transcripts (measured with the E2N probe) over E6-E7 transcripts was plotted for CIN2, CIN3, SCC and HPV16-associated cell lines (Caski, Caski-E2, SiHa and IC3). C) Same as in B, except E2 transcription was measured using the E2-E4 probe. Cell lines were not included in the comparison since no E2-E4 signal was detected for SiHa or IC3. P-value were calculated using 2-tail t-test. Data are from Table 1.
Cyclin B1 is Activated Both at Transcriptional and Protein Levels in Dedifferentiated Cells
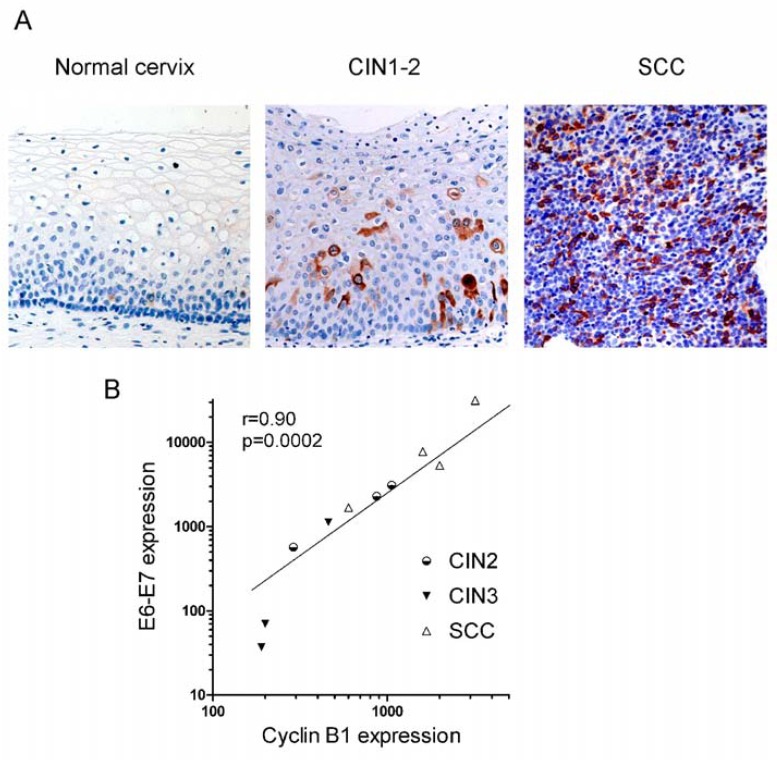
We have shown previously that, by promoting degradation of the pRB repressor, E7 induces activation of E2F and E2F target genes as well as mitotic genes in cervical carcinoma cells [20, 21]. One of these mitotic transcriptional targets is cyclin B1 whose protein expression can be detected by IHC as shown in Fig. (5A). Cyclin B1 protein is expressed at very low levels in normal cervix, being scattered in the parabasal layers; in CIN2 and CIN3, it is expressed in the cytoplasms of cells composing the lower and intermediate layers, while its protein expression extends to the whole tissue in SCC (Fig. 5A).
Fig. (5).
Cyclin B1 expression both at protein and transcription levels increases from normal cervix to SCC. A) Cyclin B1 staining obtained by IHC in normal cervix, CIN1-2 and SCC samples as indicated (20X). B) Correlation between E6-E7 and cyclin B1 transcription. Correlation coefficient and p-value were computed using 1-tail Pearson correlation test. Data are from Table 1.
We then analyzed cyclin B1 transcription. In Caski cells, ectopic expression of HPV18 E2 down-regulated cyclin B1 transcription more than 11-fold, together with repression of E6-E7 transcription, consistent with the role of E7 in cyclin B1 activation (Table 1). This E2-mediated repression is specific, since the cyclin B1 transcripts levels were standardized with 3 housekeeping genes. In addition, the p53 target gene p21 was activated 15-fold by E2 in this experiment due to repression of E6 and stabilization of p53, as previously shown [21]. Cyclin B1 was also actively transcribed in other cervical carcinoma cell lines, as well as in non-HPV-associated C33 and N-TERT, but much less in normal cervix. In patients’ samples, cyclin B1 was actively transcribed in SCC while intermediate levels of transcription were observed in CIN and transcription of CylinB1 was directly correlated to expression of the viral oncogenes in most samples (Fig. 5B and Table 1). As already noticed for E6-E7 and E2, cyclin B1 was less expressed in CIN3 than in CIN2, at least in some cases, in accordance with the fact that cyclin B1 is a downstream target of E7. Expression of p16INK4 protein is relatively high in the CIN3 samples, even though E7 could be transcribed at low levels, as in sample 18CIN3 for instance, indicating that p16 is a sensitive marker of progression while CyclinB1 transcription would more accurately reflect E7 expression.
Accumulation of the cyclin B1 protein in the cytoplasm of cells of the low and intermediate layers in CIN lesions indicates that these cells show slow activity in the G2/M phase as previously noted in in vitro differentiated raft cultures of keratinocytes expressing HPV18 [22]. Interestingly, protein expression of E2 and cyclin B1 were inversely correlated, thus confirming that another regulatory control of the E2 protein expression could indeed be its cell cycle-dependent instability, occurring in cells in G2/M phase due to concomitant over expression of the SCFskp2 ubiquitin ligase [12].
DISCUSSION
When expression of the E2 protein was compared with viral DNA replication, studied by ISH in serial sections of the same samples, a clear correlation between elevated levels of the E2 protein and viral DNA replication was seen, thereby coinciding with E2’s role as a replication factor. In addition, high expression of the E2 protein in the intermediate and upper layers of the infected epithelium also coincides with cellular differentiation associated with cell cycle arrest where E2 is stabilized [12]. In contrast, when HPV DNA was less replicative as in CIN3 and SCC, there was a clear decrease of E2 protein expression coinciding with decreased cellular differentiation. Actually the E2-E4 probe has originally been designed to detect episomal transcription compared to transcription from integrated viral DNA with disruption of the E2 ORF. Interestingly however that probe detected high RNA levels in the 10 clinical samples studied, even though at least 2 of the SCC samples contained only integrated HPV DNA. Our data thus demonstrate that transcriptional expression of the full HPV16 early region of integrated HPV DNA can occur even though the E2 and E4 proteins are not expressed [23, 24].
A typical example of high expression of E2 transcripts with no protein expression is provided by the Caski cells, which have been shown to transcribe all the HPV16 early genes previously [17] as confirmed in the present work. However, we could demonstrate that the viral E2 protein is not expressed in Caski by using an antibody which is sensitive enough to detect E2 expression in clinical samples (the present work and [10]). It was previously shown that a cDNA containing the E2 ORF isolated from Caski cells could give rise to efficient E2 translation [18]. However, the corresponding transcript has been shown to be about 100 times less abundant than the other early transcripts in Caski [17]. As discussed previously by Alloul et al. [18], it is possible that the most efficiently translated E2 transcript could be initiated at the late promoter at position 670 of the HPV16 genome, which is differentiation inducible [25]. This hypothesis would fit well with our data showing high protein expression of E2 in the intermediate and upper differentiated layers of low grade lesions where the P670 promoter is presumably active ([10] and the present work). It would also explain why the E2 protein is not expressed in higher grade lesions or in Caski cells, where the late promoter is inactive [17]. The main outcome of the present study is that the presence of transcripts containing full-length E2 does not automatically imply efficient translation of the protein. Therefore, the level of viral gene expression is of little prognosis value for grading HPV-associated neoplastic lesions as previously noted [26].
Previous studies of high grade CIN3 and SCC samples associated with different HPV types have demonstrated that integration occurs at various loci in the cellular genome [27] and that it gives rise to mixed viral cellular transcripts that, in most cases, are spliced at the E1 splice donor (nt 880 in HPV16) directly to cellular sequences [28]. We did not attempt to molecularly characterize the integration patterns in our SCC samples and we do not know whether mixed viral cellular transcripts are made. Conversely, transcription of the viral genes was not studied in the previously cited articles so that the two types of viral and mixed transcripts may coexist. Our data reveal that integration of the HPV16 genome in the cellular genome with disruption of the E2 ORF is not the only mechanism suppressing E2 protein in cancer. E2 protein expression should be down regulated to allow high E6 and E7 transcription and we have shown here that this can occur by at least one additional pathway linked to either transcriptional or translational controls or both. Interestingly the viral proteins E2, E6 and E7 are coexpressed in CIN1 and CIN2 but we have shown that this occurs in different compartments of the lesions, with E6-E7 being expressed in the basal layers while E2 is expressed in the upper layers. It is therefore quite possible that E2 actively represses early transcription in cells of the upper layers thus allowing viral DNA replication. The drastic decrease in E2 protein expression in high grade lesions is demonstrated in a limited number of samples but each of these samples has been well characterized and thereby enabled us to draw solid conclusions. More work is needed to fully comprehend regulation of transcription and translation of the viral genes in vivo. Importantly, the current study demonstrates that transcription of E2 correlates with disease progression if compared with the transcription of the viral oncogenes even in the absence of viral genomic integration. Therefore, the relative abundance of viral transcripts represents a useful marker of carcinogenesis.
ACKNOWLEDGEMENTS
We thank members of the FT lab for helpful discussion and comments. We thank John Connolly from SIgN Immunomonitoring Core for help with the NanoString technology.
ABBREVIATIONS
- HPV
= Human Papillomavirus
- ORF
= Open Reading Frame
- CIN
= Cervical Intraepithelial Neoplasia
- SCC
= Squamous Cell Carcinoma
- IHC
= Immunohistochemistry
- ISH
= In situ hybridization
- ADV
= Adenovirus
- T1-9
= Transcripts 1-9
- MW
= Molecular weight
- GFP
= Green Fluorescent Protein
- m.o.i.
= Multiplicity of infection
- HSIL
= High grade Squamous Intraepithelial Lesion
- LSIL
= Low grade Squamous Intraepithelial Lesion
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1. Kadaja M, Silla T, Ustav E, Ustav M. Papillomavirus DNA replication - from initiation to genomic instability. Virology. 2009;384:360–8. doi: 10.1016/j.virol.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 2. Abbate EA, Berger JM, Botchan MR. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 2004;18:1981–96. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thierry F. Transcriptional regulation of the papillomavirus oncogenes by cellular and viral transcription factors in cervical carcinoma. Virology. 2009; 384:375–9. doi: 10.1016/j.virol.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 4. Bellanger S, Tan CL, Xue YZ, Teissier S, Thierry F. Tumor suppressor or oncogene? A critical role of the human papillomavirus (HPV) E2 proten in cervical cancer progression. Am J Cancer Res. 2011; 1:373–89. [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006; 11:2286–302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romanczuk H, Thierry F, Howley PM. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–59. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schneider-Gädicke A, Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 1986;5:2285–92. doi: 10.1002/j.1460-2075.1986.tb04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwarz E, Freese UK, Gissmann L, Mayer W, Roggenbuck B, Stremlau A. Z H H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–4. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 9. Klaes R, Woerner SM, Ridder R, et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6. [PubMed] [Google Scholar]
- 10. Xue Y, Bellanger S, Zhang W, et al. HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 2010;70:5316–25. doi: 10.1158/0008-5472.CAN-09-3789. [DOI] [PubMed] [Google Scholar]
- 11. Bellanger S, Demeret C, Goyat S, Thierry F. Stability of the human papillomavirus type18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J Virol. 2001; 75:7244–51. doi: 10.1128/JVI.75.16.7244-7251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bellanger S, Tan CL, Nei W, He PP, Thierry F. The human papillomavirus type 18 E2 protein is a cell cycle-dependent target of the SCFSkp2 ubiquitin ligase. J Virol. 2010; 84:437–44. doi: 10.1128/JVI.01162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gray E, Pett MR, Ward D, et al. In vitro progression of human papillomavirus 16 episome-associated cervical neoplasia displays fundamental similarities to integrant-associated carcinogenesis. Cancer Res. 2010; 70:4081–91. doi: 10.1158/0008-5472.CAN-09-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peter M, Stransky N, Couturier J, et al. Frequent genomic structural alterations at HPV insertion sites in cervical carcinoma. J Pathol. 2010; 221:320–30. doi: 10.1002/path.2713. [DOI] [PubMed] [Google Scholar]
- 15. Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008; 26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 16. Baker CC, Phelps WC, Lindgren V, Braun MJ, Gonda MA, Howley PM. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987; 61:962–71. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmitt M, Pawlita M. The HPV transcriptome in HPV16 positive cell lines. Mol Cell Probes. 2011; 25:108–13. doi: 10.1016/j.mcp.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 18. Alloul N, Sherman L. The E2 protein of human papillomavirus type 16 is translated from a variety of differentially spliced polycistronic mRNAs. J Gen Virol. 1999; 80:29–37. doi: 10.1099/0022-1317-80-1-29. [DOI] [PubMed] [Google Scholar]
- 19. Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001; 92:276–84. doi: 10.1002/ijc.1174. [DOI] [PubMed] [Google Scholar]
- 20. Thierry F, Benotmane MA, Demeret C, Mori M, Teissier S, Desaintes C. A genomic approach reveals a novel mitotic pathway in papillomavirus carcinogenesis. Cancer Res. 2004; 64:895–903. doi: 10.1158/0008-5472.can-03-2349. [DOI] [PubMed] [Google Scholar]
- 21. Teissier S, Ben K Y, Mori M, Pautier P, Desaintes C, Thierry F. A new E6/P63 pathway, together with a strong E7/E2F mitotic pathway, modulates the transcriptome in cervical cancer cells. J Virol. 2007; 81:9368–76. doi: 10.1128/JVI.00427-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banerjee NS, Wang HK, Broker TR, Chow LT. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J Biol Chem. 2011; 286:15473–82. doi: 10.1074/jbc.M110.197574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Middleton K, Peh W, Southern S, et al. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J Virol. 2003; 77:10186–201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doorbar J. The papillomavirus life cycle. J Clin Virol. 2005;32(1 ) Suppl:S7–15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 25. Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996; 70:2339–49. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hafner N, Driesch C, Gajda M, et al. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. 2008; 27:1610–7. doi: 10.1038/sj.onc.1210791. [DOI] [PubMed] [Google Scholar]
- 27. Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004; 64:3878–84. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 28. Kraus I, Driesch C, Vinokurova S, et al. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 2008;68:2514–22. doi: 10.1158/0008-5472.CAN-07-2776. [DOI] [PubMed] [Google Scholar]
- 29. Blachon S, Bellanger S, Demeret C, Thierry F. Nucleo-cytoplasmic shuttling of high risk human Papillomavirus E2 proteins induces apoptosis. J Biol Chem. 2005;280:36088–98. doi: 10.1074/jbc.M505138200. [DOI] [PubMed] [Google Scholar]