Abstract
5-HT neurons are topographically organized in the hindbrain, and have been implicated in the etiology and treatment of psychiatric diseases such as depression and anxiety. Early studies suggested that the raphe 5-HT neurons were a homogeneous population showing similar electrical properties, and feedback inhibition mediated by 5-HT1A autoreceptors. We utilized histochemistry techniques in ePet1-eGFP and 5-HT1A-iCre/R26R mice to show that a subpopulation of 5-HT neurons do not express the somatodendritic 5-HT1A autoreceptor mRNA. In addition, we performed patch-clamp recordings followed by single-cell PCR in ePet1-eGFP mice. From 134 recorded 5-HT neurons located in the dorsal, lateral, and median raphe, we found lack of 5-HT1A mRNA expression in 22 cells, evenly distributed across raphe subfields. We compared the cellular characteristics of these neuronal types and found no difference in passive membrane properties and general excitability. However, when injected with large depolarizing current, 5-HT1A-negative neurons fired more action potentials, suggesting a lack of autoinhibitory action of local 5-HT release. Our results support the hypothesis that the 5-HT system is composed of subpopulations of serotonergic neurons with different capacity for adaptation.
The 5-hydroxytryptamine (5-HT, serotonin) neurotransmitter system has been strongly implicated in the etiology and treatment of psychiatric diseases such as depression and anxiety.1,2 5-HT neurons are topographically organized in the hindbrain where distinct groups of neurons receive and send synaptic inputs from/to specific brain regions, suggesting that the neuronal activity of subpopulation of 5-HT neurons is under discrete spatiotemporal control.3
Given the occurrence of neurochemically diverse neurons in the raphe nuclei, early studies proposed a series of features or “landmarks”, common to all 5-HT neurons, that would help in the identification of putative 5-HT cells.4−6 These included the regular firing of broad action potentials (clocklike activity) followed by a large afterhyperpolarization potential, high input resistance, and suppression of firing by 5-hydroxytryptamine receptor 1A (5-HT1A) agonists.4−6 However, this dogma is progressively being questioned. Recent studies found that the electrophysiological characteristics of chemically identified 5-HT neurons in the raphe are more diverse than originally thought. Using juxtacellular labeling techniques, it was found that, besides the population of slow-firing clocklike cells, in vivo discharge of 5-HT neurons also includes subpopulations of fast-firing and bursting neurons.7,8 Moreover, in vitro patch-clamp studies conducted in brain slices demonstrated a high degree of variability in the passive and active membrane properties of 5-HT neurons.9,10 Finally, anatomical and electrophysiological studies showed that 5-HT1A receptors are not only expressed in 5-HT neurons, but also in other neuronal classes in the raphe nuclei.10−12 Together these evidence call to re-evaluate the definition of serotonergic cell, and to determine which methods are most appropriate for their identification.
In this work, we tested the hypothesis that subpopulations of 5-HT neurons do not express the 5-HT1A autoreceptor. It is currently believed that all serotonergic neurons express this receptor subtype.13−16 5-HT1A receptors are present in the soma and dendrites of 5-HT neurons, and provide strong autoinhibition.17,18 Activation of this receptor type inhibits neuronal firing, and subsequently decreases the release of 5-HT in target areas. Repeated activation of this receptor has been shown to induce a desensitization phenomenon, a mechanism that has been associated to the delayed therapeutic action of selective 5-HT reuptake inhibitors.17,19 Previous anecdotal reports on the absence of 5-HT1A expression in serotonergic neurons can be found in the literature, but most studies lack convincing proof and/or thorough quantification. To address this question, we used a combination of histochemistry techniques, and molecular detection with single-cell resolution.
ePet1-Cre mice were crossed with a reporter mouse line, RCE:loxP, previously shown to drive a strong expression of eGFP protein in serotonergic cells.20 In these mice, all the GFP-expressing neurons contained 5-HT. We used this model to evaluate the expression of 5-HT1A receptors in serotonergic cells. Brain sections including the raphe were subjected to combined GFP-immunohistochemistry and 5-HT1A in situ hybridization detection (see Methods). Typical distribution of 5-HT-eGFP expressing cells was found across the different raphe subnuclei (Figures 1 and 4J). As expected, the majority of eGFP+ neurons also expressed 5-HT1A receptors (Figures 1 and 2). However, even at low magnifications, a group of eGFP+ neurons that did not express 5-HT1A mRNA was detectable (Figure 1). High magnification images confirmed this observation (Figure 2). No other evident features could be observed in the 5-HT neurons lacking 5-HT1A receptor mRNA. The eGFP-positive/5-HT1A-negative cells were neighbored by 5-HT1A-positive cells (Figure 2). Because low mRNA expression could escape HIS detection protocols, we further assessed a genetic labeling of the 5-HT1A-expressing cells, using the 5-HT1A-iCre/R26R mice.21 In these mice, all the cells that have an active promoter of the 5-HT1A receptor gene drive an enhanced expression of Cre recombinase (iCre22) that mediate the specific expression of β-galactosidase protein, and its activity is then easily visualized with a highly sensitive X-Gal staining. This genetic reporter method generally yields a much more sensitive readout for gene expression than HIS, because of the stability of B-gal and the sensitivity of the histochemical detection.
Figure 1.
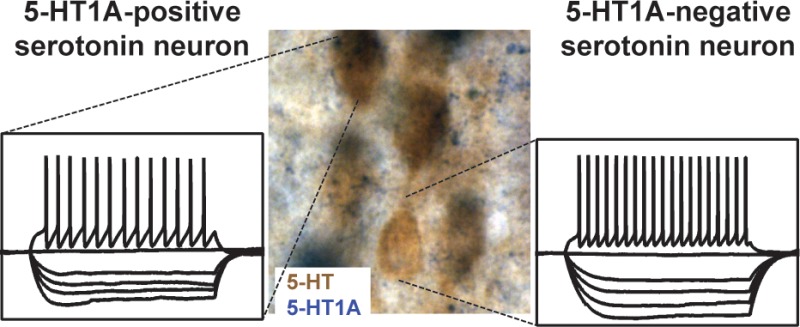
Colocalization of 5-HT neurons and 5-HT1A receptors in the mouse raphe. 5-HT1A mRNA expression (blue) and eGFP expression (brown) were detected using combined in situ hybridization and immunohistochemistry in ePet1-eGFP mice. Images depict the presence of 5-HT-positive neurons that do not express the 5-HT1A receptor (white arrows). Image magnification is 10× (A, scale bar 200 μm), 20× (B, scale bar 150 μm), and 40× (A, scale bar 50 μm).
Figure 4.
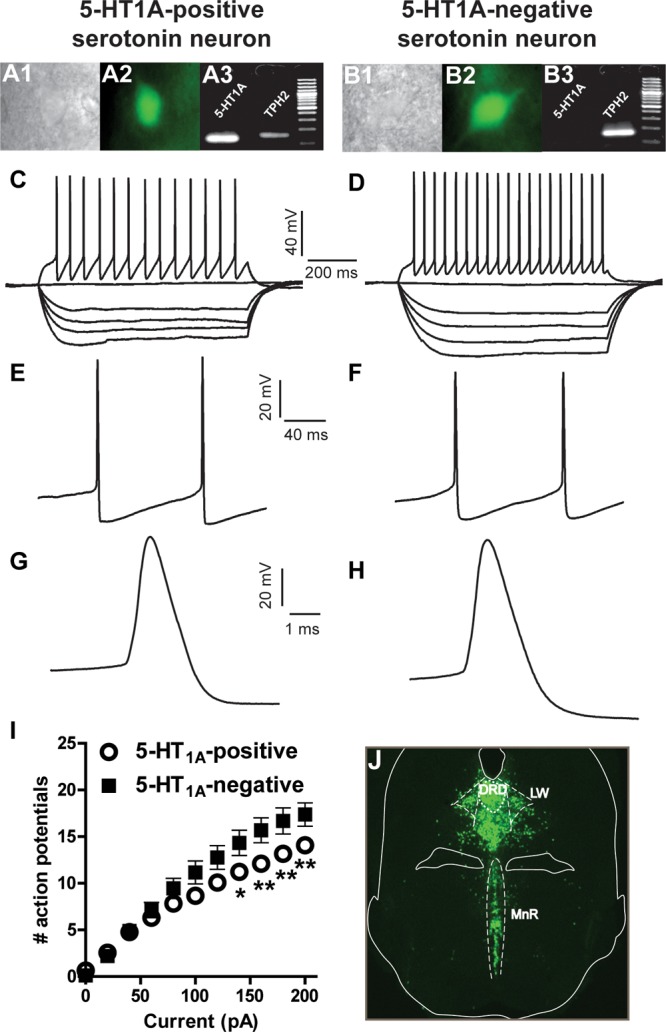
Intrinsic membrane properties of 5-HT1A-positve and -negative serotonergic neurons. Individual eGFP-tagged 5-HT neurons visualized in the electrophysiology rig under infrared interference contrast (A1 and B1) and epifluorescence (A2 and B2). Single-cell PCR was conducted to assess the expression of TPH2 and 5-HT1A mRNA. Agarose gels showing the presence of PCR products and DNA ladder for the two neuronal types (A3 and B3). Increasing current injections were applied to the cells to evaluate passive and active membrane properties. (C and D) Representative voltage traces after injection of −100, −80, −60, −40, 0, and +200 pA. (E and F) Representative voltage traces depicting two action potentials after injection of +100 pA; no significant differences were found in the amplitude and duration of afterhyperpolarization potentials. (G and H) Representative voltage traces depicting the shape of an action potential in a 5-HT1A-positive and -negative neuron, respectively. No significant differences were found in the width or amplitude of action potentials in these two neuronal populations. (I) Number of spikes elicited at each step of current injection. Serotonergic neurons that do not express the 5-HT1A autoreceptor discharge significantly higher number of spikes at current injections of 140, 160, 180, and 200 pA. (J) Fluorescent microscopy image showing the distribution of eGFP-tagged neurons in ePet1-eGFP mice. *P < 0.05, **P < 0.01, after two-way ANOVA and Bonferroni’s post hoc tests.
Figure 2.
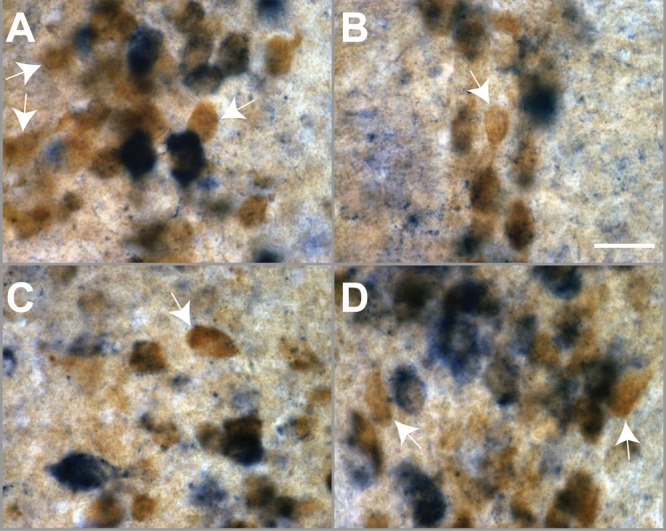
High magnification images depicting the existence of 5-HT1A-negative serotonergic neurons (white arrows). 5-HT1A mRNA expression (blue) and eGFP expression (brown) were detected using combined in situ hybridization and immunohistochemistry in ePet1-eGFP mice. Images were taken at 60× magnification (scale bar 25 μm).
A careful analysis of X-Gal/5-HT double-staining in the dorsal raphe nucleus of 5-HT1A-iCre/R26R mice revealed a large number of neurons positive for both markers, confirming that a large majority of 5-HT neurons expresses the 5-HT1A receptor (Figure 3). However, again, we found a number of 5-HT-IR neurons with no X-gal expression (Figure 3), adding supplementary proof that a population of 5-HT neurons did not activate the 5-HT1A promoter sequence during their life span. Previous studies aimed at mapping the distribution of 5-HT1A receptors in the brain have yielded some controversy on whether all serotonergic neurons express this receptor.11,16,23,24 For example, using a commercially available antibody, Kirby and collaborators11 described the presence of histochemically identified 5-HT neurons that do not express 5-HT1A receptors. The authors reported that this cell group was as abundant as 40% of all the serotonergic cells in some regions of the raphe, with a considerable variability of occurrence throughout the rostro-caudal extend of the dorsal raphe.11
Figure 3.
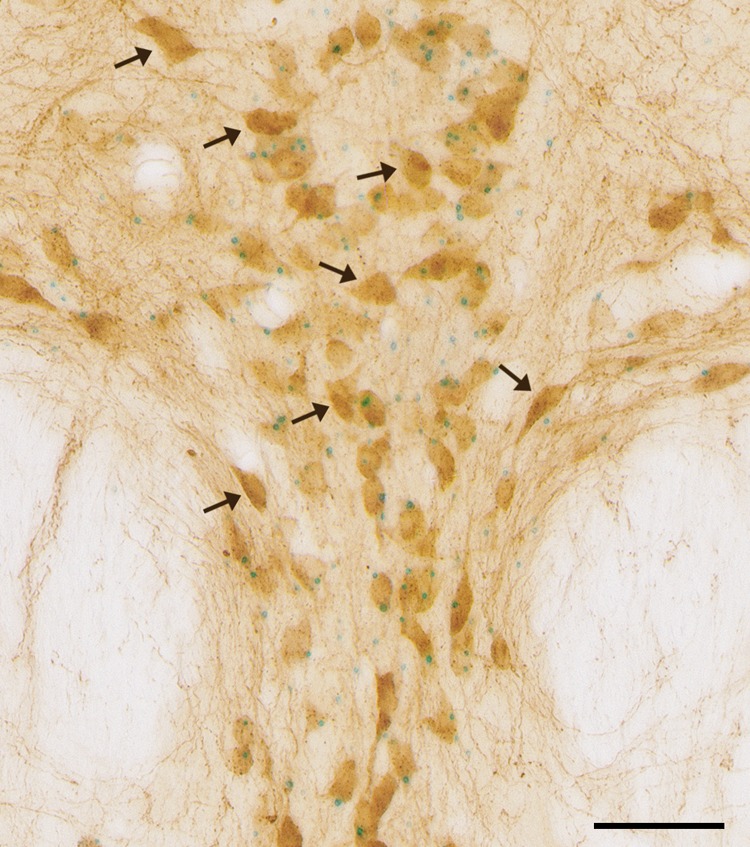
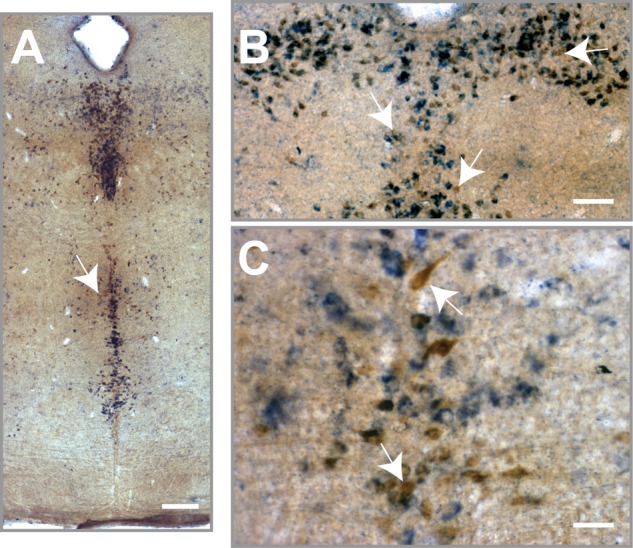
High magnification image depicting the existence of 5-HT1A-negative neurons in the dorsal raphe of 5-HT1A-iCre/R26R mice. 5-HT1A receptor mRNA expression (blue dots) and 5-HT-IR (brown) were detected using combined X-Gal staining and immunohistochemistry in 5-HT1A-iCre/R26R mice. The black arrows points to 5-HT-IR single-labeled neurons. Images were taken at 40× magnification (scale bar 50 μm).
In order to corroborate the histochemical results obtained, we prepared acute slices from ePet1-eGFP mice and conducted patch-clamp recordings followed by single-cell PCR, to detect the presence of tryptophan hydroxylase (TPH2) and 5-HT1A mRNAs. All the recorded neurons were positive for TPH2, confirming that eGFP expression driven by ePet1 occurs only in serotonergic neurons. The recordings were conducted in three different subfields of the raphe, namely, the dorsal raphe dorsal, dorsal raphe lateral (lateral wings), and the median raphe. From 134 recorded 5-HT cells, we found a total of 22 cells that did not express 5-HT1A mRNA (16%), as revealed by detection on agarose gel electrophoresis. The 5-HT1A-negative cells were evenly distributed across the raphe subfields (6 dorsal raphe dorsal, 8 lateral wings, 8 in median raphe). These data thus confirm that a subpopulation of serotonergic neurons do not express 5-HT1A mRNA. We then compared the electrophysiological profile of 5-HT1A-positive and -negative serotonergic neurons. We performed a fast characterization in current-clamp mode and determined a series of electrical parameters (Table 1). We did not find any significant differences in passive membrane properties such as resting potential, input resistance, membrane time constant, and capacitance (Table 1). The amount of current injected needed to elicit an action potential, that is, rheobase, was also similar in both cell populations, as it was the spike threshold (Table 1). We observed no significant differences in the width and amplitude of action potential, or in the duration and amplitude of the afterhyperpolarization potential (Table 1, Figure 4). Depolarizing current steps were injected to the cells until a maximum of 200 pA, and the number of spikes discharged at each step was scored. This parameter was not different for the two cell populations in the early current steps, but as more current was injected serotonergic cells that do not express the 5-HT1A receptor yielded more action potentials than those cells expressing it. This effect reached statistical significance at current injections of 140, 160, 180, and 200 pA (Figure 4I).
Table 1. Comparison of Electrophysiological Properties of Serotonin Neuronsa.
| 5-HT1A-positive
serotonin neurons (n = 112) |
5-HT1A-negative
serotonin neurons (n = 22) |
||||
|---|---|---|---|---|---|
| parameter | mean | SEM | mean | SEM | t test comparison (P values) |
| membrane potential (mV) | –58.79 | 0.41 | –60.73 | 1.1 | 0.1707 |
| input resistance (mohm) | 1023.10 | 40.58 | 910.96 | 61.04 | 0.1291 |
| time constant (ms) | 83.06 | 3.86 | 78.35 | 6.61 | 0.5408 |
| capacitance (pF) | 84.17 | 3.40 | 84.43 | 4.39 | 0.9629 |
| rheobase (pA) | 31.15 | 2.44 | 37.33 | 7.83 | 0.4560 |
| action potential threshold (mV) | –39.88 | 0.38 | –40.08 | 0.83 | 0.8304 |
| action potential amplitude (mV) | 75.67 | 0.85 | 74.03 | 2.14 | 0.4802 |
| action potential duration (ms) | 1.55 | 0.04 | 1.77 | 0.12 | 0.0922 |
| afterhyperpolarization potential amplitude (mV) | –17.77 | 0.33 | –16.71 | 0.77 | 0.2111 |
| Afterhyperpolarization potential time to peak (ms) | 25.93 | 1.03 | 30.76 | 2.94 | 0.1304 |
Electrophysiological parameters were measured as described (see Methods). P values reported after Student’s t test with Welch’s correction.
The convergence of results obtained by histological and molecular approaches strongly support the hypothesis that not all serotonergic neurons of the raphe express 5-HT1A receptor mRNA, a finding that is in accordance with reports showing that some 5-HT neurons do not express 5-HT1A protein.11,16 However, since the proof of an absence of expression is always subject to debate, we relied on the different methodologies utilized to increase the certitude of the estimate yielded by each technique alone. The occurrence of false negatives cannot be ruled out, and it is possible that in some cells the levels of 5-HT1A transcripts are extremely low and escape detection. We have established a detection limit of 0.12 pg of cRNA using our multiplex nested PCR methods; however, it is difficult to establish how much mRNA is necessary to yield a signal detectable by in situ hybridization. It is then possible that, in some cells, 5-HT1A protein turnover is rather low and therefore the synthesis of mRNA is minimal. However, this interpretation is not supported by our findings using the β-galactosidase reporter, since in this construct all cells which expressed even low levels of the 5-HT1A receptor gene will have a stable and highly active expression of iCre22 driving β-galactosidase activity. This is consistent with immunohistochemical studies that showed a significant percentage of 5-HT cells that do not present immunoreactivity to a 5-HT1A antibody, indicating lack of protein expression.11,16 In addition, electrophysiological studies where hyperpolarizing responses elicited by superfusion of a 5-HT1A agonist were assessed, found a considerable number of nonresponding identified serotonergic cells.9,10,12,25 We hypothesized that while following the classic dogma to identify 5-HT neurons, many investigators have discarded data involving 5-HT1A-negative serotonergic neurons.
The subpopulation of 5-HT1A-negative serotonergic cells described in this study did not present significant differences in cellular features such as resting potential, resistance, and capacitance. Also, the excitability of these cells, as measured by rheobase and threshold values, was not different. However, when a series of increasing current steps where injected, 5-HT1A-negative neurons fired more action potentials at large current injections. One possible explanation is that the depolarization induced by current injection caused local release of 5-HT which activates somatodendritic 5-HT1A, causing autoinhibition. In neurons where 5-HT1A receptors are not expressed, the local release of 5-HT would not induce autoinhibition through the activation of these receptors. However, the difference in spike frequency between both populations of 5-HT neurons is not large, which may be related with technical limitation to evaluate 5-HT autoinhibition and/or the occurrence of compensatory mechanisms involving other inhibitory autoreceptors such as 5-HT1B/1D.26,27
In vivo electrophysiological recordings in the dorsal raphe nucleus have shown that 5-HT neurons respond rapidly and transiently to specific aversive and rewarding stimuli, indicating that 5-HT release can modulate even the earliest stages of sensory processing. One remarkable observation from these studies is that neighboring 5-HT neurons have a high degree of diversity to respond to discrete stimuli, suggesting that 5-HT neurons are not a homogeneous population.28,29 Our findings support this hypothesis, and confirm the existence of a subpopulation of serotonergic neurons that do not express the 5-HT1A receptor. It is difficult to speculate whether these cells correspond to any of the firing behaviors described for neurons recorded in vivo, or whether they could respond differently to external stimuli. However, our results showed that given the same excitatory input, cells that do not express 5-HT1A reach higher discharge frequencies. It is then possible, that these cells could correspond to the subpopulation of fast-firing,7,30 or even bursting 5-HT neurons.28,31
In conclusion, we showed here that, despite the classic dogma, not all 5-HT neurons in the mouse raphe express the 5-HT1A autoreceptor mRNA, and that 5-HT cells lacking 5-HT1A mRNA are prone to fire at higher discharge frequencies. Our results support the hypothesis that the 5-HT system is composed of subpopulations of functionally relevant serotonergic neurons.32 The ongoing investigations unraveling the molecular heterogeneity of serotonergic neurons is an exciting development in the characterization of serotonergic neurotransmission and offers the hope of more precisely understanding the pathophysiology of disorders such as major depression and schizophrenia. Indeed, it may be speculated that patients suffering from these disorders may have imbalances in the proportions of neurons that do/do not express receptors such as 5-HT1A.33 Is it possible that 5-HT1A receptors expression levels fluctuate during development, and external challenges such as stress or exposure to pharmacological agents, such as SSRIs, can have an influence on this process? Despite these speculations, more work will be needed to better understand the correlation between the molecular heterogeneity and the functional organization of the serotonergic system.
Methods
All experiments performed in mice were in compliance with the standard ethical guidelines (European Community Guidelines and French Agriculture and Forestry Ministry Guidelines for Handling Animals—decree 87849). ePet1-EGFP mice were generated by crossing an ePet1-Cre mouse line34 with a RCE:loxP indicator strain.35 Brains obtained from ePet1-EGFP mice were examined for EGFP expression and colocalization with 5-HT. Strong EGFP expression was detected across all subfield of the raphe nuclei, with an approximate 95% colocalization with 5-HT immunoreactivity.20 Double transgenic mice (5-HT1A-iCre/R26R; C57B/L6 background) were obtained by crossing 5-HT1A-iCre transgenic mice21 and R26R reporter mice harboring a LacZ transgene.36 Brains obtained from 5-HT1A-iCre/R26R mice were examined for β-galactosidase expression, monitored by X-Gal staining, and colocalization with 5-HT. X-Gal staining in different brain regions showed an expression pattern that overlaps with that of 5-HT1A receptor.21 To identify 5-HT1A-iCre/R26R mice, PCR DNA amplifications were performed on genomic DNA using iCre-F (5′-CCAGCTCAACATGCTGCACA-3′) and iCre-R (5′-TTCTCCACACCAGCTGTGGA-3′) primers. The LacZ transgene in R26R mice was detected by PCR DNA amplification using LacZ-F (5′-GTCGTTTTACAACGTCGTGACT-3′) and LacZ-R (5′-GATGGGCGATCGTAACCGTGC-3′) primers.
In Situ Hybridization Combined with Immunohistochemistry in ePet1-eGFP Mice
Adult ePet1-EGFP mice were anesthetized and perfused transcardially with 4% paraformaldehyde in 0.12 M phosphate buffer (PB), pH 7.4. Brains were removed from the skull and postfixed overnight in the same fixative. After cryoprotection (PB with 30% sucrose for 48 h), serial 50-μm-thick coronal sections were made on a freezing microtome and collected in PB.
Specific antisense RNA probes for 5-HT1A (IMAGE:8861702) gene were used for free-floating in situ hybridization. DIG-probes were synthesized with a labeling kit according to the manufacturer’s instructions (Roche, France). Frozen sections were rinsed 3 × 5 min in PBS + 0.1% Tween-20 (PBTw), dehydrated in graded ethanol/PBTw (25, 50, 75, and 100%, 5 min), bleached with 1% H2O2 in 100% ethanol (1 h), and rehydrated through a reverse ethanol/PBTw washing series (100, 75, 50, 25%). After two washes in PBTw (5 min), sections were treated with proteinase K (10 μg/mL) for 1 min, which was then blocked in PBTw + glycine (2 mg/mL). Sections were then rinsed in PBTw and postfixed in a mixture of 4% paraformaldehyde, 0.2% glutaraldehyde, and 0.1% Tween (20 min), and then rinsed in PBTw. A prehybridization step is performed at 70 °C (1 h) in the hybridization buffer (50% formamide, 5× SSC, 50 μg/mL yeast RNA, 50 μg/mL heparin, 0.1% Tween-20), before adding the DIG-labeled probes diluted 1/100 (overnight incubation). The next day, sections were sequentially washed in solution 1 (50% formamide, 5× SSC, 0.1% Tween-20 for 1.5 h at 70 °C), then in solution 2 (0.5 M NaCl, 10 mM Tris pH 7.6, 0.1% Tween-20 for 1 h at room temperature), and finally in solution 3 (50% formamide, 2× SSC, 0.1% Tween-20 for 1.5 h at 70 °C).
For immunological detection of DIG-labeled hybrids, sections were first blocked in TBST solution (1.5 mM NaCl, 0.03 mM KCl, 0.025 M Tris-HCl, pH 7.6, 0.1% Tween-20) with 10% normal goat serum for 2 h. Sections were then incubated overnight at 4 °C in a solution containing sheep anti-DIG-alkaline phosphatase-conjugated Fab fragments (Roche, France) diluted 1/2000 in TBST buffer plus 1% NGS and levamisole (5 mg/10 mL). The third day sections were washed 2 × 15 min in TBST buffer and 30 min in NTMT buffer (100 mM NaCl, 100 mM Tris-HCl, pH 9.5, 50 mM MgCl2, 0.1% Tween-20). The alkaline phosphatase chromogen reaction was performed in NTMT buffer containing 100 mg/mL nitroblue tetrazolium (Roche, France) and 50 mg/mL 5-bromo-4-chloro-3-indolyl phosphate (Roche, France) at room temperature for 5–6 h and stopped with Tris-EDTA buffer.
The sections were rinsed several times in PBS and PGTx (PBS with 0.2% gelatin and 0.25% Triton X-100) and then processed for immunohistochemistry. The 5HT 1A ISH detection was combined with anti-GFP-immunohistochemistry (rabbit polyclonal anti-GFP antibodies, 1/5000, Molecular Probes). The reaction was revealed in a solution containing 1% diaminobenzidine, 0.005% H2O2 in 0.1 M Tris buffer, pH 7.6. Sections were mounted on superfrost slides, dried, dehydrated in graded ethanol solutions, cleared in xylene, and coverslipped in Eukitt.
Combined Immunohistochemistry with X-Gal Staining in 5-HT1A-iCre/R26R Mice
5-HT1A-iCre/R26R mice were deeply anesthetized by an intraperitoneal injection of sodium pentobarbital (73 mg/kg) and perfused transcardially with 100 mL of 4% paraformaldehyde (PF; Sigma, St. Louis, MO) in phosphate-buffered saline (PBS, pH 7.4; Invitrogen, Carlsbad, CA). The brains were removed and immersed in the same fixative for 17 h at 4 °C. Then, coronal ponto-mesencephalic sections were cut at 30 μm thickness using a Vibratome (VT 1000, Leica Microsystèmes, Rueil-Malmaison, France). Brain sections of 5-HT1A-iCre/R26R mice were then used to detect both β-galactosidase expression and 5-HT. For β-galactosidase staining, sections were incubated overnight in X-Gal solution (5 mM potassium ferricyanide, 5 mM ferrocyanide, 2 mM MgCl2, 0.02% NP-40, 0.01% sodium deoxycholate and 1 mg/mL X-Gal) at 37 °C. β-Galactosidase staining was visualized as blue dot staining in the cytoplasm of labeled cells. For the second step (5-HT immunolabeling), nonspecific antigens were blocked by incubating sections in PBS containing 4% bovine serum albumin (BSA) and 0.1% Triton X-100 for 1 h at RT. Sections were then incubated overnight at 4 °C with a rat monoclonal anti-5-HT antibody (1:50 dilution; Sera Lab, West Sussex, U.K.). After several rinses in PBS supplemented with 0.1% Triton X-100, sections were incubated for 2 h in the blocking buffer with biotinylated antirat secondary antibodies (1:200 dilution with ABC Vectastain Elite kit, Vector, Burlingame, CA). Then further rinses were followed by incubation in avidin–biotin–horseradish-peroxidase solution (ABC Vectastain Elite kit, Vector) for 1 h. For staining, sections were treated with 0.04% diaminobenzidine (DAB, Sigma) and supplemented with increasing concentrations of H2O2 (from 0.00015% to 0.0048%; Sigma), yielding a brown cytoplasmic product in 5-HT-immunoreactive (IR) neurons. Stained sections were finally mounted on gelatin-coated slides, dehydrated, and coverslipped with Eukitt mounting medium (Kindler).
Image Acquisition
Bright field histology images were captured with a Cool Snap FX camera fitted to an Olympus Provis microscope using 10×/0.40 and 63×/0.103 objectives (magnification/numerical aperture). Stitching of multiple micrographs into a single frame was done with Metamorph software. The same settings of brightness/contrast were used within a given experiment.
For double X-Gal/5-HT staining, photomicrographs under brightfield illumination were made by using a TRI CCD camera (JVC KY-F50; resolution 576 × 768 pixels), which sent a tricolor red, green, and blue (RGB) output to computer. Images at different focal planes were captured by using a 40× objective and digitized by using a 3 × 8-bit color-scale Openlab software (Improvision, Coventry, U.K.). An operator allowed the combination, pixel-by-pixel, of images in different focal planes. These operations resulted in the production of one image by incorporating the darker value of the corresponding pixel in each focal plane for each red, green, and blue color plane. Images were exported to Adobe Photoshop CS2 (version 9.0; Adobe Systems, Mountain View, CA) in order to mount adjacent digitized images as a final large-field high-resolution image. Then brightness, contrast, and image scale were adjusted. Finally, additional indications and/or anatomical landmarks were incorporated.
Electrophysiological Recordings and Single-Cell PCR Amplification
ePet1-EGFP mice aged 3–4 weeks old were anesthetized with ketamine/xylaxine and decapitated, and the brain was rapidly dissected out. Coronal slices (250 μm) containing DR and MnR were prepared in a vibratome and placed in aCSF containing (in mM) 11 glucose, 2.5 KCl, 26.2 NaHOC3, 1 NaH2PO4, 124 NaCl, 2 CaCl2, and 2 MgCl2 bubbled with a mixture of 95% O2/5% CO2. After a 1 h recovery period, individual slices were placed in an electrophysiology chamber continuously perfused with aCSF maintained at 31 °C. Neurons were visualized by combined epifluorescent and infrared/differential interference contrast visualization using an Olympus BX51WI upright microscope holding 5× and 40× objectives. The methodology involving harvesting of cytoplasmic content and subsequent single-cell PCR amplification has been described previously.37 Briefly, borosilicate glass pipettes (3–5 MΩ) were made in a HEKA PIP5 puller and filled with 8 μL of autoclaved RT-PCR internal solution (in mM): 144 K-gluconate, 3 MgCl2, 0.5 EGTA, 10 HEPES, pH 7.2 (285/295 mOsm). Single EGFP-positive neurons were approached with a pipet, and whole-cell recordings were obtained using a Multiclamp 700B instrument (Molecular Devices, Sunnyvale, CA). Signals were collected and stored using a Digidata 1440A converter and pCLAMP 10.2 software (Molecular Devices, CA). After the recording, the cytoplasmic content of the cell was harvested by applying gentle negative pressure to the pipet. Cell content was expelled into a tube where a reverse transcription reaction was performed in a final volume of 10 μL. cDNA sequences were thereafter amplified by conducting a multiplex nested PCR, designed to simultaneously detect mRNAs encoding TPH2 and 5-HT1A. Initially, genes were simultaneously amplified in a single tube using 10 μL of cDNA, 200 nM of each primer, and 2.5 U of Taq polymerase in a final volume of 100 μL. PCR reaction was carried out using 6 min hot start at 94 °C, followed by a 21-cycles program (94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s). Subsequently, 2 μL of the amplified cDNA was used as the template for the second amplification step. Here, each gene was individually amplified in a separate tube submitted to a 35-cycles PCR program using the same conditions as mentioned above, in a final volume of 100 μL. The products of the second PCR were analyzed by electrophoresis in 2.5% agarose gels using SYBR safe gel stain (Invitrogen, Oregon). The size of the PCR-generated fragments was as predicted by the mRNA sequences. The primer pairs utilized were as follows: TPH2 (NM_173391.2) external sense 5′-CCCGGAACCAGATACATGCC-3′, antisense 5′-TTTCACACACGCCTTGTCGG-3′; TPH2 internal sense 5′-TGCGGATCCCAAGTTTGCTC-3′, antisense 5′-TAACCCTGCTCCATACGCCC-3′. 5-HT1A (NM_008308.4) external sense 5′-CAGCCAGGTAGTGGGGACTG-3′, antisense 5′-GTCTTCCTCTCACGGGCCAA-3′; 5-HT1A internal sense 5′-CGGTGAGACAGGGTGAGGAC-3′, antisense 5′-GCGGGGACATAGGAGGTAGC-3′.
Electrophysiological Analysis
In order to describe the electrophysiological features of 5-HT neurons, the following electrophysiological parameters were determined for each cell from recordings made in current-clamp mode. Resting membrane potential defined as the voltage at which I = 0; input resistance (Rm), and membrane time constant (τm) were measured on responses to a −20 pA hyperpolarizing current pulse. Membrane capacitance (Cm) was calculated according to the equation Cm = τm/Rm. Rheobase was defined as the first current pulse to elicit an action potential. Action potential amplitude was measured from threshold to the positive peak, and the duration was measured at half amplitude. The amplitude and time to negative peak of the first spike’s afterhyperpolarization potential were also measured. Finally, at each step of current injection, the number of spikes was scored. Data are expressed as mean ± SEM, and statistical analysis was performed using GraphPad Prism v5 software (GraphPad, San Diego, CA).
Acknowledgments
We are indebted to Jean-Christophe Poncer and Carolina Cabezas for use of electrophysiology equipment and expertise. We thank Gord Fishell (NYU Neuroscience Institute) for the gift of RCE:LoxP mice.
This research was funded by the European Commission (FP7-health-2007-A-201714), the Fondation Jerome Lejeune and the Agence Nationale pour la Recherche (ANR605-neur-046). The Fondation pour la Recherche Medicale funds S.P.F.
The authors declare no competing financial interest.
References
- Ressler K. J.; Nemeroff C. B. (2000) Role of serotonergic and noradrenergic systems in the pathophysiology of depression and anxiety disorders. Depression Anxiety 12(Suppl 1), 2–19. [DOI] [PubMed] [Google Scholar]
- Graeff F. G.; Zangrossi H. Jr (2010) The dual role of serotonin in defense and the mode of action of antidepressants on generalized anxiety and panic disorders. Cent. Nerv. Syst. Agents Med. Chem. 10, 207–217. [DOI] [PubMed] [Google Scholar]
- Hale M. W.; Lowry C. A. (2011) Functional topography of midbrain and pontine serotonergic systems: implications for synaptic regulation of serotonergic circuits. Psychopharmacology (Berlin, Ger.) 213, 243–264. [DOI] [PubMed] [Google Scholar]
- Aghajanian G. K.; Vandermaelen C. P. (1982) Intracellular recordings from serotonergic dorsal raphe neurons: pacemaker potentials and the effect of LSD. Brain Res. 238, 463–469. [DOI] [PubMed] [Google Scholar]
- Aghajanian G. K.; Vandermaelen C. P. (1982) Intracellular identification of central noradrenergic and serotonergic neurons by a new double labeling procedure. J. Neurosci. 2, 1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermaelen C. P.; Aghajanian G. K. (1983) Electrophysiological and pharmacological characterization of serotonergic dorsal raphe neurons recorded extracellularly and intracellularly in rat brain slices. Brain Res. 289, 109–119. [DOI] [PubMed] [Google Scholar]
- Kocsis B.; Varga V.; Dahan L.; Sik A. (2006) Serotonergic neuron diversity: identification of raphe neurons with discharges time-locked to the hippocampal theta rhythm. Proc. Natl. Acad. Sci. U.S.A. 103, 1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweimer J. V.; Mallet N.; Sharp T.; Ungless M. A. (2011) Spike-timing relationship of neurochemically-identified dorsal raphe neurons during cortical slow oscillations. Neuroscience 196, 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S. G.; Pan Y.-Z.; Akanwa A. C.; Kirby L. G. (2004) Median and dorsal raphe neurons are not electrophysiologically identical. J. Neurophysiol. 91, 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calizo L. H.; Akanwa A.; Ma X.; Pan Y.-Z.; Lemos J. C.; Craige C.; Heemstra L. A.; Beck S. G. (2011) Raphe serotonin neurons are not homogenous: electrophysiological, morphological and neurochemical evidence. Neuropharmacology 61, 524–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby L. G.; Pernar L.; Valentino R. J.; Beck S. G. (2003) Distinguishing characteristics of serotonin and non-serotonin-containing cells in the dorsal raphe nucleus: electrophysiological and immunohistochemical studies. Neuroscience 116, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. R.; Soffin E. M.; Roberts C.; Kew J. N. C.; de la Flor R. M.; Dawson L. A.; Fry V. A.; Coggon S. A.; Faedo S.; Hayes P. D.; Corbett D. F.; Davies C. H.; Hagan J. J. (2006) SB-699551-A (3-cyclopentyl-N-[2-(dimethylamino)ethyl]-N-[(4′-{[(2-phenylethyl)amino]methyl}-4-biphenylyl)methyl]propanamide dihydrochloride), a novel 5-ht5A receptor-selective antagonist, enhances 5-HT neuronal function: Evidence for an autoreceptor role for the 5-ht5A receptor in guinea pig brain. Neuropharmacology 51, 566–577. [DOI] [PubMed] [Google Scholar]
- Barnes N. M.; Sharp T. (1999) A review of central 5-HT receptors and their function. Neuropharmacology 38, 1083–1152. [DOI] [PubMed] [Google Scholar]
- Jacobs B. L.; Azmitia E. C. (1992) Structure and function of the brain serotonin system. Physiol. Rev. 72, 165–229. [DOI] [PubMed] [Google Scholar]
- Day H. E. W.; Greenwood B. N.; Hammack S. E.; Watkins L. R.; Fleshner M.; Maier S. F.; Campeau S. (2004) Differential expression of 5HT-1A, alpha 1b adrenergic, CRF-R1, and CRF-R2 receptor mRNA in serotonergic, gamma-aminobutyric acidergic, and catecholaminergic cells of the rat dorsal raphe nucleus. J. Comp. Neurol. 474, 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo C.; Cholley B.; El Mestikawy S.; Gozlan H.; Hamon M. (1990) Direct Immunohistochemical Evidence of the Existence of 5-HT1A Autoreceptors on Serotoninergic Neurons in the Midbrain Raphe Nuclei. Eur. J. Neurosci. 2, 1144–1154. [DOI] [PubMed] [Google Scholar]
- Hensler J. G. (2003) Regulation of 5-HT1A receptor function in brain following agonist or antidepressant administration. Life Sci. 72, 1665–1682. [DOI] [PubMed] [Google Scholar]
- Lanfumey L.; Hamon M. (2000) Central 5-HT(1A) receptors: regional distribution and functional characteristics. Nucl. Med. Biol. 27, 429–435. [DOI] [PubMed] [Google Scholar]
- Hjorth S.; Bengtsson H. J.; Kullberg A.; Carlzon D.; Peilot H.; Auerbach S. B. (2000) Serotonin autoreceptor function and antidepressant drug action. J. Psychopharmacol. (Oxford,U.K.) 14, 177–185. [DOI] [PubMed] [Google Scholar]
- Kiyasova V.; Fernandez S. P.; Laine J.; Stankovski L.; Muzerelle A.; Doly S.; Gaspar P. (2011) A genetically defined morphologically and functionally unique subset of 5-HT neurons in the mouse raphe nuclei. J. Neurosci. 31, 2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly I.; Fabre V.; Vyas S.; Milet A.; Rouzeau J.-D.; Hamon M.; Lazar M.; Tronche F. (2007) 5-HT1A-iCre, a new transgenic mouse line for genetic analyses of the serotonergic pathway. Mol. Cell. Neurosci. 36, 27–35. [DOI] [PubMed] [Google Scholar]
- Shimshek D. R.; Kim J.; Hübner M. R.; Spergel D. J.; Buchholz F.; Casanova E.; Stewart A. F.; Seeburg P. H.; Sprengel R. (2002) Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26. [DOI] [PubMed] [Google Scholar]
- Bonnavion P.; Bernard J.-F.; Hamon M.; Adrien J.; Fabre V. (2010) Heterogeneous distribution of the serotonin 5-HT(1A) receptor mRNA in chemically identified neurons of the mouse rostral brainstem: Implications for the role of serotonin in the regulation of wakefulness and REM sleep. J. Comp. Neurol. 518, 2744–2770. [DOI] [PubMed] [Google Scholar]
- Pompeiano M.; Palacios J. M.; Mengod G. (1992) Distribution and cellular localization of mRNA coding for 5-HT1A receptor in the rat brain: correlation with receptor binding. J. Neurosci. 12, 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss D. A.; Li Y. W.; Talley E. M. (1997) Effects of serotonin on caudal raphe neurons: activation of an inwardly rectifying potassium conductance. J. Neurophysiol. 77, 1349–1361. [DOI] [PubMed] [Google Scholar]
- Hjorth S.; Tao R. (1991) The putative 5-HT1B receptor agonist CP-93,129 suppresses rat hippocampal 5-HT release in vivo: comparison with RU 24969. Eur. J. Pharmacol. 209, 249–252. [DOI] [PubMed] [Google Scholar]
- Piñeyro G.; de Montigny C.; Blier P. (1995) 5-HT1D receptors regulate 5-HT release in the rat raphe nuclei. In vivo voltammetry and in vitro superfusion studies. Neuropsychopharmacology 13, 249–260. [DOI] [PubMed] [Google Scholar]
- Schweimer J. V.; Ungless M. A. (2010) Phasic responses in dorsal raphe serotonin neurons to noxious stimuli. Neuroscience 171, 1209–1215. [DOI] [PubMed] [Google Scholar]
- Miyazaki K.; Miyazaki K. W.; Doya K. (2011) Activation of dorsal raphe serotonin neurons underlies waiting for delayed rewards. J. Neurosci. 31, 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers K. A.; Sharp T. (2003) Neurochemical and anatomical identification of fast- and slow-firing neurones in the rat dorsal raphe nucleus using juxtacellular labelling methods in vivo. Neuroscience 122, 193–204. [DOI] [PubMed] [Google Scholar]
- Hajós M.; Gartside S. E.; Villa A. E.; Sharp T. (1995) Evidence for a repetitive (burst) firing pattern in a sub-population of 5-hydroxytryptamine neurons in the dorsal and median raphe nuclei of the rat. Neuroscience 69, 189–197. [DOI] [PubMed] [Google Scholar]
- Gaspar P.; Lillesaar C. (2012) Probing the diversity of serotonin neurons. Philos. Trans. R. Soc. London, Ser. B: Biol. Sci. 367, 2382–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonde S.; Turecki G.; Bakish D.; Du L.; Hrdina P. D.; Bown C. D.; Sequeira A.; Kushwaha N.; Morris S. J.; Basak A.; Ou X.-M.; Albert P. R. (2003) Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J. Neurosci. 23, 8788–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. M.; Wylie C. J.; Lerch J. K.; Murphy R.; Lobur K.; Herlitze S.; Jiang W.; Conlon R. A.; Strowbridge B. W.; Deneris E. S. (2005) A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc. Natl. Acad. Sci. U.S.A. 102, 16472–16477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa V. H.; Miyoshi G.; Hjerling-Leffler J.; Karayannis T.; Fishell G. (2009) Characterization of Nkx6–2-derived neocortical interneuron lineages. Cereb. Cortex 19(Suppl 1), i1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Cauli B.; Porter J. T.; Tsuzuki K.; Lambolez B.; Rossier J.; Quenet B.; Audinat E. (2000) Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc. Natl. Acad. Sci. U.S.A. 97, 6144–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]


