Abstract
Based on the crystal structure of bovine β4Gal-T1 enzyme, mutation of a single amino acid Y289 to L289 (Y289L) changed its donor specificity from Gal to N-acetyl-galactosamine (GalNAc). A chemoenzymatic method that uses GalNAc analogues like GalNAz or 2-keto-Gal as sugar donors with the enzyme Y289LGal-T1 has identified hundreds of cytosolic and nuclear proteins that have O-GlcNAc modifications. To avoid potential cytotoxicity at Mn2+ concentrations required to selectively modify GlcNAc residues on the surface of live cells, we have engineered a Mg2+-dependent enzyme. Earlier, we have found that the mutation of the metal-binding residue Met-344 to His-344 in bovine β4Gal-T1 enzyme altered its meta-lion specificity in such a way that the M344H-β4Gal-T1 enzyme exhibits better catalytic activity with Mg2+ than with Mn2+. Here, we find that when these two mutations are combined, the double mutant, Y289L-M344H-β4Gal-T1, transfers GalNAc and its analogue sugars to the acceptor GlcNAc in the presence of Mg2+. Using this mutant enzyme, we have detected free GlcNAc residues on the surface glycans of live HeLa cells and platelets. The specific transfer of a synthetic sugar with a chemical handle to the terminal GlcNAc residues on the surface of live cells provides a novel tool for selective modification, detection, and isolation of GlcNAc-ending glycans present on the cellular surface.
INTRODUCTION
The surface of a cell is covered with a dense layer of glycans, called a glycocalyx. They are the oligosaccharides that form part of the plasma membrane-bound glycoproteins, proteoglycans, and glycolipids. Glycoproteins at the cell surface contain N-linked, bi-, tri-, or tetra-antennary oligosaccharides, and/or O-GalNAc-linked mucin-type oligosaccharides1, and/or a recently identified O-GlcNAc single sugar attachment.2 Distinct sets of glycans, synthesized by a superfamily of enzymes called glycosyltransferases (GTs) (http://www.cazy.org/)3, form the glycosylation phenotype of a cell. These glycans change during cellular growth, development, and differentiation, as well as under pathological conditions such as inflammation and cancer.4–8
In the growing glycomics field, new strategies for detecting and identifying the glycan moieties associated with cellular status are needed. Monosaccharides with chemical handles have been shown to be incorporated through cellular metabolic pathways by the glycosyltransferases in the glycans of glycoconjugates.9 The presence of these sugars with chemical handles on the cell surface glycans can be detected by many bioconjugation techniques.10
In our laboratory, we have studied the structure and function of the β1,4-galactosyltransferase 1 (β4Gal-T1). This enzyme is a member of a subfamily that, in the presence of Mn2+ ion, transfers galactose from UDP-Gal to an acceptor substrate, N-acetylglucosamine (GlcNAc), that is present at the nonreducing ends of glycans, creating a β1-4 disaccharide linkage. These have led to the structure-based design of several mutant enzymes with novel sugar donor substrate specificities.11 For example, the mutation of the Tyr-289 residue to Leu289 in bovine β4Gal-T1 creates a cavity in the catalytic pocket of the enzyme that can accommodate a UDP-Gal molecule carrying a chemical handle at C2, such as 2-keto-Gal or GalNAz.12 This mutant enzyme, Y289L-β4Gal-T1, has been used for in vitro detection of O-GlcNAc residues on proteins13, even at the cellular level.14 In addition, the Y289L-Gal-T1 enzyme has been used to detect the presence of a terminal GlcNAc moiety on the cell surface glycans of normal and malignant tumor tissues embedded in paraffin.15
The Mn2+ ion is a cofactor required for β4Gal-T1 catalytic activity and we have previously shown that three residues, Asp-254, Met-344, and His-347 coordinate the metal ion Mn2+.16 When Met-344 is substituted with His-344, the resulting mutant enzyme, M344H-β4Gal-T1, loses 98% of its Mn2+-dependent activity, but shows 25–30% activity in the presence of Mg2+.16 For enzymatic in vitro galactosylation assays, usually a 5–10 mM concentration of Mn2+ is used for optimal activity. It has been shown that these concentrations of Mn+2 have potential cytotoxic effects.17–19 To overcome these effects, we have combined the two mutations, Y289L and M344H, in β4Gal-T1 to generate a double mutant, Y289L-M344H- β4Gal-T1, that transfers GalNAc or Gal with a chemical handle at C2 in the presence of Mg2+.
Using the double mutant enzyme, Y289L-M344H- β4Gal-T1, we can detect GlcNAc in the presence of Mg2+ on the surface of live human cervical cancer cells (HeLa). In addition, with the described method we show that there is an increase in exposed free GlcNAc on the cell surface of chilled human platelets, as reported previously.20 We also show that cell surface glycoprotein molecules carrying transferred GalNAz after conjugation with BCN-biotin molecules can be captured with and eluted from streptavidin-magnetic beads for further characterization.
EXPERIMENTAL PROCEDURES
Protein expression and folding
The β4Gal-T1 gene containing the double mutation Y289L and M344H (Y289L-M344H-β4Gal-T1) was generated by combining the genes containing the corresponding single mutations from Y289L-β4Gal-T1 and M344H-β4Gal-T1 plasmids that were described previously.12,16 Briefly, the catalytic domain of the bovine β4Gal-T1 gene is constructed in the pET23a vector between the Bam HI and Eco RI restriction sites and the gene fragment has a Mlu I restriction endonuclease site between the Tyr-289 and Met-344 codons. The DNA fragment in the Y289L-β4Gal-T1 plasmid between the Mlu I and EcoRI restriction sites, was exchanged with the corresponding fragment in the M344H-β4Gal-T1 plasmid that contains the mutation M344H between the MluI and EcoRI sites. The resulting plasmid DNA, Y289L-M344H-β4Gal-T1, which carries both mutations, was transfected into BL21(DE3) pLysS cells as previously described.12,16 The recombinant protein was expressed, folded, and purified as described earlier.21 The protein concentration was estimated using Bio-Rad protein assay dye reagent (Bio-Rad Laboratories), based on the method of Bradford, using Gal-T1 as a standard for protein determination. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized using Brilliant Blue G (Sigma-Aldrich) according to standard protocols.
GalNAc-T enzyme assay
The reactions were performed at 37 °C for 10 min in a 100 µl volume containing 20 mM MgCl2, 20 mM Tris HCl (pH 8.0), from 0 to 2 mM UDP-GalNAc, 0.05 µCi 3H-labeled sugar nucleotide, and 25mM β-benzyl-GlcNAc with 1.5 µg of enzyme. The reactions were stopped by the addition of 200 µl of ice-cold water, and the mixture was passed through a 0.5-ml bed volume column of AG1-X8 cation resin (BioRad) to remove any unreacted UDP-GalNAc. The column was washed twice with 500 µl of water, and the column flow-through was diluted with Ecoscint A scintillation solution. Radioactivity was measured using a Beckman counter. A reaction without the acceptor was used as a control. Experiments were done in triplicate and the values were averaged. Transfer of C2 keto-galactose from its UDP derivative to GlcNAc residues on a glycoacceptor was performed as previously described.22 The apparent Km value for UDP-GalNAc was calculated based on the enzyme activity curve, fitted with SIGMA plot using an equation defining ligand binding to one-site saturation without inhibition.
Mass spectrometry
The transfer of GalNAc or 2-keto-Gal to a GlcNAc moiety on a biantennary N-glycan acceptor substrate linked to a tetra-peptide, that is desialylated and degalactosylated, was followed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry.
Cell culture
Human cervix adenocarcinoma (HeLa) cells (ATCC CCL-2) were grown in DMEM with high glucose (Gibco) with 10% fetal bovine serum (FBS) (Gibco) and 100 IU/ml penicillin-100 µg /ml streptomycin (HyClone). Cells were harvested using trypsin (Gibco).
Cell viability assay
After labeling, the cells were resuspended in 100 µl of DPBS (Gibco), and mixed with an equal volume of 0.4% Trypan blue stain (Gibco). Clear, viable cells were counted microscopically on KOVA Glasstic slides with grid chambers (Hycor, Garden Grove, CA). Cell number is expressed as a percentage of the control cells (without treatments).
Cell surface labeling and detection of GlcNAc on HeLa cells for confocal microscopy
A suspension of 104 HeLa cells was plated on an 18-well slide (ibiTreat, Ibidi Integrated BioDiagnostics, Munich, Germany) and grown overnight in DMEM supplemented with 10% FBS. Cells were washed once with media and pretreated with 1.5 mU of recombinant β1,4-galactosidase from Streptococcus pneumoniae (Calbiochem) and 2.5 mU of recombinant α2-3,6,8,9-neuraminidase from Arthrobacter ureafaciens (Calbiochem) for 30 min at 37 °C in DMEM. Cells were washed three times with media and incubated with a cocktail containing 0.5 mM UDP-GalNAz (Life Technology), 10 µg of Y289L-M344H-β4Gal-T1 enzyme, 20mM MgSO4, and 3 mM HEPES in HBSS buffer for 30 min at 37 °C. Cells were washed twice and incubated with 100 µM DIBO-Alexa Fluor 488 (Life Technologies) in DMEM for 1 h at 28.5 °C. Cells were washed six times with DMEM and fixed for 15 min with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, Hartfield, PA) in PBS. Nuclei were stained with 5 µg/ml Hoechst 33342 dye (Invitrogen) for 5 min and slides were mounted with mounting media (Ibidi). Cells were examined using an Olympus FluoView 1000 inverted microscope with 405 LD and 488 Multi-Argon lasers. A 60× 1.42 oil-immersion objective was used. Images were acquired at 4 µs/pixel using Olympus FV100 2.0c software. For analysis, FV10-ASW 1.6 Viewer and ImageJ (Wayne Rasband, National Institutes of Health) were used.
Cell surface labeling and detection of GlcNAc on HeLa cells for flow cytometry
HeLa cells in suspension were pretreated with 2.5 mU of recombinant α2-3,6,8,9-neuraminidase from Arthrobacter ureafaciens (Calbiochem) and 1.5 mU of recombinant β1,4-galactosidase from Streptococcus pneumoniae (Calbiochem) for 30 min at 37 °C. Following two washes with media, cells were incubated with a cocktail containing 0.5 mM UDP-GalNAz, 10 µg of Y289L-M344H-β4Gal-T1 enzyme, 20mM MgSO4, and 3 mM HEPES in HBSS buffer for 30 min at 37 °C. Cells were washed twice and incubated with 100 µM DIBO-Alexa Fluor 488 in DMEM for 1 h at 28.5 °C. Cells were washed three times and 10,000 events were acquired with a Becton Dickinson FACScalibur cytometer using CellQuest software.
Detection of cell surface GlcNAc by Western blot analysis
HeLa cells in suspension were pretreated with glycosidases as described above, followed by enzymatic transfer with Y289L-M344H-β4Gal-T1 enzyme. After two washes, the cells were incubated with 100 µM BCN-biotin conjugate (SynAffix, AJ Nijamen, The Netherlands) in DMEM for 1 h at 28.5 °C. After three washes, the cells were resuspended in lysis buffer (20 mM HEPES, 0.5% SDS). Following a 1-min sonication, the cell lysate was boiled for 10 min and centrifuged for 15 min at 21450g. Lysates were stored at −80 °C for further analysis. Protein estimation was performed with the Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL). Two micrograms of total protein were resolved in a 4–20% SDS-PAGE gel and transferred to a nitrocellulose membrane (Invitrogen, Carlsbad, CA) by electroblotting. The membrane was blocked in a solution of 5% milk and 0.2% Tween 20 for 30 min at room temperature. After three 5-min washes with water, the membrane was incubated with 1:3000 SA-HRP in PBS-BSA buffer (3% BSA, 0.02% Tween 20) for 30 min at room temperature. The membrane was then washed four times (5 min each) with blocking solution, incubated with substrate from the ECL Western Blotting Analysis System (GE Healthcare, UK), and exposed to HyBlot CL autoradiography film (Denville Scientific, Metuchen, NJ).
Streptavidin purification of biotinylated cell protein extracts
To purify biotin-labeled proteins, SDS was removed from 10 µg of glycosidase pretreated and BCN-biotinylated HeLa lysate proteins using Calbiosorb resin (Calbiochem) for 15 min on ice. The resin was allowed to settle. The supernatant was collected and incubated with 20 µl of Dynabeads MyOne Streptavidin C1suspension (Invitrogen) with gentle agitation for at least 24 h at room temperature. Unbound protein was collected and the beads were washed twice with PBS containing 0.01% Tween 20. To elute the biotinylated proteins, the beads were boiled for 5 min in 0.1% SDS. Proteins were resolved on 4–20% SDS-PAGE gels and detection was performed as described above.
N-glycan release from labeled cell extracts
The SDS in the eluted fraction from the streptavidin-coated beads that contained BCN-biotinylated HeLa cell proteins was removed using Calbiosorb resin (Calbiochem) for 15 min on ice. Briefly, the sample was denatured by boiling for 10 min in 1X glycoprotein denaturing buffer. A portion of the sample was treated with 1X G7 reaction buffer, 1% NP40, and PNGase F (New England Biolabs); in another portion the PNGase F enzyme was omitted. Protein samples were resolved in SDS-PAGE gels and analyzed by Western blot analysis.
Cell surface labeling and detection of GlcNAc on human platelets for flow cytometry
Human whole blood in 3.8% trisodium citrate was spun for 20 min at 900 rpm. Platelet-rich plasma (PRP) was obtained from the supernatant and 0.2 U/ml of apyrase and 2 µl/ml of PGE1 were added. The PRP was kept at room temperature unless otherwise indicated. To study the effect of platelet chilling, 2×107 platelets were incubated overnight at room temperature or at 8 °C. After the treatment, the platelets were divided into two tubes, spun down for 5 min at 3000 rpm, resuspended in galactosylation cocktail (0.5 mM UDP-GalNAz, 10 µg of Y289L-M344H-β4Gal-T1 enzyme, 20mM MgSO4, and 3 mM HEPES in HBSS buffer), and incubated for 30 min at 37 °C. The platelets were washed twice in buffer containing 140 mM NaCl, 3 mM KCl, 1 mM MgCl2, 5 mM NaHCO3, 10 mM glucose, 10 mM HEPES, pH 7.4, and 5 mM EDTA. The platelets were then conjugated with 100 µM DIBO-Alexa Fluor 488 for 1 h at 28.5 °C. Following two washes, the platelets were fixed in 2% PFA in PBS for 20 min at room temperature, washed with PBS, and analyzed by flow cytometry. Fifty thousand gated events were acquired in a Becton Dickinson FACScalibur flow cytometer using CellQuest software.
Labeling of α-crystallin
Twenty micrograms of α-crystallin from bovine eye lens were incubated for 3 h at 30 °C with 2 µg of Y289L or Y289LM344H in the presence of 7.5 mM MnCl2 or MgCl2, respectively, in a buffer containing 1 mM UDP-keto-Gal, 50 mM NaCl, 20 mM HEPES, and 2% NP-40 in a 20 µl final volume reaction. In control reactions, the UDP-2 keto-galactose was omitted. Biotinylation was carried out overnight at room temperature in a 60 µl volume reaction containing 50 mM sodium acetate buffer, pH 3.9, and 3 mM aminooxy-biotin (ARP, Dojindo Laboratories). One hundred nanograms of protein were resolved in a 14% SDS-PAGE gel and transferred to nitrocellulose membranes. The membranes were blocked with 5% BSA, 250 mM NaCl, and 0.3% Tween 20. Detection with streptavidin-HRP was performed as described above.
RESULTS
Expression and activity of the double mutant enzyme Y289L-M344H-β4Gal-T1
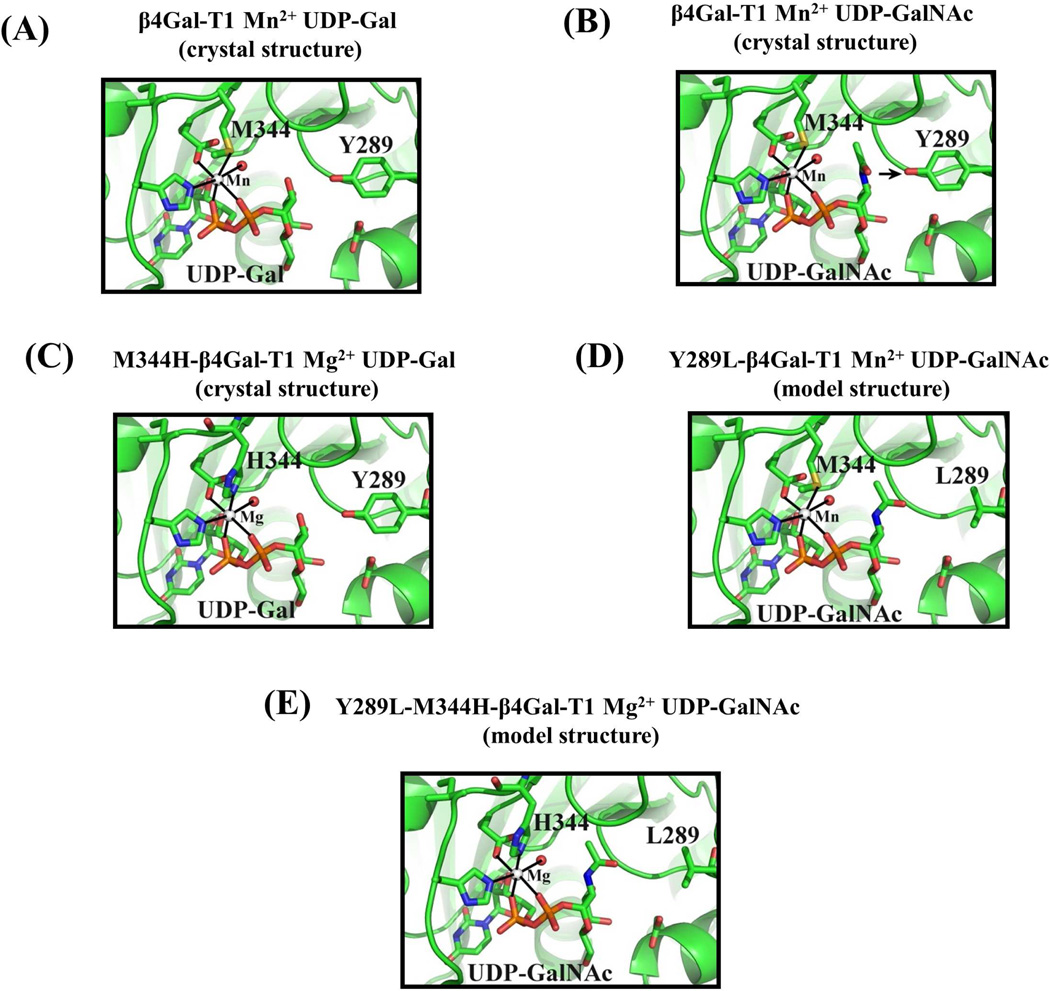
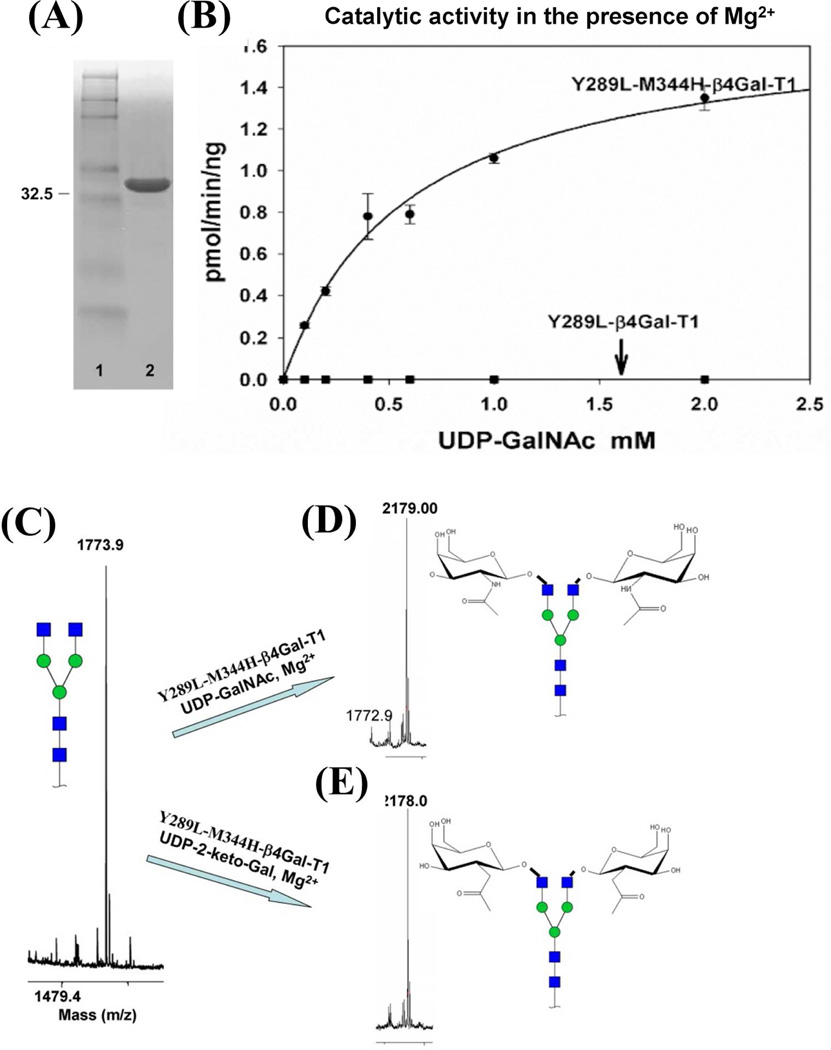
The double mutant enzyme Y289L-M344H-β4Gal-T1 was constructed from two single bovine β4Gal-T1 mutants, Y289L and M344H. The structures of the catalytic pocket are shown (Fig. 1) of the wild-type β4Gal-T1 with either bound UDP-Gal (Fig. 1A) or UDP-GalNAc (Fig. 1B), or M344H-β4Gal-T1 with bound UDP-Gal (Fig. 1C) or the modeled UDP-GalNAc-bound structure for the Y289L-β4Gal-T1 (Model, Fig. 1D) and Y289L-M344H-β4Gal-T1 (Model, Fig. 1E). The double mutant Y289L-M344H-β4Gal-T1 was expressed in E. coli as inclusion bodies, as were the single mutants. The refolding of the inclusion bodies, using the methods previously described (21), yielded about 37 mg of active protein from 80 mg of inclusion bodies. The protein remained in a monomeric form as judged by SDS-PAGE analysis under nonreducing conditions, which showed a single 33 kDa band (Fig. 2A). The enzymatic activity of the Y289L-M344H-β4Gal-T1 protein was measured at fixed concentrations of MgCl2 and increasing concentrations of UDP-GalNAc, showing a sigmoidal behavior with an apparent Km of 590 µM for UDP-GalNAc (Fig. 2B). In the absence of MnCl2, the wild-type or the Y289L-β4Gal-T1 mutant enzyme does not transfer Gal to the GlcNAc residue. The MALDI mass spectrometric analysis of a commercially available biantennary glycan (Fig. 2C) shows one peak at 1773.9 m/z. This biantennary glycan was used as an acceptor substrate. After UDP-GalNAc transfer by the mutant enzyme Y289L-M344H-β4Gal-T1 in the presence of Mg2+ (Fig. 2D), a peak appears at 2179.0 m/z, corresponding to the addition of two GalNAc moieties. Similarly, after UDP-C2 keto-Gal transfer, a peak at 2178.0 m/z appears that corresponds to the addition of two C2 keto-Gal moieties to the terminal GlcNAc residues of the biantennary glycan (Fig. 2E).
Figure 1.
A: In the crystal structure of the β4Gal-T1 complex with Mn2+ and UDP-Gal (pdb 1TVY), the side chain sulfur atom Sδof the Met-344 residue forms a coordination bond with the bound Mn2+ ion. B: In the crystal structure of the bovine β4Gal-T1 molecule that is in complex with UDP-GalNAc (pdb 1OQM), the N-acetyl moiety of the GalNAc sugar exhibits steric hindrance with the side chain of Tyr289. When the Tyr residue is mutated to Leu, resulting in the mutant Y289L-Gal-T1, it creates a cavity that can accommodate the N-acetyl moiety of GalNAc. The mutant enzyme Y289L-β4 GalT1 transfers GalNAc to GlcNAc (12). C: When the residue Met-344 is mutated to a His, a Mg2+ ion, in place of the Mn2+ ion, coordinates with the His-344 residue and activates the Met-344-His (M344H) mutant enzyme. D: Based on the crystal structure, the Y289L mutation has been modeled to show the space created between the L289 residue and the N-acetyl moiety of the GalNAc residue. E: The combination of both mutations, Y289L and M344H, has been modeled. The double mutant Y289L-M344H-β4 GalT1 is expected to transfer GalNAc sugars in the presence of Mg2+.
Figure 2.
The expression and the catalytic activity of the double mutant enzyme Y289L-M344H-β4GalT1. A: Nonreducing SDS-PAGE gel stained with Brilliant Blue G showing 10 µg of active in vitro refolded Y289L-M344-β4GalT1 mutant protein in lane 2, with the molecular weight marker in lane 1. B: Catalytic activity of the Y289L-β4Gal-T1 and Y289L-M344H-β4Gal-T1 proteins in the presence of MgCl2, UDP-GalNAc, and a 25 mM β-benzyl-GlcNAc as an acceptor substrate. The single mutant enzyme does not show any catalytic activity in the presence of Mg2+.MALDI-TOF spectra of the G0 form of the N-glycan acceptor substrate, C: and its G2 form of the product with GalNAc, D: and 2-keto-Gal sugars, E, in the catalytic reaction with the double mutant enzyme Y289L-M344-β4Gal-T1 in the presence of Mg2+. Major peaks are annotated with the carbohydrate structure that is shown in symbols for monosaccharides, according to the nomenclature adopted by the Consortium for Functional Glycomics.
Cell surface GlcNAc detection using the double mutant enzyme Y289L-M344H-β4Gal-T1
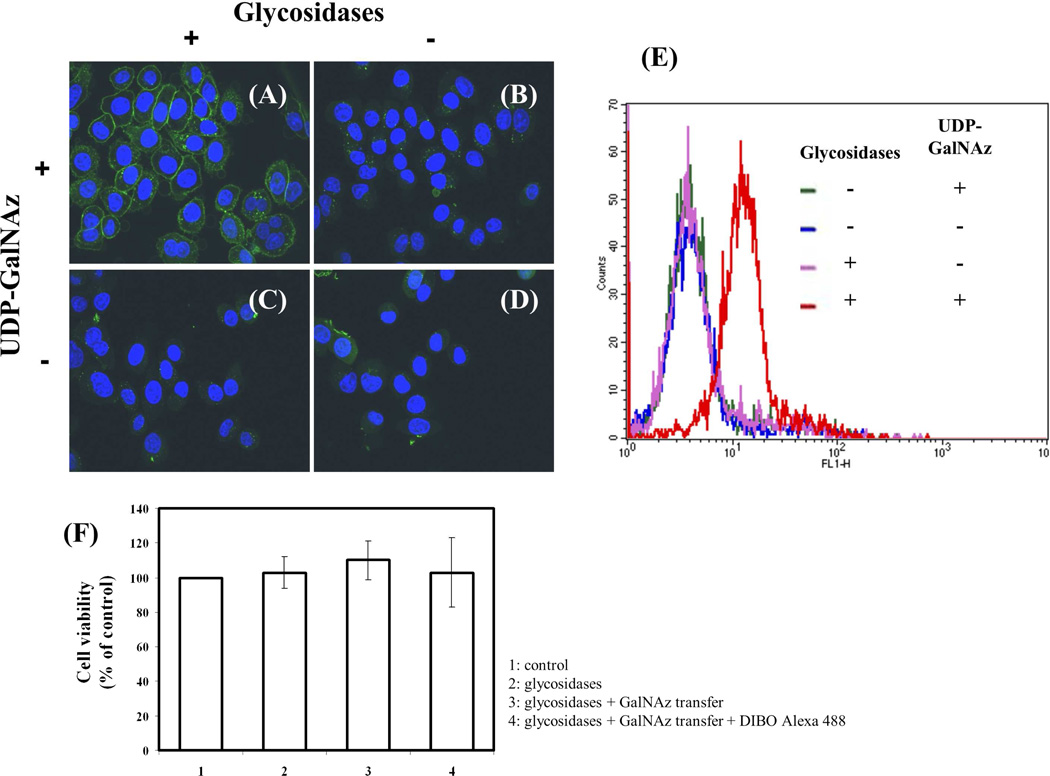
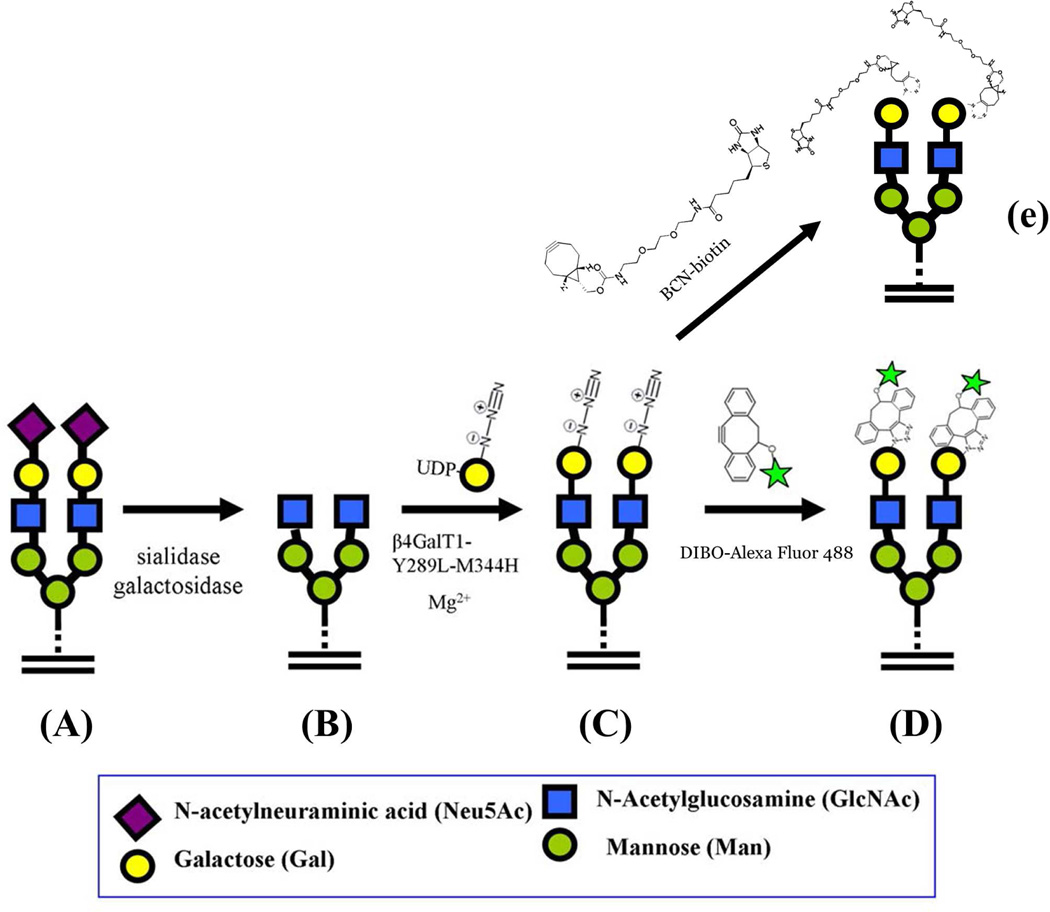
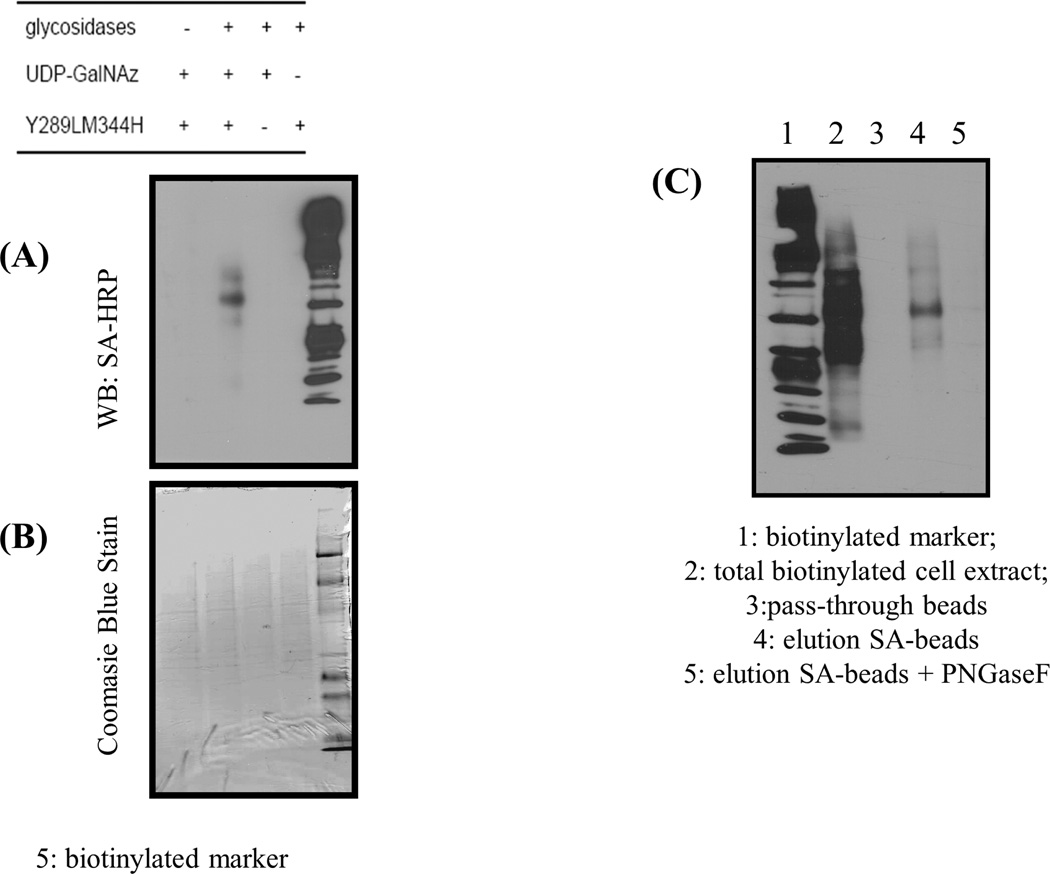
To look for truncated glycans with terminal GlcNAc at their nonreducing ends on the surface of human cervix adenocarcinoma (HeLa) cells, we used confocal microscopy on attached cells (Fig. 4A–D) and FACS analysis (Fig. 4F) to evaluate the expression of these glycans in suspended cells. Glycosylation reactions were performed using the Y289L-M344H-β4Gal-T1 enzyme in the presence of Mg2+. To expose the potential oligosaccharide acceptor substrate with the GlcNAc moiety at the nonreducing end, the cells were pretreated with the recombinant α2-3,6,8,9-neuraminidase enzyme and β1,4-galactosidase enzyme (schematic, Fig. 3). HeLa cells grown on slides showed green fluorescent membrane signal (corresponding to coupled DIBO-Alexa Fluor 488) only when the cells were pretreated with glycosidases, then treated with the Y289L-M344H-β4Gal-T1 enzyme and the UDP-GalNAz donor sugar (Fig. 4A). When the cells were not pretreated with glycosidase enzymes (Fig. 4B), or when UDP-GalNAz was omitted from the transfer reaction buffer (Fig. 4, C and D), no detectable fluorescence membrane signal was observed. Similarly, HeLa cells in suspension, with and without glycosidase pretreatment, were enzymatically galactosylated in the presence of the synthetic sugar nucleotide UDP-GalNAz and Mg2+. The transferred GalNAz was coupled with DIBO-Alexa Fluor 488. The cells were analyzed by FACS analysis (Fig. 4E). In agreement with the results from the confocal microscopy experiments, we observed a shift only in the cases where cells had been enzymatically pretreated, indicating that GlcNAc moieties are not freely exposed on the surface of HeLa cells. Viability of the cells was determined using the trypan blue method. Compared to control cells, which did not go through any treatment, no significant differences in cellular viability found after cells were labeled (Fig. 4F), indicating that the labeling protocol is suitable for detection of GlcNAc on live cells. These results indicate that free terminal GlcNAc residues could be detected by this methodology. In addition, labeled proteins could also be detected by Western blot analysis of the cell extract, which was obtained after GalNAz transfer and conjugation with BCN-biotin23 on the HeLa cell surface (Fig. 5C). BCN-biotin reactivity was detected only in samples that were pretreated with glycosidases, therefore exposing their GlcNAc moieties.
Figure 4.
Detection of cell surface terminal free GlcNAc residues using Y289L-M344H-β4Gal-T1 enzyme. After pretreatment with or without glycosidases, live cells were labeled with the Y289L-M344H-β4Gal-T1 enzyme in the presence or absence of UDP-GalNAz. After conjugation with DIBO-Alexa Fluor 488 (green fluorescent signal), the cells were washed and fixed. Nuclei were stained with Hoechst 33342 and HeLa cell images were obtained by confocal fluorescence microscopy. Treatments were performed as follows: A: Pretreatment with glycosidases and UDP-GalNAz present; B: No pretreatment with glycosidases and UDP-GalNAz present; C: Pretreatment with glycosidases and UDP-GalNAz omitted; and D: No pretreatment with glycosidases and UDP-GalNAz omitted. E: Cell surface GlcNAc detection by flow cytometry using the mutant enzyme β4Gal-T1-M344H-Y289L. FACS analysis of HeLa cells with and without pretreatment with the enzymes sialidase and galactosidase. Negative controls are cells incubated in the absence of the donor sugar UDP-GalNAz. F: Estimation of HeLa cell viability using trypan blue method. No significant differences in cell viability were found, indicating that the labeling protocol is suitable for detection of GlcNAc on live cells.
Figure 3.
Schematic diagram showing the GlcNAc specific transfer of GalNAz and its conjugation with Alexa Fluor 488 fluoroprobe. A: On the HeLa cell surface, the GlcNAc moiety in glycoconjugates is galactosylated and sialylated. A representative cell surface glycan is shown as a biantennary N-glycan that is in fully glycosylated form. B: When the cells are treated with the α1-3,6,8 sialidase and β1-4-galactosidase, the terminal sialic acid and β1-4-linked galactose are sequentially removed from the glycans, thereby exposing the GlcNAc moiety at the nonreducing end. C: The pretreated cells are incubated with the Y289L-M344H-β4Gal-T1 enzyme in the presence of UDP-GalNAz and Mg2+, transferring GalNAz to the non-reducing end of the GlcNAc moiety. D: When the glycan modified cells are treated with DIBO Alexa Fluor 488, the fluoroprobe molecules selectively conjugate with the GalNAz moieties, forming a covalent bond with them. Now the cells are site-specifically labeled; the fluorescence detected on the cell surface corresponds to the amount of the free GlcNAc moiety present on them. E: Copper-free click reaction with BCN-biotin to detect free GlcNAc residues by Western blot analysis or streptavidin-magnetic bead isolation.
Figure 5.
Western blot analysis of biotinylated cell extracts. A: Control samples are incubated without UDP-GalNAz, or double mutant enzymes or pretreatment with glycosidases. Coupling was performed using BCN-biotin. B: Loading control using Coomasie Blue Stain. C: Western blot analysis of streptavidin-magnetic bead purification from total cell lysate.
Capture and isolation of biotinylated proteins
We wanted to further determine whether the biotinylated proteins would be efficiently captured and eluted from the magnetic streptavidin beads. The Western blot analysis (Fig. 5C, lane 4) showed that the proteins can be purified by this method, indicating that glycoproteins with exposed GlcNAc termini could be isolated for identification purposes. The treatment of eluted biotinylated proteins from the cell extract with PNGase F made the major biotinylated protein bands disappear, suggesting that transfer occurred mostly on N-glycans (Fig. 5C, lane 5).
GlcNAc detection on human platelets
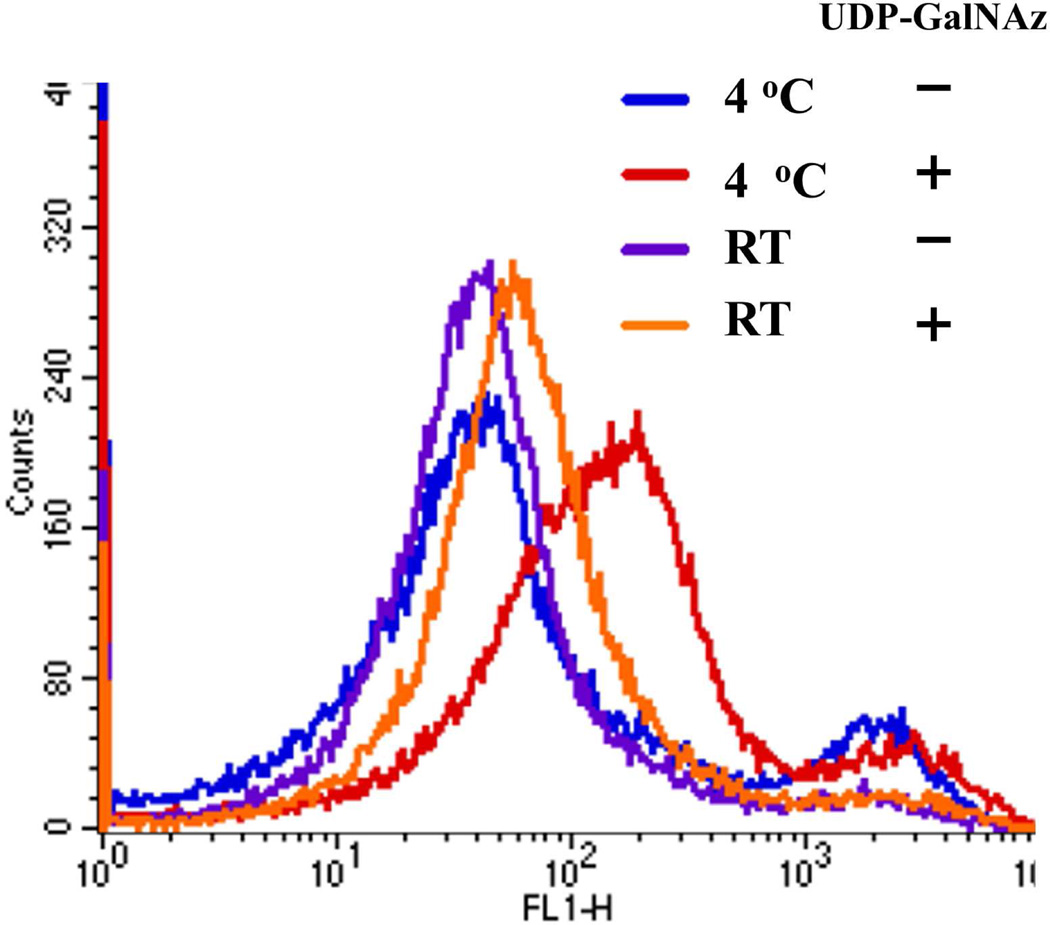
Human platelets show an increase in free GlcNAc residues when incubated in the cold.20 To demonstrate that our method can detect free GlcNAc residues on chilled human platelets, we incubated human platelets overnight in the cold or at room temperature and then transferred GalNAz from UDP-GalNAz in the presence of Mg2+ using the double mutant enzyme Y289L-M344H-β4Gal-T1. These platelets were subsequently coupled to the DIBO Alexa Fluor 488 fluoroprobe. FACS analysis revealed (Fig. 6) a small shift corresponding to the presence of GlcNAc residues on the platelets’ cell surfaces in the samples incubated at room temperature overnight, indicating the free GlcNAc residues already available on the cell surface. Upon overnight incubation in the cold, the fluorescence shift increased, indicating that chilling induces an increase in free GlcNAc residues, as has been reported earlier.20
Figure 6.
Detection of terminal free GlcNAc residues in human platelets using the mutant enzyme M344H-Y289L-β4Gal-T1. Terminal free GlcNAc residues are very rare on human cell surfaces, but they are known to be present in platelets. Platelets express GPIb — glycoprotein Ib– which contains b-GlcNAc-terminating immature glycans. Upon short-term cooling, these receptors cluster at 4 °C (20).
O-GlcNAc detection using Y289LM344H mutant
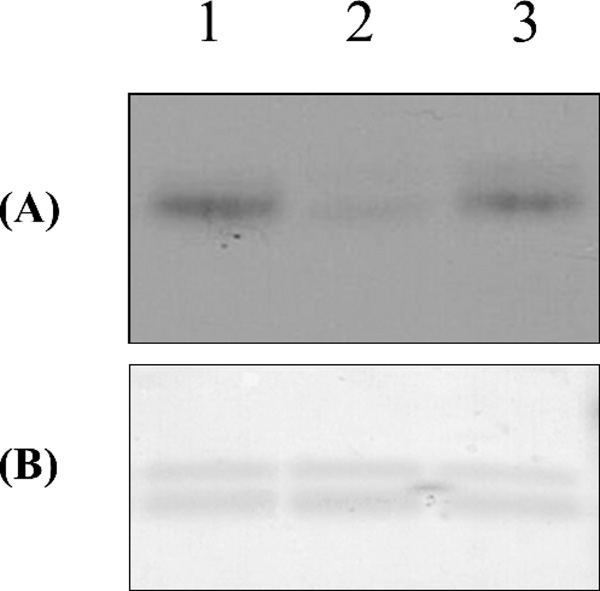
It has been shown that the Y289L Mn2+-dependent enzyme can efficiently detect O-GlcNAc modifications on bovine lens α-crystallin protein.13 To determine whether the Mg2+-dependent Y289L-M344H enzyme can also detect this modification, we performed the enzymatic transfer of the modified sugar 2-keto-Gal from its sugar nucleotide donor to the O-GlcNAc residues present in this protein (Fig. 7). Western blot analysis shows aminooxy-biotin-reactive bands in the Mg2+-dependent Y289L-M344H enzyme sample (Fig. 7A, lane 3), as well as in the Mn2+-dependent Y289L-Gal-T1 enzyme sample (Fig. 7A, lane 1), indicating the presence of O-GlcNAc residues that can be detected with both enzymes. As a negative control, UDP-2-keto-Gal was omitted from one reaction, which yielded a barely detectable band (lane 2).
Figure 7.
O-GlcNAc detection using the double mutant enzyme M344H-Y289L-β4Gal-T1. UDP-keto-galactose was transferred to a-crystallin protein using the Y289L-M344H mutant. A: Detection of the coupled biotin was performed using streptavidin conjugated with HRP and detected by Western blot analysis (lane 5). B,: Coomasie Blue staining. Lane 1: positive control using Y289L; lane 2: negative control where UDP-2-keto-Gal was omitted from transfer reaction; lane 3: Y289LM344H transfers UDP-2-keto Gal to alpha crystallin.
DISCUSSION
The surface of a cell is decorated with glycans, which are highly complex molecules. This complexity is due, in part, to a greater combinatorial diversity of monosaccharides compared to nucleotides or amino acids. Because of their location, they are the first type of molecule encountered by neighboring cells, microorganisms, and antibodies, as well as by the extracellular matrix. In addition, these glycans compose part of the glycoconjugates, which change dynamically during physiological and pathological conditions. The role of glycans can be structural as well as functional. Glycans have been studied at the cellular level (without cell disruption) using different approaches, including through the use of lectins, antibodies, and metabolic labeling.24–28 However, lectins have a low affinity and they require multivalent interactions for high-avidity binding.29 In addition, they possess a range of specificities since they have a higher affinity for the monosaccharide than for the total glycan. Monoclonal antibodies could be more specific than lectins as glycan determinants, but they are less characterized with respect to binding specificities.1 Therefore, there is an increasing need to develop new, highly specific techniques to study the glycome.
Glycan biosynthesis is not template driven, but rather the result of the actions of many enzymes, including glycosyltransferases and glycosidases. Since each glycosyltransferase transfers the donor sugar to a specific acceptor they can be unique reagents for labeling individual cell surface glycans. Bovine milk galactosyltransferase has been used to label cell surface GlcNAc residues of lymphocytes in the presence of Mn2+ with H3-galactose.30 However, this labeling method requires days for exposure to detect H3-label and also Mn2+ may be toxic to some cells.18,19 Structure-function studies on glycosyltransferases have enabled us to design mutant enzymes that can transfer sugars possessing a chemical handle, which can then be utilized for bioconjugation and detection of their respective acceptors.22 We have previously shown that mutations in a few residues in the sugar donor-binding site of glycosyltransferases broaden their sugar donor specificities.12,31–32 Mutation of Tyr289 to Leu289 in bovine β4Gal-T1 enzyme, the Y289L-β4Gal-T1 mutant, accommodates a Gal with a C2-N-acetyl group (GalNAc) or a chemical handle that is similar in size and shape to it, such as a C2-keto- or C2-azido group.22 Also, it had been shown that the metal ion specificity in β4Gal-T1 is determined by the Met-344 residue and that mutating Met-344 to a His-344 residue changes the metal ion specificity of the resulting mutant enzyme, M344H-β4Gal-T1, from Mn2+ to Mg2+.16
In the present study, we show that the combination of these two mutations, the double mutant Y289L-M344H-β4Gal-T1, transfers GalNAc to an acceptor GlcNAc in the presence of Mg2+. The double mutant enzyme also transfers Gal with a chemical handle, such as C2-keto-Gal or C2-azido (GalNAz), from their respective UDP derivatives to GlcNAc on a glycoprotein. Since the concentration of Mn2+ ion (5mM) used for the enzyme reaction is potentially toxic to cells17–19, the nontoxic, Mg2+-dependent double mutant enzyme, Y289L-M344H-β4Gal-T1, was used for the transfer of Gal with C2-keto or C2-azido (GalNAz) handles to GlcNAc residues on glycans on the surface of live cells. The transferred GalNAz could be conjugated with the fluoroprobe, DIBO-Alexa Fluor 488, in a copper-free reaction. The fluorescence signal, corresponding to labeled GlcNAc residues on the glycans at the surface of the live cells, was detected by two methodologies: confocal microscopy and flow cytometry. This study shows that the amount of free GlcNAc at the reducing end of the cell surface glycans in cultured HeLa cells is nearly undetectable. However, when HeLa cells are treated with sialidase and galactosidase, removing the terminal sialic acid and galactose, an abundant amount of GlcNAc is detected on the cell surface. Interestingly, in a similar, separate study using a α1,3-galactosyltransferase mutant enzyme, 280SGG282-α1-3Gal-T, which detects the free terminal LacNAc moiety31, we also found an abundant amount of the LacNAc moiety present on the HeLa cell surface without pretreatment with a sialidase enzyme (data not shown).
It has been shown that, during short-term incubation in the cold, platelets increase the irreversible exposure of the free GlcNAc residues that are part of their GPIb complex.20, 33 Upon short-term cooling, this process has been shown to be responsible for the removal of chilled platelets by interacting with the integrin receptors on macrophages. In view of this, we used our methods to investigate the changes in the cell surface free GlcNAc moiety upon exposure to cold temperatures. We have observed by FACS analysis that there was a small but detectable amount of the free GlcNAc moiety on their cell surface before incubating them at a cold temperature. However, upon cooling, a significant amount of GlcNAc present on the cell surface was detected, which is in line with previous observations from lectin-binding studies.20 These findings indicate that the present technique can be used for cell surface detection of GlcNAc residues, as well as cell surface modifications, on naturally (platelets) or artificially (HeLa cells) exposed acceptors.
We have furthermore shown that the double mutant enzyme is also able to transfer modified UDP-keto-Gal to O-GlcNAc-modified α-crystallin protein. This indicates that the magnesium-dependent Y289L-M344H-β4Gal-T1 mutant enzyme could also be used for the detection of recently reported O-GlcNAc post-translational modifications of the extracellular domain of cell surface proteins.2, 34, 35 For this reason, the mutant enzyme Y289L-M344H-β4Gal-T1 could be a potentially useful tool for the detection of GlcNAc residues from N-glycans, as well as from O-GlcNAcylated extracellular proteins.
Site-specific in vitro labeling using glycosyltransferases has many advantages for studying cell surface glycans. Since glycosyltransferases have little substrate promiscuity, the substrate specificity is high compared to the range of lectin specificities. Under optimal reaction conditions, every acceptor substrate on the cell surface can be labeled through a covalent bond, with a very small detecting agent, allowing an accurate quantitation of the epitopes. However, this method is limited to the mutations that can be introduced in the enzyme to accommodate a chemical handle in the donor sugar. The use of enzymes for site-specific detection is an emerging technique. Biotin ligase has been used to detect an acceptor peptide on a cell surface protein.36 Transglutaminase has been employed to detect a Q-tag protein on the cell surface.37 LacNAc disaccharide on the cell surface has recently been detected using a recombinant Helicobacter pylori α (1,3)-fucosyltransferase.38
In conclusion, using site-specific labeling we have developed a novel methodology for the detection of a single monosaccharide on the cell surface. This methodology could be a useful tool to investigate the cell’s glycophenotype through the detection, isolation, and further identification of ligands.
ACKNOWLEDGMENTS
We thank Dr Lockett and Ms Kim Peifley for confocal microscopy assistance, Dr Mc Vicar and Ms Laura Quigley for platelets isolation, and Dr Floris L van Delft from Nijamen, The Netherlands for BCN-biotin reagent. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the view or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported [in part] by the Intramural Research Program of the NIH, Frederick National Laboratory for Cancer Research.
ABBREVIATIONS
- β 4Gal-T1
β 1,4-galactosyltransferase
- FACS
Fluorescence Activated Cell Sorting
- PNGase F
Peptide N-glycosidase F
- Gal
galactose
- GlcNAc
N-acetylglucosamine; N-acetylazidogalactosamine (Gal-2-NHCO-CH2-N3)
- UDP-2-keto-Gal
Uridine 5’-diphospho-2-acetonyl-2-deoxy-galactose
- UDP-GalNAc
Uridine 5’-diphospho-N-acetylgalactosamine
- UDP-GalNAz
Uridine 5’-diphospho-N-acetyl-azido-galactosamine
- MALDI
Matrix-Assisted Laser Desorption Ionization
- MS
Mass Spectra
- AOB
N-aminooxymethylcarbonylhydrazino-D-biotin
- HRP
horseradish peroxidase
- FBS
Fetal Bovine Serum
- BSA
Bovine Serum Albumin
- PBS
Phosphate Buffer Saline
REFERENCES
- 1.Varki A, Lowe JB. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG, 2nd, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F, Smith RD. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci U.S.A. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakomori S. Cancer-associated glycosphingolipid antigens: Their structure, organization, and function. Acta Anat. 1998;161:79–90. doi: 10.1159/000046451. [DOI] [PubMed] [Google Scholar]
- 5.Fuster MM, Esko JD. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 6.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 7.Kiessling LL, Pohl NL. Strength in numbers: non-natural polyvalent carbohydrate derivatives. Chem. Biol. 1996;3:71–77. doi: 10.1016/s1074-5521(96)90280-x. [DOI] [PubMed] [Google Scholar]
- 8.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim. Biophys. Acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs CL, Yarema KJ, Mahal LK, Nauman DA, Charters NW, Bertozzi CR. Metabolic labeling of glycoproteins with chemical tags through unnatural sialic acid biosynthesis. Methods Enzymol. 2000;327:260–275. doi: 10.1016/s0076-6879(00)27282-0. [DOI] [PubMed] [Google Scholar]
- 10.Laughlin ST, Bertozzi CR. Imaging the glycome. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qasba PK, Ramakrishnan B, Boeggeman E. Structure and function of beta-1,4-galactosyltransferase. Curr Drug Targets. 2008;9:292–309. doi: 10.2174/138945008783954943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramakrishnan B, Qasba PK. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity. J. Biol. Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 13.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem. Soc. 2002;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 14.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satomaa T, Heiskanen A, Leonardsson I, Angström J, Olonen A, Blomqvist M, Salovuori N, Haglund C, Teneberg S, Natunen J, Carpén O, Saarinen J. Analysis of the human cancer glycome identifies a novel group of tumor-associated N-acetylglucosamine glycan antigens. Cancer Res. 2009;69:5811–5819. doi: 10.1158/0008-5472.CAN-08-0289. [DOI] [PubMed] [Google Scholar]
- 16.Ramakrishnan B, Boeggeman E, Qasba PK. Effect of the Met344His mutation on the conformational dynamics of bovine beta-1,4-galactosyltransferase: crystal structure of the Met344His mutant in complex with chitobiose. Biochemistry. 2004;43:12513–12522. doi: 10.1021/bi049007+. [DOI] [PubMed] [Google Scholar]
- 17.El Mchichi B, Hadji A, Vazquez A, Leca G. p38 MAPK and MSK1 mediate caspase-8 activation in manganese-induced mitochondria-dependent cell death. Cell Death Differ. 2007;14:1826–1836. doi: 10.1038/sj.cdd.4402187. [DOI] [PubMed] [Google Scholar]
- 18.Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, Wohlgemuth R. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat Med. 2008;14:181–187. doi: 10.1038/nm1703. [DOI] [PubMed] [Google Scholar]
- 19.Oubrahim H, Stadtman ER, Chock PB. Mitochondria play no roles in Mn(II)-induced apoptosis in HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9505–9510. doi: 10.1073/pnas.181319898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmeister KM, Josefsson EC, Isaac NA, Clausen H, Hartwig JH, Stossel TP. Glycosylation restores survival of chilled blood platelets. Science. 2003;301:1531–1534. doi: 10.1126/science.1085322. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishnan B, Shah PS, Qasba PK. alpha-Lactalbumin (LA) stimulates milk beta-1,4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine. Crystal structure of beta 4Gal-T1 × LA complex with UDP-Glc. J. Biol. Chem. 2001;276:37665–37671. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]
- 22.Boeggeman E, Ramakrishnan B, Kilgore C, Khidekel N, Hsieh-Wilson LC, Simpson JT, Qasba PK. Direct identification of nonreducing GlcNAc residues on N-glycans of glycoproteins using a novel chemoenzymatic method. Bioconjugate Chem. 2007;18:806–814. doi: 10.1021/bc060341n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dommerholt J, Schmidt S, Temming R, Hendriks LJ, Rutjes FP, van Hest JC, Lefeber DJ, Friedl P, van Delft FL. Readily accessible bicyclononynes for bioorthogonal labeling and three-dimensional imaging of living cells. Angew. Chem., Int. Ed. Engl. 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 25.Ota H, Hayama M, Nakayama J, Hidaka H, Honda T, Ishii K, Fukushima M, Uehara T, Kurihara M, Ishihara K, Hotta K, Katsuyama T. Cell lineage specificity of newly raised monoclonal antibodies against gastric mucins in normal, metaplastic, and neoplastic human tissues and their application to pathology diagnosis. Am. J. Clin. Pathol. 2001;115:69–79. doi: 10.1309/AMUR-K5L3-M2DN-2DK5. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M, Mitoma J, Hoshino H, Yu SY, Shimojo Y, Suzawa K, Khoo KH, Fukuda M, Nakayama J. Prominent expression of sialyl Lewis X-capped core 2-branched O-glycans on high endothelial venule-like vessels in gastric MALT lymphoma. J. Pathol. 2011;224:67–77. doi: 10.1002/path.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharon N, Lis H. Lectins. 2nd Ed. The Netherlands: Springer; 2007. [Google Scholar]
- 30.Torres C, Hart GW. Topography and Polypeptide Distribution of Terminal N-Acetylglucosamine Residues on the Surfaces of Intact Lymphocytes. J. Biol. Chem. 1984;277:20833–20839. [PubMed] [Google Scholar]
- 31.Pasek M, Ramakrishnan B, Boeggeman E, Manzoni M, Waybright TJ, Qasba PK. Bioconjugation and detection of lactosamine moiety using alpha1,3-galactosyltransferase mutants that transfer C2-modified galactose with a chemical handle. Bioconjugate Chem. 2009;20:608–618. doi: 10.1021/bc800534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasek M, Ramakrishnan B, Boeggeman E, Mercer N, Dulcey AE, Griffiths GL, Qasba PK. The N-acetyl-binding pocket of N-acetylglucosaminyltransferases also accommodates a sugar analog with a chemical handle at C2. Glycobiology. 2012;22:379–388. doi: 10.1093/glycob/cwr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rumjantseva V, Grewal PK, Wandall HH, Josefsson EC, Søensen AL, Larson G, Marth JD, Hartwig JH, Hoffmeister KM. Dual roles for hepatic lectin receptors in the clearance of chilled platelets. Nat. Med. 2009;15:1273–1280. doi: 10.1038/nm.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J. Biol. Chem. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann BR, Liu Y, Mosher DF. Modification of EGF-like module 1 of thrombospondin-1, an animal extracellular protein, by O-linked N-acetylglucosamine. PLoS One. 2012;7:e32762. doi: 10.1371/journal.pone.0032762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 37.Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng T, Jiang H, Gros M, del Amo DS, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, Wu P. LacNAc disaccharide has recently been detecting using a recombinant Helicobacter pylori a(1,3)-fucosyltransferase. Angew. Chem., Int. Ed. Engl. 2011;50:4113–4118. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]