Abstract
Neurogenesis in the nervous system is regulated by both protein coding genes and non-coding RNA molecules. microRNAs (miRNAs) are endogenous small non-coding RNAs and usually negatively regulate gene expression by binding to the 3′ untranslated region (3′UTR) of target messenger RNAs (mRNAs). miRNAs have been shown to play an essential role in neurogenesis, regulating neuronal proliferation, differentiation, maturation, and migration. An important strategy used to reveal miRNA function is the manipulation of their expression levels and patterns in specific regions and cell types in the nervous system. In this review we will systemically highlight established and new approaches used to achieve gain-of-function and loss-of-function of miRNAs in vitro and in vivo, and will also summarize miRNA delivery techniques. As the development of these leading edge techniques come online, more exciting discoveries of the roles miRNAs play in neural development and function will be uncovered.
Keywords: neurogenesis, microRNAs, miRNA inhibitor, miRNA sponge, mRNA protector
The proper development of the nervous system relies on precisely programmed regulation of gene expression. The discovery of the non-coding microRNAs (miRNAs) has revealed a new paradigm of gene expression control, and the manipulation of miRNA expression levels and patterns in the nervous system has demonstrated the critical role they play in Neurogenesis. In this review, we will highlight various leading edge approaches that have been used to investigate miRNA function in neural development.
miRNA discovery and biogenesis
miRNAs are endogenous, small non-coding, single stranded RNA molecules of ~22 nucleotide (nt) in length. Most miRNAs act as repressors of target messenger RNAs (mRNAs) by a posttranslational regulatory mechanism. Since the first identification of miRNA lin-4 in Caenorhabditis. elegans, the database of published miRNA sequences has rapidly expanded, with 1600 miRNA precursors and 2042 mature miRNAs in humans, 855 precursors and 1281 mature miRNAs in mice, 238 precursors and 426 mature miRNAs in Drosophila, and 223 precursors and 368 mature miRNAs in C. elegans (The miRBase Sequence Database Release 19, http://www.mirbase.org) (Kozomara and Griffiths-Jones, 2011).
In most cases, miRNAs are transcribed as single-stranded primary miRNA (pri-miRNA) from intragenic or intergenic genomic regions by RNA polymerase II (Pol II) (Lee et al., 2004; Rodriguez et al., 2004). The pri-miRNAs are further cleaved by the RNase III-type nuclease Drosha and its co-factor DiGeorge Syndrome Critical Region Gene 8 (DGCR8/Pasha) to produce ~70 bp precursor miRNAs (pre-miRNAs) (Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004). A different biogenesis pathway exists for miRNAs that are transcribed from intronic regions of protein-coding genes, termed mirtrons that are processed by the spliceosome and then by lariat debranchase activity to generate pre-miRNAs in various species (Berezikov et al., 2007; Okamura et al., 2007; Ruby et al., 2007). The hairpin shaped pre-miRNAs from both the canonical and mirtron biogenesis pathways are transported from nucleus to cytoplasm, and further cleaved by another RNase III member Dicer into imperfect complementary double stranded mature miRNAs of ~18–25 bp (Hammond et al., 2000; Grishok et al., 2001; Hutvagner et al., 2001). In addition, a number of alternative miRNA biogenesis pathways such as Drosha/DGCR8 independent and Dicer dependent miRNA biogenesis pathways have been reported (Yang and Lai, 2011). And the maturation of miR-451 has been shown to require argonaute 2 (Ago2) but not Dicer (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010).
For most miRNAs, one strand from the mature miRNA duplex is loaded into the RNA−induced silencing complex (RISC) (Hutvagner and Zamore, 2002; Chendrimada et al., 2005; Bartel, 2009). The RISC, guided by miRNA, binds to the 3′UTR of target mRNAs through specific complementarity of the 2–7 nt at the 5′$ end of the miRNA, which is termed the seed sequence (Lewis et al., 2003, 2005). Translation of mRNAs targeted in this manner is suppressed and the mRNAs may undergo degradation (Bartel, 2009) (Figure 1A). The RISC-miRNA pathway thus represents a novel and important gene silencing mechanism that exists in many organisms.
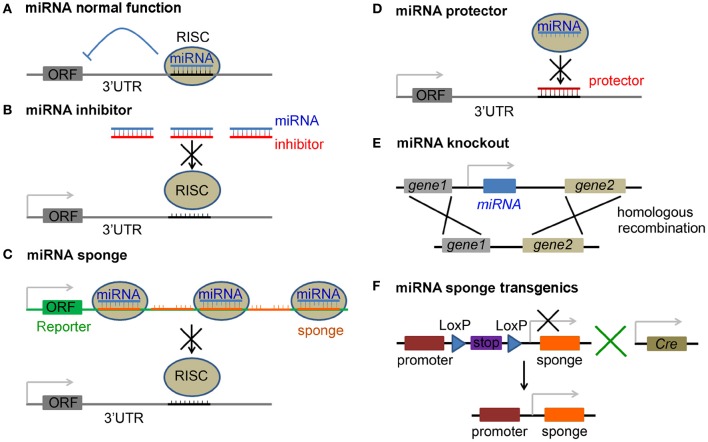
Figure 1.
Approaches to inhibition of miRNA function. (A) Normal function of miRNAs is to suppress translation of the target mRNA with an open reading frame (ORF), or cause mRNA degradation, by guiding RNA-induced silencing complex (RISC) to the 3′ untranslated region (3′UTR) of the mRNA. (B) miRNA inhibitors are antisense miRNA oligonucleotides (AMOs), including 2′-O-methyl modified AMO, antagomir, locked nucleic acid (LNA), phosphorodiamidate morpholino oligonucleotide (PMO) and peptide nucleic acid (PNA), and block miRNA silencing activity by a complimentary binding to the mature miRNA. (C) miRNA is saturated by miRNA sponges that carry tandem multiplex of complementary sequences, which usually imperfectly match the target miRNA and are inserted in the 3′UTR of a reporter gene. (D) mRNA protector functions by a perfect binding to the 3′UTR of a mRNA and protects it from being bound by its miRNA. (E) Loss-of-function of a miRNA is achieved by direct miRNA knockout from the genome. (F) Tissue specific blockage of miRNA activity is achieved by breeding floxed miRNA sponge transgenic mice (with a stop signal flanked by two LoxP sites) with a proper Cre line.
Approaches to manipulating miRNAs in the nervous system
In the nervous system, the process of neurogenesis involves precise regulation of neuronal proliferation, differentiation, maturation, and migration. Accumulating evidence has highlighted the critical role that miRNAs play in neurogenesis (Kosik and Krichevsky, 2005; Kosik, 2006; Fineberg et al., 2009; Liu and Zhao, 2009; Lau and Hudson, 2010; Shi et al., 2010; Bian and Sun, 2011; Cochella and Hobert, 2012; Luikart et al., 2012). We here summarize approaches that have been used to manipulate miRNA expression in order to investigate their functions.
Approaches to blocking miRNA biogenesis
miRNA biogenesis has been blocked in different regions or cell types in the nervous system by tissue specific ablation of Dicer using various Cre lines such as the Emx1-Cre line in the embryonic cortex and the CamKII-Cre line in the postnatal brain and adult hippocampus (Cuellar et al., 2008; Davis et al., 2008; De Pietri Tonelli et al., 2008; Kawase-Koga et al., 2009; Shin et al., 2009; Soukup et al., 2009; Andersson et al., 2010; Budde et al., 2010; Huang et al., 2010; Kawase-Koga et al., 2010; Konopka et al., 2010; Zehir et al., 2010; Zhao et al., 2010b,c; Zheng et al., 2010; Iida et al., 2011; Li et al., 2011; Liu et al., 2011; Nowakowski et al., 2011; Tao et al., 2011; Chen and Wichterle, 2012; Repetto et al., 2012; Rosengauer et al., 2012). Mice with tissue specific Dicer deletion show various neural defects, suggesting an important role of miRNAs in neural development and function. Moreover, a mouse model with a microdeletion syntenic to the human chromosome 22q11 deletion, which includes DGCR8 gene, has shown altered miRNA biogenesis, suggesting a role of miRNAs in behavioral and cognitive deficits in humans with 22q11 deletion (Stark et al., 2008). Postmitotic neurons in the brain have shown more severe defects in Dicer knockout mice than DGCR8 knockout mice, which are both ablated by the CamKII-Cre line, suggesting that a subpopulation of miRNAs generated by Dicer but bypassed by DGCR8 processing may play an important role in postmitotic neuron development (Babiarz et al., 2011). In Drosophila, Dicer1 and Pasha mutations but not Ago1 and Ago2 mutations have shown disrupted olfactory projection neuron phenotypes (Berdnik et al., 2008).
Ablation of molecules in the miRNA biogenesis pathway has proven important roles of miRNAs in neural development. However, the weakness of such studies should also be noticed. First, a large number of miRNAs is affected when miRNA biogenesis is blocked, making it critical to distinguish the role of individual miRNAs in neurogenesis. Second, Dicer is also required for maintaining the heterochromatin assembly, likely by the short interfering RNA (siRNA) pathway, and cleaving long strand Alu transcripts (Fukagawa et al., 2004; Kanellopoulou et al., 2005; Yang and Lai, 2011). Thus, phenotypes of Dicer deletion need to be carefully interpreted.
Approaches to overexpression of miRNAs
miRNA mimics
Gain-of-function for a specific miRNA can be achieved by overexpressing the mature sequence of the miRNA. miRNA mimics can be chemically synthesized as oligonucleotides according to sequences of the endogenous miRNA. Double stranded miRNA mimics, with the sequence of one strand identical to the endogenous mature miRNA, are usually used to increase the efficiency of augmenting miRNA expression. The strand identical to the endogenous miRNA will be loaded into the RISC complex and silence target genes as the endogenous (Martinez et al., 2002). When designing a miRNA mimic, one needs to be cautious to avoid formation of a new miRNA from the complementary strand of the mimic duplex. Since there is no additional sequence introduced in the target system, the synthesized miRNA mimic has no vector-based toxicity. Studies have shown that infusion of miR-132 mimics to the mouse visual cortex blocks ocular dominance plasticity produced by monocular deprivation (Tognini et al., 2011), and miR-137 mimics negatively regulates neural stem cell proliferation (Sun et al., 2011). However, the transfection efficiency of miRNA mimics is low, especially in neurons, and the transfection is usually transient, which has limited its application.
miRNA precursors
A vector-based technique has been used to obtain a long-term stable expression of miRNAs by inserting the miRNA precursor sequence downstream of the RNA polymerase III (Pol III)-driven or Pol II-driven promoters in a vector system (Takamizawa et al., 2004; Chung et al., 2006; McLaughlin et al., 2007). Plasmid and virus-based overexpression of miRNA precursors allows ectopic expression of a miRNA in specific regions and cell types in the nervous system, for instance introduction of the miR-9 precursor plasmid in spinal motor neurons and the miR-124 precursor in mouse subventricular zone stem cells (Cheng et al., 2009; Otaegi et al., 2011b). However, the amount of overexpression needs to be carefully optimized to minimize side effects, for example consistent expression of miRNAs and large amount of short hairpin RNAs (shRNAs) can induce serious liver injury and lead to mouse lethality due to oversaturation of the endogenous small RNA pathways (Grimm et al., 2006).
miRNA biogenesis enhancement
Instead of manipulating miRNAs themselves, miRNA expression levels can be modified by modulating the miRNA processing pathway. The fluoroquinolone antibiotic, enoxacin, has been found to promote the biogenesis of endogenous miRNAs. Among the upregulated miRNAs, a number of them such as miR-124a, miR-125a, miR-23a, and let-7b are highly involved in neural development (Shan et al., 2008). Moreover, enoxacin has been shown to enhance the production of tumor suppressor miRNAs and miRNAs associated with neurogenesis such as let-7, miR-125, and miR-7 by binding to the miRNA biosynthesis protein TAR RNA-binding protein 2 (Melo et al., 2011). To manipulate biogenesis of individual miRNAs, small molecule libraries have been screened to identify a miR-122 activator in hepatocellular carcinoma cells (Young et al., 2010). While mechanisms by which small molecules activate miRNA expression are largely unclear, these studies have allowed development of novel tools to manipulate miRNA expression such as screening small molecules for activating miRNA expression in neurogenesis.
The gain-of-function approach by overexpressing miRNAs has both advantages and disadvantages (Table 1). miRNA mimics can be directly synthesized and delivered, making them convenient to use. Plasmid or virus based miRNA precursors are easy for delivery in tissues and cells that miRNA mimics are difficult to introduce such as neurons. However, overexpression of miRNAs may cause side effects such as oversaturation of the miRNA processing pathway. The efficiency of processing mature miRNAs from their precursors can vary and may affect their function. Using miRNA precursors flanking with longer genomic DNA sequences can improve the efficiency miRNA processing. The miRNA biogenesis enhancement strategy using small molecules is easy to administrate in vitro and in vivo, and is able to elevate levels of multiple miRNAs simultaneously, but it still requires optimization to be able to elevate specific miRNAs. Nevertheless, overexpression of miRNAs is a powerful approach to manipulate miRNAs in neural development.
Table 1.
Advantages and drawbacks of miRNA gain-of-function technologies.
| Technology | Advantages | Drawbacks |
|---|---|---|
| miRNA mimics | Convenient; time saving | Expensive; non-specific effect; difficult to introduce into neurons, toxicity |
| miRNA precursors | flexible for in vitro and in vivo studies | Unpredicted processing efficiency; vector-based side effect; toxicity for mass overexpression; non-specific effect |
| miRNA biogenesis enhancement | Small molecules, easy to administrate; upregulating multiple miRNAs simultaneously | Unknown tissue distribution; unclear mechanisms; low miRNA specificity |
Approaches to knockdown of miRNAs
miRNA inhibitors
Antisense miRNA oligonucleotides (AMOs) have been used as miRNA inhibitor to knock down endogenous miRNA activity. The sequences of these oligonucleotide analogs are complementary to the endogenous mature miRNAs, and function by directly binding to the single strand mature miRNA to block miRNA silencing (Figure 1B). We here highlight inhibitor technologies developed to block miRNA silencing activities.
2′-O-methyl group modified AMOs. Native oligonucleotides are sensitive to degradation in serum or by endogenous cellular exonucleases and endonucleases, and their efficiency to cross cell membranes is low, making them ineffective in C. elegans (Hutvagner et al., 2004). Therefore, chemically modified AMOs has been generated. 2′-O-methyl group modified AMOs have been shown to significantly increase resistance to nuclease degradation, display enhanced binding to miRNAs and inhibit endogenous miRNA activities (Hutvagner et al., 2004). For example knocking down let-7b and miR-124 using 2′-O-methyl AMO can promote proliferation of neural stem cells (Cheng et al., 2009; Zhao et al., 2010a). Although 2′-O-methyl group modified AMOs show longer survival time in the intracellular environment than common DNA oligonucleotides, they are still unstable in serum (Lennox and Behlke, 2010). Since nuclease degrades nucleic acids by breaking the phosphodiester bonds between nucleotides, replacing these bonds by phosphorothioate linkage in AMOs has been effective in reducing degradation. The drawback of this modification, however, is that it decreases the binding of the AMO to its target miRNA.
Antagomirs. Antagomirs are 22–23 nt 2′-O-methyl modified, 3′ end cholesterol-conjugate RNA analogs that have a complementary sequence to a miRNA. Some phosphodiester linkages in the phosphate backbone of Antagomir are substituted by phosphorothioate and the cholesterol conjugation facilitates the in vivo delivery of this AMO. Studies have shown that the antagomir for miR-122 exhibits specific inhibition of miR-122 in the mouse liver by tail vein injection, and the antagomir for miR-16 inhibits miR-16 expression in multiple tissues, including direct injection of miR-16 antagomir into mouse brains (Krützfeldt et al., 2005, 2007). The advantage of antigomirs is that they are nuclease resistant, and can be delivered into cells directly without any vector assistant, which avoids complication of using delivery vehicles. The drawbacks of antagomirs are high usage dose and possible off-target effects, which has limited their application as a therapeutic reagent in humans (Krützfeldt et al., 2005; Morrisey, 2010; Patrick et al., 2010).
Other 2′ site modifications of AMOs have been developed to produce more powerful miRNA inhibitors. Introduction of 2′-O-methoxyethyl groups increases affinity and specificity of binding to miRNAs compared to 2′-O-methyl analogs (Hutvagner et al., 2004). Moreover, AMOs with both 2′Flouro (2′F) and phosphorothioate backbone modifications have been shown to display high efficiencies of inhibition of miRNAs, with an effect further enhanced by addition of 2′-O-methoxyethyl groups to both ends (Davis et al., 2006, 2009).
Locked nucleic acids (LNAs). In the LNA molecule, the 2′-O and 4′-C of the ribose are bridged by a methylene group, which results in a bicyclic nucleotide with a locked conformation. This specially locked structure stabilizes the LNA/RNA duplex and makes them strongly resistant to nuclease degradation (Kaur et al., 2006). LNA probes have been extensively used to detect spatiotemporal miRNA expression, in such techniques as Northern blot analysis (Valoczi et al., 2004), in situ hybridization (Wienholds et al., 2005; Kloosterman et al., 2006; Nelson et al., 2006) and miRNA expression profiling assays (Castoldi et al., 2006). In addition to their use as probes, LNA-based miRNA inhibitors have been designed by forming an LNA core sequence with flanking DNA sequences, generating a DNA-LNA-DNA chimeric molecule. The DNA-LNA-DNA oligonucleotides have been shown to inhibit activities of miR-21 in glioblastoma cell lines (Chan et al., 2005), and miR-181 in the differentiation process of mouse myoblasts (Naguibneva et al., 2006). Moreover, LNA-only AMOs can suppress miRNA bantam in cultured Drosophila cells (Orom et al., 2006). Phosphorothioate modified LNA-based miR-122 inhibitor can markedly reduce miR-122 silencing activities at lower dosages compared to the miR-122 antagomir (Krützfeldt et al., 2005; Elmén et al., 2008). Inhibiting miR-219 by intracerebralventricular deliver of LNA AMO can increase its target gene expression in prefrontal cortex (Kocerha et al., 2009). Because of their high efficiency blocking of miRNA activities, LNA-based AMOs have been developed for use as therapeutic tools: an LNA-based miR-122 AMO is under phase II clinical trail for anti-hepatitis C therapy (Lanford et al., 2010).
Phosphorodiamidate morpholino oligonucleotides (PMOs). PMOs are designed as a strategy in which the riboses of the nucleic acid are substituted by 6-membered morpholine rings, and the phosphodiester bonds are substituted by phosphorodiamidates to produce stronger steric blocking of nucleases and prevent degradation. miR-124 has been shown to regulate muscle cell fate in zebrafish by knocking down miR-124 activity using PMOs (Flynt et al., 2007). Microinjections of PMOs targeting miR-183 family members in zebrafish embryos have demonstrated their roles in sensorineuron fate determination (Li et al., 2010). To improve bioavailabilities, further modifications of PMOs have been developed, called vivo-Morpholinos, which show more efficient penetration in multiple tissues in vivo (Morcos et al., 2008).
Peptide nucleic acids (PNAs). PNAs are artificial DNA/RNA mimics that have peptides flanking the nucleic acid sequences to increase target affinity, specificity, nucleases resistance and/or cell penetration. Delivering PNAs directly by either electroporation or conjugation of PNA with cell penetrating peptides, or by linkage of PNAs with four lysine residues has demonstrated inhibition of miR-122 activity in human liver cancer cells (Huh7) and rat primary hepatocyte cells with high efficiency (Fabani and Gait, 2008). PNA AMOs has also been shown to inhibit miR-155 function in mice in vivo (Fabani et al., 2010). By screening 11 cell-penetrating peptides, Oh et al., have shown that a Tat-modified-conjugated PNA is the most effective for delivering into cells and inhibiting miRNA function without the assistant of transfection reagents (Oh et al., 2010). PNA AMOs delivered into neurons have been shown to suppress miR-326 activities and cause an increased expression of a synaptic plasticity-related gene Arc, indicating a promising approach in neuroscience research (Wibrand et al., 2012).
The effects of AMOs need to be carefully interpreted. Some studies have shown anti-heart failure effect of miR-21 antagomirs, but some have shown no effect, which is also supported by genetic deletion study of miR-21 (Thum et al., 2008, 2011; Morrisey, 2010; Patrick et al., 2010). The difference is likely caused by high concentration of delivered antagamirs that can bind to target tissues and induce non-specific disruptions of gene expression. The delivery methods for antagomirs such as vein injections may interfere with tissue distribution and metabolisms of AMOs, or cause toxicities. Optimizing AMO design will reduce the amount of AMO usage in vivo, and in turn reduce AMO toxicity and off-target effects (Park et al., 2011).
miRNA sponges
The transient nature of current AMO miRNA inhibitors has driven the development of new approaches, such as miRNA sponge, to achieve long-term miRNA loss-of-function. The miRNA sponge contains multiple binding sequences complementary to a mature miRNA and can bind to endogenous miRNAs and block their silencing activity (Figure 1C). The most common miRNA sponge design involves the insertion of a tandem multiplex artificial miRNA binding sequence into the 3′UTR of a Pol II-driven reporter gene, such as green fluorescence protein (GFP) or luciferase. The miRNA binding sequence of the sponge can be perfectly complementary to the seed sequence of the miRNA and then bulged, or kept complementary at positions 9–12 (Ebert et al., 2007; Gentner et al., 2009; Loya et al., 2009). The bulge is constructed on purpose to protect against RNA interference-type cleavage and degradation of the sponge RNA by the Ago2 component of the RISC. Sponges with exact complementary to the target miRNA also have been shown to have inhibitory effects (Ebert et al., 2007; Ebert and Sharp, 2010). In addition, there is a second miRNA sponge system that utilizes the Pol III promoter to drive sponge expression, in which 5′ and 3′ stem-loop structures are added to stabilize the RNA products to substitute the reporter gene in the Pol II promoter system. Both Pol II- and Pol III-driven sponges with bulged or perfect miR-20 binding sites have been shown to successfully rescue miR-20 binding to its target sites (Ebert et al., 2007). An advantage of using the sponge approach is that binding sequences for a family of miRNAs (due to the conserved seed sequences) or even completely unrelated miRNAs can be constructed together in one vector. The silencing activities of these miRNAs can be repressed simultaneously to facilitate efficient suppression of multiple miRNAs.
A miR-9 sponge has been shown to modify motor neuron subtypes in the developing spinal cord (Otaegi et al., 2011a). By optimizing the sponge design, Otaegi et al., have shown that a shorter spacing between binding sequences and the inclusion of a coding gene improve the sponge effect. Moreover, optimizing the number of binding sequences also affects sponge activity (Otaegi et al., 2011b).
Similar to the sponge, tough decoy (TuD) RNA driven by RNA Pol III promoter (U6) has been developed to achieve long term suppression of miRNAs using lentiviral based vectors (Haraguchi et al., 2009). TuD RNAs containing two miRNA binding sites with four extra nts inserted in each and flanked by 3 nts linkers and two stem structures, show the strongest miRNA inhibition (Haraguchi et al., 2009). The use of recombinant adeno-associated virus (rAAV) to deliver the TuD RNAs for let-7 and miR-122 has shown strong, long term inhibition of let-7 and miR-122 expression in mice (Xie et al., 2012).
The miRNA sponge and TuD RNA techniques have shown promising inhibition of miRNAs in vitro and in vivo. However, the design and efficiency of sponge and TuD RNAs, as well as their delivery by either plasmid or viral vectors, require careful examination for individual miRNAs in the nervous system.
mRNA protectors
The approaches described above are focused on direct manipulations of miRNAs, which may affect expression of all target genes of these miRNAs. To evaluate the direct silencing activities of miRNAs on their specific target mRNAs, the mRNA protector technique has been developed. The mRNA protector has a perfect complementary sequence to the miRNA binding site on the 3′UTR of a putative target mRNA and can compete with endogenous miRNAs for the binding site, with higher affinity to prevent miRNA/RNA association (Figure 1D). For example, Choi et al., have used a PMO-based target mRNA protector in vivo to prevent the action of miR-430 on its target genes squint and lefty in zebrafish (Choi et al., 2007). Another morpholino target protector has been used to identify hairy1 as a primary target for miR-9 in neural progenitors in Xenopus (Bonev et al., 2011). Current mRNA protectors using morpholinos are designed to bind to the region of the target mRNA complementary to the miRNA seed sequence and to 3′ or 5′ flanking sequences in the 3′UTR (Choi et al., 2007; Bonev et al., 2011; Staton and Giraldez, 2011). Whether oligo-based mRNA protectors have a long term effect in vivo, and the challenge of delivering mRNA protector oligos especially in mammals have limited their application. Developing a plasmid- or virus-based mRNA protector strategy is an option to achieve long-term and tissue specific exogenous protector expression. Moreover, as a miRNA often functions to regulate multiple target genes, protecting only one target mRNA may not reveal the full miRNA function. Nevertheless, due to the specificity of mRNA protectors, they can be used to screen major targets for a miRNA in neurogenesis.
Small molecule inhibitors
Small molecules serving as inhibitors of miRNAs have been explored. Using a luciferase-based assay, azobenzene was identified to selectively inhibit miR-21 transcription from more than 1000 compound screened (Gumireddy et al., 2008). Studies have demonstrated that inhibitors for miR-122 show specific blocking activity on miR-122 but no other miRNAs, suggesting that small molecules can be used as inhibitors for specific miRNAs (Young et al., 2010; Connelly et al., 2012). These data further indicate the feasibility of using small molecules to inhibit miRNA function in neurogenesis.
Delivery techniques to manipulate miRNAs in vivo
The success of manipulating miRNA expression in the nervous system relies on efficient delivery systems. We here highlight the delivery techniques that can be used for examining miRNA function in neurogenesis.
Microinjection
Microinjection is one of the simplest methods for gene delivery. miRNA mimics or AMO inhibitors can be injected into the yolk or cytoplasm of early embryonic cells, using fine glass needles with the assistant of a pressure microinjector and micromanipulator, to interrupt miRNA function. For example, microinjection of miR-124a precursors at the 8-cell stage revealed the role of miR-124a in early eye development in Xenopus, and double-stranded miRNA let-7 was injected into the zygotes of zebrafish and frogs (Kloosterman et al., 2004; Qiu et al., 2009). Injections of miR-183, miR-96 and miR-182 LNA inhibitors in zebrafish embryos have demonstrated the roles of these miRNAs in the development of sensorineurons (Li et al., 2010). The miRNA microinjection technique was also applied to study the development of Drosophila (Berry et al., 2009). However, microinjection in mammalian embryos and postnatal tissues is difficult, especially in the nervous system, which has limited its application.
Virus infection
Virus infection is a common tool to transport exogenous transgenes into target cells and tissues in vitro and in vivo. Adenoviral (Ad) vectors infect both dividing and non-dividing cells and can be used in vitro and in vivo (Xia et al., 2002; Xu et al., 2011). However, Ad vectors cannot integrate into the host genome, and are found to elicit the immune response, which has limited their use for long term studies of miRNA effects (Bessis et al., 2004). Adeno-associated viruses (AAVs) work in both dividing and non-dividing cells and unlike the Ad vector, the AAV does not induce strong immune reaction and can integrate into the host genome (Rutledge and Russell, 1997; Grimm and Kay, 2003). The AAV carried miRNA-based hairpins have been shown to efficiently knock down the photoreceptor-specific gene Peripherin-2 in the mouse retina (Georgiadis et al., 2010). miR-183/96/182 sponges in the AAV have been shown to inhibit activity of this cluster in the photoreceptor of mouse eyes in vivo (Krol et al., 2010). Retroviral vectors (RVs) including the lentiviral vectors (LVs) are genome-integrating vectors. The lentiviruses can express a gene of interest in both dividing and non-dividing cells, stem cells, zygotes and their differentiated progeny, to maintain persistent gene expression (Rubinson et al., 2003). Mice infected by miR-326 or its sponge LVs have been used to examine the pathogenesis of multiple sclerosis (Du et al., 2009). Studies have used RVs and LVs to overexpress and knock down miR-124 in mice, and demonstrated miR-124 function in postnatal and adult neurogenesis (Cheng et al., 2009; Åkerblom et al., 2012).
Virus infection has the potential to target cells and tissues other than the nervous system and causes side effects when delivered. Some viruses such as RVs and LVs may exhibit unexpected exogenous gene transcriptional silencing, which hinders highly efficient transgene expression. These are the factors that should be considered when using virus infection to study miRNA function in neurogenesis.
In utero/in ovo electroporation
To conduct quick Gain-of-function and loss-of-function studies for a gene of interest in the developing nervous system, approaches such as in utero or in ovo electroporation have been widely used. This technique utilizes electric field pulses to drive nucleic acid molecules through pores generated in the cell membrane to introduce the exogenous transgene into cells. The in utero electroporation technique allows passage of the target gene plasmid DNA into the lateral ventricle of mouse embryonic brains through the uterus wall of the pregnant mouse, and introduction of DNA constructs into proliferative ventricular zone progenitor cells (Nishimura et al., 2012; Pacary et al., 2012). For example, functions of a brain-enriched miR-137 in the developing mouse brain have been tested using in utero electroporation (Sun et al., 2011). In utero electroporation of miR-9 and let-7 into embryonic mouse brains was used to study neural stem cell fate determination (Zhao et al., 2009a, 2010a). The role of miR-9 in motor neuron subtype specification has been examined in the chick embryonic spinal cord using in ovo electroporation (Otaegi et al., 2011a,b). Inhibition and overexpression of miR-124 by in ovo electroporation were applied to explore its function in chick neural tube development (Cao et al., 2007; Visvanathan et al., 2007). Studies have reported successful expression of an exogenous gene in primitive streak stage embryos using electroporation (Yasuda et al., 2000; Kobayashi et al., 2002). Electroporation may cause variations of expression levels and positions of an exogenous gene in neural tissues, and a large number of samples are required to draw a reliable conclusion. Nevertheless, in utero/in ovo electroporation is a powerful tool to manipulate miRNA expression in specific regions in the nervous system.
miRNA transgenic animals
Although the procedure to construct transgenic animals is time consuming, it is still an effective strategy to test miRNA functions in vivo. miRNA gain-of-function can be achieved by miRNA overexpression transgenic constructs and loss-of-function can be achieved in miRNA gene knockout or sponge expression animals. Similar to coding genes, miRNAs can be ablated from the genome using DNA recombination, and also depleted in specific tissues via, for instance the Cre-LoxP system, to generate miRNA conditional knockout mice or tissue specific miRNA sponge transgenic mice (Figures 1E,F).
To study the function of miR-132 in hippocampal-dependant learning and memory, a doxycycline-regulated miR-132 mouse line has been generated to show activity-dependant regulation of miR-132 in cognition (Hansen et al., 2012). Conditional over-expression of miR-9* and miR-124 under the control of the nestin promoter in neural progenitors has been shown to suppress their target gene BAF53a and lead to progenitor proliferation defects (Yoo et al., 2009). Transgenic mice with a lymphocyte specific miR-17–92 overexpression construct developed lymphoproliferative disease and autoimmunity and died prematurely (Xiao et al., 2008).
miRNA knockout mice have been used to reveal many aspects of miRNA functions (Park et al., 2010). Functions of miR-208 in heart growth, miR-155 in immune system and miR-1-2 in heart development have been demonstrated in miRNA knockout mice (Rodriguez et al., 2007; Thai et al., 2007; Van Rooij et al., 2007; Zhao et al., 2007). Knockout mice of the miR-17-92 cluster have revealed their critical function in embryogenesis (Ventura et al., 2008). miR-133 knockout mice have proven subtle function of miR-133 in midbrain dopaminergic neuron development (Heyer et al., 2012). In the double knockout mice in which miR-9-2 and miR-9-3 precursors are ablated, the brain development is severely affected due to proliferation defects in neural progenitors (Shibata et al., 2011). Conditional deletion of the miR212/132 locus using retrovirus-based Cre expression has shown dendrite growth and arborization defects in adult hippocampal neurons (Magill et al., 2010). In other species, the gene knockout technique is also utilized to explore the function of miRNAs in C. elegans and Drosophila (Clark et al., 2010; Sun et al., 2012).
Mature miRNA normally has several precursors with similar seed sequences and other miRNA precursors may functionally compensate the knockout of a single miRNA precursor, unless all precursors are ablated. For example, less severe phenotypes of miR-182 ablation in retina are likely caused by functional redundancy (Jin et al., 2009). miRNAs are often localized closely as clusters in the genome. Ablation of a single miRNA without disturbing other miRNAs is technically challenging. Generating sponge transgenic animals can overcome these drawbacks (Figure 1F). miRNA sponge transgenic mouse model that blocks the activity of the miR-183/96/182 cluster simultaneously and selectively in the retina has revealed their important role in acute light-induced retinal degeneration (Zhu et al., 2011). miR-29 sponge transgenic mice have demonstrated the role of miR-29 in suppressing intracellular pathogen-induced immune responses (Ma et al., 2011). Moreover, fly miRNA sponge transgenics has revealed the function of miR-8 in neuromuscular junction formation (Loya et al., 2009).
Liposome, polymer, hydrogel, microsphere, and nanoparticle
A recent study has shown that liposome encapsulated plasmids or oligonucleotides can pass into the phospholipid bilayer, and cationic liposome protects oligonucleotides from nuclease degradation and facilitates cell uptake (Zhao et al., 2009b). However, liposome reagents are usually toxic, initiate immune responses, prone to accumulate in the reticuloendothelium system and have short half-life, all of which have limited their applications (Chiarantini et al., 2005; Aagaard and Rossi, 2007; Zhao et al., 2009b). Alternatively, a study using lipid-like materials called lipidoids to transfer 2′-O-methyl AMOs into cells has been proven to be safe and efficient in mice, rat and non-human primates models (Akinc et al., 2008). Moreover, biodegradable polymers are a solution to the problems of using liposomes. Polymer delivery of oligonucleotides has been shown to exhibit sustained release and better tissue distribution (Chirila et al., 2002). Hydrogels, microspheres and nanoparticles have also been evaluated for their suitability to transport nucleic acids into target cells and have been proved to be efficient, stable and of low toxicity (Rosi et al., 2006; Bisht et al., 2008; Zhao et al., 2009b). Moreover, nanoparticles have potential to allow the manipulation of miRNA expression by using cell type specific targeting molecules, such as peptides, ligands, antibodies and other bioactive molecules in neuronal cells. For example, increased accumulation of miR-124 has been detected in brains of mice with tail vein injection of a miR-124 nanocarrier (Hwang Do et al., 2011).
Conclusions
Accumulating evidence has demonstrated the importance of miRNAs in neural development and function. Both gain-of-function and loss-of-function approaches have been applied to uncover the role of miRNAs in neurogenesis. We here have systematically summarized new techniques used to manipulate miRNA expression in vitro and in vivo and each approach has technical advantages and disadvantages. Combination of multiple approaches may be necessary to further advance the investigation of miRNA functions. Dysregulation of miRNAs has been found to be associated with several neurological disorders (Bian and Sun, 2011; Fiore et al., 2011; Sibley and Wood, 2011; Junn and Mouradian, 2012; Saito and Saito, 2012; Tan et al., 2012). Taking advantage of miRNA synthesis and delivery techniques, miRNA manipulations are becoming a promising means of gene therapy to treat human neurological diseases.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Hirschl/Weill-Caulier Trust (Tao Sun), an NPRP grant (09-1011-3-260) from the Qatar National Research Fund (Benjamin Shykind and Tao Sun) and an R01-MH083680 grant from the NIH/NIMH (Tao Sun).
Key Concept
- microRNAs (miRNAs)
~22 nucleotide endogenous small non-coding RNAs that exist in many organisms such as plants, invertebrates and vertebrates. Mature miRNAs regulate gene expression by binding to the 3′untranslated region (3′UTR) of target protein coding genes, and affect stability of messenger RNAs (mRNAs) and/or reduce protein translation.
- Neurogenesis
A process of generating neurons from neural stem cells and neural progenitors. It involves tightly controlled regulation of proliferation, differentiation, migration and maturation.
- miRNA inhibitor
Chemically synthesized antisense miRNA oligonucleotides designed to knock down endogenous miRNA silencing activity by directly binding to the single strand mature miRNA using complementary sequences.
- Antagomir
A form of miRNA inhibitors that is a 22–23 nucleotide, 2′-O-methyl modified, 3′ end cholesterol-conjugate RNA analog with a complementary sequence to a mature miRNA.
- miRNA sponge
An RNA transcript contains binding sites designed to be targeted by a specific miRNA or a family of miRNAs to block the silencing activity of the endogenous miRNAs to their putative targets.
- mRNA protector
An RNA transcript or oligonucleotide contains a perfect complementary sequence to the miRNA binding site on the 3′UTR of a putative target mRNA. It competes with endogenous miRNAs for the binding site and prevents miRNA/RNA association to block the endogenous miRNA silencing activity.
- Gain-of-function and loss-of-function
Approaches to altering function of a coding gene or a non-coding RNA by increasing and decreasing its expression levels in specific cells or tissues, respectively.
Biography
 Tao Sun received his Ph.D. in Neurobiology from University College London, England. He then performed his postdoctoral training in Neurogenetics at Harvard Medical School at Boston, USA. He is now an Associate Professor at Weill Medical College of Cornell University at New York City, USA. His research interests include neurogenesis, neural development, neural stem cells, neurological diseases, behaviors, microRNA, and non-coding RNA.
Tao Sun received his Ph.D. in Neurobiology from University College London, England. He then performed his postdoctoral training in Neurogenetics at Harvard Medical School at Boston, USA. He is now an Associate Professor at Weill Medical College of Cornell University at New York City, USA. His research interests include neurogenesis, neural development, neural stem cells, neurological diseases, behaviors, microRNA, and non-coding RNA.
 Haijun Zhang received his medical degree from Weifang Medical College, and his Ph.D. in Biochemistry and Molecular Biology from Peking University in China. After finishing his Ph.D. project, he studied in Masonic Cancer Center at University of Minnesota as a Postdoctoral Associate. Then he was recruited to Dr. Tao Sun's Lab at Weill Medical College of Cornell University to conduct postdoctoral training in neural development by examining the roles of microRNAs in the development of the cerebral cortex.
Haijun Zhang received his medical degree from Weifang Medical College, and his Ph.D. in Biochemistry and Molecular Biology from Peking University in China. After finishing his Ph.D. project, he studied in Masonic Cancer Center at University of Minnesota as a Postdoctoral Associate. Then he was recruited to Dr. Tao Sun's Lab at Weill Medical College of Cornell University to conduct postdoctoral training in neural development by examining the roles of microRNAs in the development of the cerebral cortex.
References
- Aagaard L., Rossi J. J. (2007). RNAi therapeutics: principles, prospects and challenges. Adv. Drug Deliv. Rev. 59, 75–86 10.1016/j.addr.2007.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerblom M., Sachdeva R., Barde I., Verp S., Gentner B., Trono D., et al. (2012). MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 32, 8879–8889 10.1523/JNEUROSCI.0558-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A., Zumbuehl A., Goldberg M., Leshchiner E. S., Busini V., Hossain N., et al. (2008). A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 26, 561–569 10.1038/nbt1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson T., Rahman S., Sansom S. N., Alsio J. M., Kaneda M., Smith J., et al. (2010). Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE 5:e13453 10.1371/journal.pone.0013453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz J. E., Hsu R., Melton C., Thomas M., Ullian E. M., Blelloch R. (2011). A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA 17, 1489–1501 10.1261/rna.2442211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D., Fan A. P., Potter C. J., Luo L. (2008). MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr. Biol. 18, 1754–1759 10.1016/j.cub.2008.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Chung W. J., Willis J., Cuppen E., Lai E. C. (2007). Mammalian mirtron genes. Mol. Cell 28, 328–336 10.1016/j.molcel.2007.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry B., Deddouche S., Kirschner D., Imler J. L., Antoniewski C. (2009). Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS ONE 4:e5866 10.1371/journal.pone.0005866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessis N., Garciacozar F. J., Boissier M. C. (2004). Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 11(Suppl. 1), S10–S17 10.1038/sj.gt.3302364 [DOI] [PubMed] [Google Scholar]
- Bian S., Sun T. (2011). Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 44, 359–373 10.1007/s12035-011-8211-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht S., Feldmann G., Koorstra J. B., Mullendore M., Alvarez H., Karikari C., et al. (2008). In vivo characterization of a polymeric nanoparticle platform with potential oral drug delivery capabilities. Mol. Cancer Ther. 7, 3878–3888 10.1158/1535-7163.MCT-08-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Pisco A., Papalopulu N. (2011). MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 20, 19–32 10.1016/j.devcel.2010.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde H., Schmitt S., Fitzner D., Opitz L., Salinas-Riester G., Simons M. (2010). Control of oligodendroglial cell number by the miR-17-92 cluster. Development 137, 2127–2132 10.1242/dev.050633 [DOI] [PubMed] [Google Scholar]
- Cao X., Pfaff S. L., Gage F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536 10.1101/gad.1519207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M., Schmidt S., Benes V., Noerholm M., Kulozik A. E., Hentze M. W., et al. (2006). A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA 12, 913–920 10.1261/rna.2332406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. A., Krichevsky A. M., Kosik K. S. (2005). MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033 10.1158/0008-5472.CAN-05-0137 [DOI] [PubMed] [Google Scholar]
- Cheloufi S., Dos Santos C. O., Chong M. M. W., Hannon G. J. (2010). A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. A., Wichterle H. (2012). Apoptosis of limb innervating motor neurons and erosion of motor pool identity upon lineage specific dicer inactivation. Front. Neurosci. 6:69 10.3389/fnins.2012.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., et al. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarantini L., Cerasi A., Fraternale A., Millo E., Benatti U., Sparnacci K., et al. (2005). Comparison of novel delivery systems for antisense peptide nucleic acids. J. Control. Release 109, 24–36 10.1016/j.jconrel.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Chirila T. V., Rakoczy P. E., Garrett K. L., Lou X., Constable I. J. (2002). The use of synthetic polymers for delivery of therapeutic antisense oligodeoxynucleotides. Biomaterials 23, 321–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. Y., Giraldez A. J., Schier A. F. (2007). Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274 10.1126/science.1147535 [DOI] [PubMed] [Google Scholar]
- Chung K. H., Hart C. C., Al-Bassam S., Avery A., Taylor J., Patel P. D., et al. (2006). Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 34:e53 10.1093/nar/gkl143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D., Xue H., Taylor D. W., Patnode H., Mishima Y., Cheloufi S., et al. (2010). A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. M., Goldstein L. D., Tevlin M., Tavare S., Shaham S., Miska E. A. (2010). The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res. 38, 3780–3793 10.1093/nar/gkq083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L., Hobert O. (2012). Diverse functions of microRNAs in nervous system development. Curr. Top. Dev. Biol. 99, 115–143 10.1016/B978-0-12-387038-4.00005-7 [DOI] [PubMed] [Google Scholar]
- Connelly C. M., Thomas M., Deiters A. (2012). High-throughput luciferase reporter assay for small-molecule inhibitors of microRNA function. J. Biomol. Screen. 17, 822–828 10.1177/1087057112439606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar T. L., Davis T. H., Nelson P. T., Loeb G. B., Harfe B. D., Ullian E., et al. (2008). Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 105, 5614–5619 10.1073/pnas.0801689105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Lollo B., Freier S., Esau C. (2006). Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 34, 2294–2304 10.1093/nar/gkl183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Propp S., Freier S. M., Jones L. E., Serra M. J., Kinberger G., et al. (2009). Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 37, 70–77 10.1093/nar/gkn904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. H., Cuellar T. L., Koch S. M., Barker A. J., Harfe B. D., McManus M. T., et al. (2008). Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330 10.1523/JNEUROSCI.4815-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D., Pulvers J. N., Haffner C., Murchison E. P., Hannon G. J., Huttner W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921 10.1242/dev.025080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Liu C., Kang J., Zhao G., Ye Z., Huang S., et al. (2009). MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10, 1252–1259 10.1038/ni.1798 [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Neilson J. R., Sharp P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M. S., Sharp P. A. (2010). MicroRNA sponges: progress and possibilities. RNA 16, 2043–2050 10.1261/rna.2414110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., et al. (2008). Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36, 1153–1162 10.1093/nar/gkm1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabani M. M., Abreu-Goodger C., Williams D., Lyons P. A., Torres A. G., Smith K. G., et al. (2010). Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 38, 4466–4475 10.1093/nar/gkq160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabani M. M., Gait M. J. (2008). miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 14, 336–346 10.1261/rna.844108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg S. K., Kosik K. S., Davidson B. L. (2009). MicroRNAs potentiate neural development. Neuron 64, 303–309 10.1016/j.neuron.2009.10.020 [DOI] [PubMed] [Google Scholar]
- Fiore R., Khudayberdiev S., Saba R., Schratt G. (2011). MicroRNA function in the nervous system. Prog. Mol. Biol. Transl. Sci. 102, 47–100 10.1016/B978-0-12-415795-8.00004-0 [DOI] [PubMed] [Google Scholar]
- Flynt A. S., Li N., Thatcher E. J., Solnica-Krezel L., Patton J. G. (2007). Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 39, 259–263 10.1038/ng1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., et al. (2004). Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6, 784–791 10.1038/ncb1155 [DOI] [PubMed] [Google Scholar]
- Gentner B., Schira G., Giustacchini A., Amendola M., Brown B. D., Ponzoni M., et al. (2009). Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods 6, 63–66 10.1038/nmeth.1277 [DOI] [PubMed] [Google Scholar]
- Georgiadis A., Tschernutter M., Bainbridge J. W., Robbie S. J., McIntosh J., Nathwani A. C., et al. (2010). AAV-mediated knockdown of peripherin-2 in vivo using miRNA-based hairpins. Gene Ther. 17, 486–493 10.1038/gt.2009.162 [DOI] [PubMed] [Google Scholar]
- Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., et al. (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- Grimm D., Kay M. A. (2003). From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 3, 281–304 10.2174/1566523034578285 [DOI] [PubMed] [Google Scholar]
- Grimm D., Streetz K. L., Jopling C. L., Storm T. A., Pandey K., Davis C. R., et al. (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 10.1038/nature04791 [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., Ha I., et al. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- Gumireddy K., Young D. D., Xiong X., Hogenesch J. B., Huang Q., Deiters A. (2008). Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. Int. Ed. Engl. 47, 7482–7484 10.1002/anie.200801555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S. M., Bernstein E., Beach D., Hannon G. J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296 10.1038/35005107 [DOI] [PubMed] [Google Scholar]
- Hansen K. F., Karelina K., Sakamoto K., Wayman G. A., Impey S., Obrietan K. (2012). miRNA-132: a dynamic regulator of cognitive capacity. Brain Struct. Funct. [Epub ahead of print]. 10.1007/s00429-012-0431-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Ozaki Y., Iba H. (2009). Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37:e43 10.1093/nar/gkp040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer M. P., Pani A. K., Smeyne R. J., Kenny P. J., Feng G. (2012). Normal midbrain dopaminergic neuron development and function in miR-133b mutant mice. J. Neurosci. 32, 10887–10894 10.1523/JNEUROSCI.1732-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Liu Y., Huang M., Zhao X., Cheng L. (2010). Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J. Mol. Cell. Biol. 2, 152–163 10.1093/jmcb/mjq008 [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A. E., Balint E., Tuschl T., Zamore P. D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard M. J., Mello C. C., Zamore P. D. (2004). Sequence-specific inhibition of small RNA function. PLoS Biol. 2:E98 10.1371/journal.pbio.0020098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Zamore P. D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 10.1126/science.1073827 [DOI] [PubMed] [Google Scholar]
- Hwang Do W., Son S., Jang J., Youn H., Lee S., Lee D., et al. (2011). A brain-targeted rabies virus glycoprotein-disulfide linked PEI nanocarrier for delivery of neurogenic microRNA. Biomaterials 32, 4968–4975 10.1016/j.biomaterials.2011.03.047 [DOI] [PubMed] [Google Scholar]
- Iida A., Shinoe T., Baba Y., Mano H., Watanabe S. (2011). Dicer plays essential roles for retinal development by regulation of survival and differentiation. Invest. Ophthalmol. Vis. Sci. 52, 3008–3017 10.1167/iovs.10-6428 [DOI] [PubMed] [Google Scholar]
- Jin Z. B., Hirokawa G., Gui L., Takahashi R., Osakada F., Hiura Y., et al. (2009). Targeted deletion of miR-182, an abundant retinal microRNA. Mol. Vis. 15, 523–533 [PMC free article] [PubMed] [Google Scholar]
- Junn E., Mouradian M. M. (2012). MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol. Ther. 133, 142–150 10.1016/j.pharmthera.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S. A., Kung A. L., Ganesan S., Drapkin R., Jenuwein T., et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 10.1101/gad.1248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., Arora A., Wengel J., Maiti S. (2006). Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry 45, 7347–7355 10.1021/bi060307w [DOI] [PubMed] [Google Scholar]
- Kawase-Koga Y., Low R., Otaegi G., Pollock A., Deng H., Eisenhaber F., et al. (2010). RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell. Sci. 123, 586–594 10.1242/jcs.059659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y., Otaegi G., Sun T. (2009). Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 238, 2800–2812 10.1002/dvdy.22109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W. P., Wienholds E., De Bruijn E., Kauppinen S., Plasterk R. H. (2006). In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 3, 27–29 10.1038/nmeth843 [DOI] [PubMed] [Google Scholar]
- Kloosterman W. P., Wienholds E., Ketting R. F., Plasterk R. H. (2004). Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 32, 6284–6291 10.1093/nar/gkh968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D., Kobayashi M., Matsumoto K., Ogura T., Nakafuku M., Shimamura K. (2002). Early subdivisions in the neural plate define distinct competence for inductive signals. Development 129, 83–93 [DOI] [PubMed] [Google Scholar]
- Kocerha J., Faghihi M. A., Lopez-Toledano M. A., Huang J., Ramsey A. J., Caron M. G., et al. (2009). MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. U.S.A. 106, 3507–3512 10.1073/pnas.0805854106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W., Kiryk A., Novak M., Herwerth M., Parkitna J. R., Wawrzyniak M., et al. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842 10.1523/JNEUROSCI.3030-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik K. S. (2006). The neuronal microRNA system. Nat. Rev. Neurosci. 7, 911–920 [DOI] [PubMed] [Google Scholar]
- Kosik K. S., Krichevsky A. M. (2005). The elegance of the microRNAs: a neuronal perspective. Neuron 47, 779–782 10.1016/j.neuron.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Busskamp V., Markiewicz I., Stadler M. B., Ribi S., Richter J., et al. (2010). Characterizing light-fegulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631 10.1016/j.cell.2010.03.039 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J., Kuwajima S., Braich R., Rajeev K. G., Pena J., Tuschl T., et al. (2007). Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 35, 2885–2892 10.1093/nar/gkm024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., et al. (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Lanford R. E., Hildebrandt-Eriksen E. S., Petri A., Persson R., Lindow M., Munk M. E., et al. (2010). Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 10.1126/science.1178178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P., Hudson L. D. (2010). MicroRNAs in neural cell differentiation. Brain Res. 1338, 14–19 10.1016/j.brainres.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox K. A., Behlke M. A. (2010). A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 27, 1788–1799 10.1007/s11095-010-0156-0 [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Li H., Kloosterman W., Fekete D. M. (2010). MicroRNA-183 family members regulate sensorineural fates in the inner ear. J. Neurosci. 30, 3254–3263 10.1523/JNEUROSCI.4948-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Bian S., Hong J., Kawase-Koga Y., Zhu E., Zheng Y., et al. (2011). Timing specific requirement of microRNA function is essential for embryonic and postnatal hippocampal development. PLoS ONE 6:e26000 10.1371/journal.pone.0026000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhao X. (2009). MicroRNAs in adult and embryonic neurogenesis. Neuromolecular Med. 11, 141–152 10.1007/s12017-009-8077-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Huang T., Zhao X., Cheng L. (2011). MicroRNAs modulate the Wnt signaling pathway through targeting its inhibitors. Biochem. Biophys. Res. Commun. 408, 259–264 10.1016/j.bbrc.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Loya C. M., Lu C. S., Van Vactor D., Fulga T. A. (2009). Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 6, 897–903 10.1038/nmeth.1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart B. W., Perederiy J. V., Westbrook G. L. (2012). Dentate gyrus neurogenesis, integration and microRNAs. Behav. Brain Res. 227, 348–355 10.1016/j.bbr.2011.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Xu S., Liu X., Zhang Q., Xu X., Liu M., et al. (2011). The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat. Immunol. 12, 861–869 10.1038/ni.2073 [DOI] [PubMed] [Google Scholar]
- Magill S. T., Cambronne X. A., Luikart B. W., Lioy D. T., Leighton B. H., Westbrook G. L., et al. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 20382–20387 10.1073/pnas.1015691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. (2002). Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574 10.1016/S0092-8674(02)00908-X [DOI] [PubMed] [Google Scholar]
- McLaughlin J., Cheng D., Singer O., Lukacs R. U., Radu C. G., Verma I. M., et al. (2007). Sustained suppression of Bcr-Abl-driven lymphoid leukemia by microRNA mimics. Proc. Natl. Acad. Sci. U.S.A. 104, 20501–20506 10.1073/pnas.0710532105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S., Villanueva A., Moutinho C., Davalos V., Spizzo R., Ivan C., et al. (2011). Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U.S.A. 108, 4394–4399 10.1073/pnas.1014720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos P. A., Li Y., Jiang S. (2008). Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques 45, 613–614, 616, 618 passim. [DOI] [PubMed] [Google Scholar]
- Morrisey E. E. (2010). The magic and mystery of miR-21. J. Clin. Invest. 120, 3817–3819 10.1172/JCI44596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., et al. (2006). The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 8, 278–284 10.1038/ncb1373 [DOI] [PubMed] [Google Scholar]
- Nelson P. T., Baldwin D. A., Kloosterman W. P., Kauppinen S., Plasterk R. H., Mourelatos Z. (2006). RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 12, 187–191 10.1261/rna.2258506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y. V., Shinoda T., Inaguma Y., Ito H., Nagata K. (2012). Application of in utero electroporation and live imaging in the analyses of neuronal migration during mouse brain development. Med. Mol. Morphol. 45, 1–6 10.1007/s00795-011-0557-0 [DOI] [PubMed] [Google Scholar]
- Nowakowski T. J., Mysiak K. S., Pratt T., Price D. J. (2011). Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS ONE 6:e23013 10.1371/journal.pone.0023013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. Y., Ju Y., Kim S., Park H. (2010). PNA-based antisense oligonucleotides for micrornas inhibition in the absence of a transfection reagent. Oligonucleotides 20, 225–230 10.1089/oli.2010.0238 [DOI] [PubMed] [Google Scholar]
- Okamura K., Hagen J. W., Duan H., Tyler D. M., Lai E. C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89–100 10.1016/j.cell.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U. A., Kauppinen S., Lund A. H. (2006). LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 372, 137–141 10.1016/j.gene.2005.12.031 [DOI] [PubMed] [Google Scholar]
- Otaegi G., Pollock A., Hong J., Sun T. (2011a). MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 31, 809–818 10.1523/JNEUROSCI.4330-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegi G., Pollock A., Sun T. (2011b). An optimized sponge for microrna mir-9 affects spinal motor neuron development in vivo. Front. Neurosci. 5:146 10.3389/fnins.2011.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E., Haas M. A., Wildner H., Azzarelli R., Bell D. M., Abrous D. N., et al. (2012). Visualization and genetic manipulation of dendrites and spines in the mouse cerebral cortex and hippocampus using in utero electroporation. J. Vis. Exp. e4163 10.3791/4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. Y., Choi Y. S., McManus M. T. (2010). Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 19, R169–R175 10.1093/hmg/ddq367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. K., Kogure T., Nuovo G. J., Jiang J., He L., Kim J. H., et al. (2011). miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 71, 7608–7616 10.1158/0008-5472.CAN-11-1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D. M., Montgomery R. L., Qi X., Obad S., Kauppinen S., Hill J. A., et al. (2010). Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 120, 3912–3916 10.1172/JCI43604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R., Liu K., Liu Y., Mo W., Flynt A. S., Patton J. G., et al. (2009). The role of miR-124a in early development of the Xenopus eye. Mech. Dev. 126, 804–816 10.1016/j.mod.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto E., Briata P., Kuziner N., Harfe B. D., McManus M. T., Gherzi R., et al. (2012). Let-7b/c enhance the stability of a tissue-specific mRNA during mammalian organogenesis as part of a feedback loop involving KSRP. PLoS Genet. 8:e1002823 10.1371/journal.pgen.1002823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. (2004). Identification of mammalian microRNA host genes and transcription units. Genome Res. 14, 1902–1910 10.1101/gr.2722704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., et al. (2007). Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengauer E., Hartwich H., Hartmann A. M., Rudnicki A., Satheesh S. V., Avraham K. B., et al. (2012). Egr2::cre mediated conditional ablation of dicer disrupts histogenesis of Mammalian central auditory nuclei. PLoS ONE 7:e49503 10.1371/journal.pone.0049503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N. L., Giljohann D. A., Thaxton C. S., Lytton-Jean A. K., Han M. S., Mirkin C. A. (2006). Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312, 1027–1030 10.1126/science.1125559 [DOI] [PubMed] [Google Scholar]
- Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., et al. (2003). A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33, 401–406 10.1038/ng1117 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge E. A., Russell D. W. (1997). Adeno-associated virus vector integration junctions. J. Virol. 71, 8429–8436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Saito H. (2012). MicroRNAs in cancers and neurodegenerative disorders. Front. Genet. 3:194 10.3389/fgene.2012.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan G., Li Y., Zhang J., Li W., Szulwach K. E., Duan R., et al. (2008). A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 26, 933–940 10.1038/nbt.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Zhao X., Hsieh J., Wichterle H., Impey S., Banerjee S., et al. (2010). MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 30, 14931–14936 10.1523/JNEUROSCI.4280-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Nakao H., Kiyonari H., Abe T., Aizawa S. (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407–3422 10.1523/JNEUROSCI.5085-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Shin J. Y., McManus M. T., Ptacek L. J., Fu Y. H. (2009). Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol. 66, 843–857 10.1002/ana.21927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C. R., Wood M. J. (2011). The miRNA pathway in neurological and skeletal muscle disease: implications for pathogenesis and therapy. J. Mol. Med. (Berl.) 89, 1065–1077 10.1007/s00109-011-0781-z [DOI] [PubMed] [Google Scholar]
- Soukup G. A., Fritzsch B., Pierce M. L., Weston M. D., Jahan I., McManus M. T., et al. (2009). Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev. Biol. 328, 328–341 10.1016/j.ydbio.2009.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K. L., Xu B., Bagchi A., Lai W. S., Liu H., Hsu R., et al. (2008). Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 40, 751–760 10.1038/ng.138 [DOI] [PubMed] [Google Scholar]
- Staton A. A., Giraldez A. J. (2011). Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nat. Protoc. 6, 2035–2049 10.1038/nprot.2011.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Ye P., Murai K., Lang M. F., Li S., Zhang H., et al. (2011). miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2:529 10.1038/ncomms1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Westholm J. O., Tsurudome K., Hagen J. W., Lu Y., Kohwi M., et al. (2012). Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 8:e1002515 10.1371/journal.pgen.1002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., et al. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 10.1158/0008-5472.CAN-04-0637 [DOI] [PubMed] [Google Scholar]
- Tan H., Xu Z., Jin P. (2012). Role of noncoding RNAs in trinucleotide repeat neurodegenerative disorders. Exp. Neurol. 235, 469–475 10.1016/j.expneurol.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Wu H., Lin Q., Wei W., Lu X. H., Cantle J. P., et al. (2011). Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J. Neurosci. 31, 8306–8319 10.1523/JNEUROSCI.0567-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., et al. (2007). Regulation of the germinal center response by microRNA-155. Science 316, 604–608 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- Thum T., Chau N., Bhat B., Gupta S. K., Linsley P. S., Bauersachs J., et al. (2011). Comparison of different miR-21 inhibitor chemistries in a cardiac disease model. J. Clin. Invest. 121, 461–462 author reply 462–463. 10.1172/JCI45938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., et al. (2008). MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 10.1038/nature07511 [DOI] [PubMed] [Google Scholar]
- Tognini P., Putignano E., Coatti A., Pizzorusso T. (2011). Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat. Neurosci. 14, 1237–1239 10.1038/nn.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi A., Hornyik C., Varga N., Burgyan J., Kauppinen S., Havelda Z. (2004). Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32:e175 10.1093/nar/gnh171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. (2007). Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 10.1126/science.1139089 [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J., Lee S., Lee B., Lee J. W., Lee S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749 10.1101/gad.1519107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K., Pai B., Siripornmongcolchai T., Bittins M., Berentsen B., Ofte M. L., et al. (2012). MicroRNA regulation of the synaptic plasticity-related gene arc. PLoS ONE 7:e41688 10.1371/journal.pone.0041688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., De Bruijn E., et al. (2005). MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- Xia H., Mao Q., Paulson H. L., Davidson B. L. (2002). siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20, 1006–1010 10.1038/nbt739 [DOI] [PubMed] [Google Scholar]
- Xiao C., Srinivasan L., Calado D. P., Patterson H. C., Zhang B., Wang J., et al. (2008). Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9, 405–414 10.1038/ni1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Ameres S. L., Friedline R., Hung J. H., Zhang Y., Xie Q., et al. (2012). Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods 9, 403–409 10.1038/nmeth.1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Xia F., Ma L., Shan J., Shen J., Yang Z., et al. (2011). MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 310, 160–169 10.1016/j.canlet.2011.06.027 [DOI] [PubMed] [Google Scholar]
- Yang J. S., Lai E. C. (2011). Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 43, 892–903 10.1016/j.molcel.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. S., Maurin T., Robine N., Rasmussen K. D., Jeffrey K. L., Chandwani R., et al. (2010). Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 15163–15168 10.1073/pnas.1006432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Momose T., Takahashi Y. (2000). Applications of microelectroporation for studies of chick embryogenesis. Dev. Growth Differ. 42, 203–206 10.1046/j.1440-169x.2000.00502.x [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Staahl B. T., Chen L., Crabtree G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. D., Connelly C. M., Grohmann C., Deiters A. (2010). Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J. Am. Chem. Soc. 132, 7976–7981 10.1021/ja910275u [DOI] [PubMed] [Google Scholar]
- Zehir A., Hua L. L., Maska E. L., Morikawa Y., Cserjesi P. (2010). Dicer is required for survival of differentiating neural crest cells. Dev. Biol. 340, 459–467 10.1016/j.ydbio.2010.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. (2009a). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Pan F., Holt C. M., Lewis A. L., Lu J. R. (2009b). Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert Opin. Drug Deliv. 6, 673–686 10.1517/17425240902992894 [DOI] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Lang M. F., Yang S., Li W., et al. (2010a). MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 1876–1881 10.1073/pnas.0908750107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Lee M. C., Momin A., Cendan C. M., Shepherd S. T., Baker M. D., et al. (2010b). Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J. Neurosci. 30, 10860–10871 10.1523/JNEUROSCI.1980-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., He X., Han X., Yu Y., Ye F., Chen Y., et al. (2010c). MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65, 612–626 10.1016/j.neuron.2010.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ransom J. F., Li A., Vedantham V., Von Drehle M., Muth A. N., et al. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell 129, 303–317 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
- Zheng K., Li H., Zhu Y., Zhu Q., Qiu M. (2010). MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J. Neurosci. 30, 8245–8250 10.1523/JNEUROSCI.1169-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Sun W., Okano K., Chen Y., Zhang N., Maeda T., et al. (2011). Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J. Biol. Chem. 286, 31749–31760 10.1074/jbc.M111.259028 [DOI] [PMC free article] [PubMed] [Google Scholar]