Summary
Mammalian pallial (cortical and hippocampal) and striatal interneurons are both generated in the embryonic subpallium, including the medial ganglionic eminence (MGE). Herein we demonstrate that the Zfhx1b (Sip1, Zeb2) zinc finger homeobox gene is required in the MGE, directly downstream of Dlx1&2, to generate cortical interneurons that express Cxcr7, MafB and cMaf. In its absence, Nkx2-1 expression is not repressed, and cells that ordinarily would become cortical interneurons appear to transform towards a subtype of GABAeric striatal interneurons. These results show that Zfhx1b is required to generate cortical interneurons, and suggest a mechanism for the epilepsy observed in humans with Zfhx1b mutations (Mowat-Wilson syndrome).
Introduction
Cell type specification within the embryonic basal ganglia is regulated at multiple levels. Distinct subdivisions within this region generate distinct neurons. For instance, the lateral ganglionic eminence (LGE) generates striatal projection neurons whereas the medial ganglionic eminence (MGE) generates pallidal projection neurons. Domains within the MGE are biased towards generating different cell types; whereas the rostrodorsal MGE largely produces cortical and striatal interneurons, the caudoventral MGE largely produces pallidal projection neurons (Flandin et al., 2010; Nobrega-Pereira et al., 2010). Distinct MGE-derived cortical interneuron subtypes appear to be generated from the same progenitors, perhaps in a temporal sequence (Brown et al., 2011).
Cortical and striatal interneurons are both generated from the MGE (Marin et al., 2000). The Nkx2-1 homeobox transcription factor has a central role in specifying their identity. While Nkx2-1 is initially required for both of these cell types, Nkx2-1 expression is repressed soon after immature cortical interneurons tangentially migrate from the MGE, while it is maintained in striatal interneurons (Butt et al., 2008; Marin et al., 2000; Nóbrega-Pereira et al., 2008; Sussel et al., 1999).
Forced expression of Nkx2-1 in cortical interneurons changes their migration so that they settle in the striatum (Nóbrega-Pereira et al., 2008), providing additional evidence that repression of Nkx2-1 is a key step in generating cortical interneurons. How Nkx2-1 expression is repressed in these cells is unknown.
Herein, we provide evidence that the Zfhx1b (Sip1, Zeb2) zinc-finger homeobox transcription factor is required to repress Nkx2-1 in the generation of cortical interneurons. In its absence, we find a decrease in cortical interneurons concomitant with increased striatal nNOS/NPY/Sst GABAergic interneurons. We provide evidence that expression of the cMaf transcription factor is a highly specific marker of the cortical interneuron lineage, and discovered that its expression is lost in Zfhx1b mutants.
Previous analysis of Zfhx1b mouse mutants has shed light on its functions in the development of cortical projection neurons (Miquelajauregui et al., 2007; Seuntjens et al., 2009). In humans, mutations of Zfhx1b result in Mowat-Wilson syndrome, a developmental disorder characterized by mental retardation, epilepsy and defects of neural crest-derived tissues, including craniofacial and enteric nervous system (Mowat et al., 2003). Our results that demonstrate Zfhx1b is required to generate cortical interneurons suggest a mechanism for the epilepsy observed in Mowat-Wilson syndrome.
Results
Conditional Deletion of Zfhx1b in the VZ or the SVZ of the Subpallium Using Nkx2.1-Cre or DlxI12b-Cre
Zfhx1b prenatal expression has been noted in migrating cortical interneurons and the subpallial telencephalon (Batista-Brito et al., 2008; Seuntjens et al., 2003). We found that Zfhx1b RNA is expressed in E12.5 MGE-derived cells that are tangentially migrating through the LGE and into the cortex by performing fluorescent in situ hybridization (FISH) on a brain in which MGE-derived cells expressed EGFP (expressed due to Nkx2.1-Cre induced recombination of the CAG:CAT-EGFP Cre reporter allele) (Figures S1 A-A”).
To determine the role of Zfhx1b in the development of the basal ganglia, we used a conditional mutagenesis approach. Using an allele of Zfhx1b, in which exon 7 is floxed (Higashi et al., 2002), we removed Zfhx1b expression using two different Cre alleles. Deletion of exon 7 creates a frameshift mutation and premature truncation of the protein. Previous analysis failed to detect the truncated mutant protein in Zfhx1b mutant tissues, providing evidence that this is a null allele (Higashi et al., 2002).
To remove Zfhx1b in the early progenitors of the MGE, we used the Nkx2.1-Cre allele (Xu et al., 2008), which drives Cre expression in the ventricular zone (VZ) of the MGE beginning around E9.5 [later it also drives expression in the subventricular and mantle zones (SVZ and MZ)]. To differentiate between the role of Zfhx1b in the VZ and the SVZ/MZ, we used the DlxI1/2b-Cre allele (Potter et al., 2008), which drives Cre expression in the SVZ and MZ of the entire subpallium beginning around E10.5. To examine the pattern of recombination, we used an antisense riboprobe designed against Zfhx1b exon 7.
By E12.5, Cre activity from both the Nkx2.1-Cre and DlxI1/2b-Cre alleles removed Zfhx1b RNA expression in the expected patterns (Figures 1A-1C). As previously reported, the Nkx2-1 allele did not express Cre in the dorsal-most portion of the MGE, thus explaining the persistence of Zfhx1b in that location (Figure 1B). Of note, in the Nkx2.1-Cre; Zfhx1b conditional mutant brains, Zfhx1b RNA expression was not observed in the cells that appear to be migrating from the dorsal MGE into the mantle of the LGE, suggesting that the Zfhx1b+ cells in the mantle of the E12.5 LGE are likely to be MGE-derived cells (e.g. cortical and/or striatal interneurons) (X in Figure 1B). Also, note that DlxI1/2b-Cre leads to recombination in the SVZ and MZ of the LGE, MGE and CGE (white arrowhead, Figure 1C and data not shown).
Figure 1.
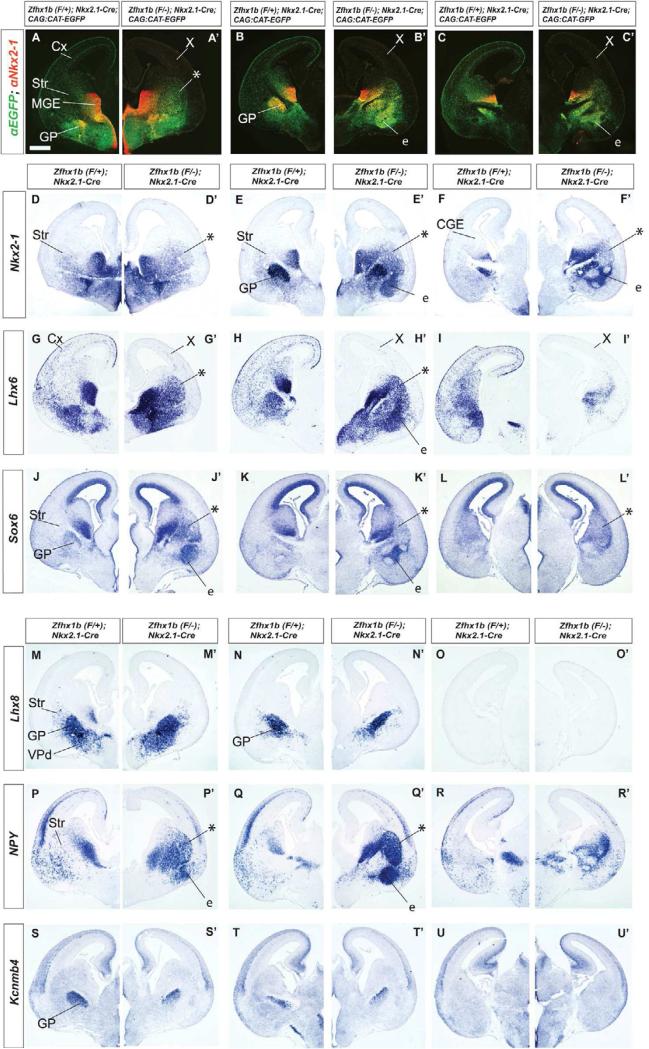
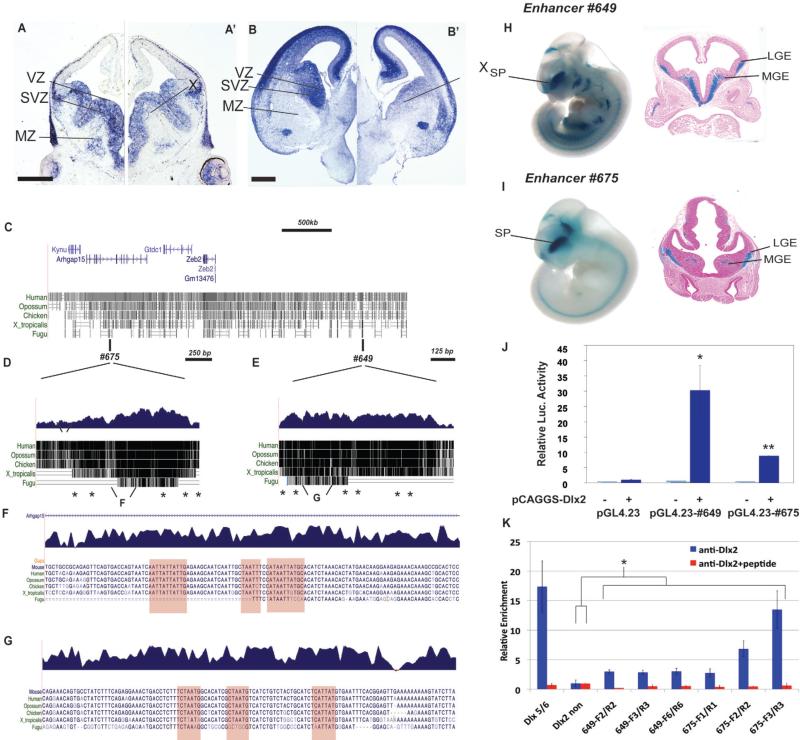
Zfhx1b expression in the MGE is required for interneuron migration at E12.5. (A-C) Zfhx1b RNA expression detected by in situ hybridization in control and conditional Zfhx1b mutant telencephalons. (B) Nkx2.1-Cre. (C) DlxI12b-Cre. Black arrowhead in B shows loss of Zfhx1b expression in the MGE VZ (except dorsal-most MGE). White arrowheads in B and C show loss of Zfhx1b expression in the SVZ of the MGE. X in B shows loss of Zfhx1b expression in the SVZ/MZ of the LGE. (D-P’) Coronal hemisections of the telencephalon comparing gene expression in three rostral-to-caudal planes of section in control (left side) and Zfhx1b Nkx2.1-Cre conditional mutants (right side). (D-F’) Two color immunofluoresence detection of EGFP (green) and NKX2-1 (red). (G,G’) Higher magnification view of two color immunofluoresence detection of EGFP (green) and Nkx2-1 (red) in control (G) and Zfhx1b mutant (G’). Solid white arrowheads show increased numbers of cells that express both EGFP (green) and NKX2-1 (red) in the mutant's LGE/Str. In the wild type cortex, black arrowheads (with white outline) show that cells express EGFP (green) and not NKX2-1. (H-P’) In situ hybridization expression analysis at E12.5 of Nkx2-1 (H-J’) and Lhx6 (K-M’), and at E13.5 of Nkx2-1 (N-P’). Asterisks in (N-P’) show increased numbers of labeled cells in striatum. X in panels (K’-M’) notes the loss of labeled cells in the cortex. Abbreviations: Cx: cortex; e: ectopia in region of the ventral striatum and central nucleus of the amygdala; GP: globus pallidus; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; MZ: mantle zone; Str: Striatum; SVZ: subventricular zone; VPd: ventral pallidum; VZ: ventricular zone. Scale bars equal 500μm (A and D), and 300 μm (G).
Next, we examined the expression of Zfhx1b's closely related homologue, Zfhx1a, in the E12.5 control and mutant telencephalon. Both Zfhx1a and Zfhx1b are expressed in the subpallial VZ, whereas only Zfhx1b is clearly expressed in the SVZ (Figures 1A and S1B). Zfhx1a's expression did not clearly change in the Nkx2.1-Cre mediated Zfhx1b mutant (Figure S1B-S1D’). Thus, in the Nkx2.1-Cre conditional Zfhx1b mutant, only the VZ of the MGE continued to strongly express a Zfhx homologue.
MGE-Derived Pallial Interneurons Migrate to the Striatum When Deleting Zfhx1b in the VZ of the MGE Using Nkx2.1-Cre
We analyzed the effect of deleting Zfhx1b, using Nkx2.1-Cre at multiple developmental stages, including E12.5, E15.5 and P0. To track the fate of Zfhx1b mutant cells, we used the CAG:CAT-EGFP Cre reporter allele (Kawamoto et al., 2000). Mutant brains had the following genotype: Nkx2.1-Cre;Zfhx1bF/-;CAG:CAT-EGFP; whereas controls had the following genotype: Nkx2.1-Cre;Zfhx1bF/+;CAG:CAT-EGFP (on occasion, some were: Nkx2.1-Cre; Zfhx1bF/+). At E12.5, while the control brain showed a robust stream of EGFP+ cells migrating into the cortex, the mutant's EGFP+ MGE derivatives failed to migrate to the cortex, and many were detected in the LGE mantle (Figures 1D-1G’).
Next, we analyzed the phenotype using molecular markers of MGE-derived cells including Nkx2-1 and Lhx6. While Nkx2-1 RNA and protein is expressed throughout the VZ and SVZ of the MGE, its expression thereafter is restricted to specific neuronal lineages. MGE-derived cortical interneurons repress Nkx2-1 expression as they migrate out of the MGE while most, but not all, classes of striatal interneurons maintain Nkx2.1 expression. (Flandin et al., 2010; Marin et al., 2000; Nóbrega-Pereira et al., 2008; Sussel et al., 1999). In the mutants, there was a subtle increase in Nkx2-1 RNA expression in the LGE and CGE (Figures 1H-1J’). This increase was more apparent at higher magnification when analyzing NKX2-1 protein expression (Figures 1G and 1G’), and at later stages (E13.5 and E15.5) (Figures 1 N-P’ and 2A-F’).
Figure 2.
Zfhx1b expression in the MGE is required for interneuron migration at E15.5. Coronal hemisections of the telencephalon comparing gene expression in three rostral-to-caudal planes of section in control (left side) and Zfhx1b;Nkx2.1-Cre conditional mutants (right side). (A-C’) Two color immunofluoresence detection of EGFP (green) and NKX2-1 (red). (D-U’) In situ hybridization analysis. Nkx2-1 (D-F’), Lhx6 (G-I’), Sox6 (J-L’), Lhx8 (M-O’), NPY (P-R’), Kcnmb4 (S-U’). Asterisks show increased numbers of labeled cells in the striatum. X shows reduced number of labeled cells in cortex. Abbreviations: CGE: caudal ganglionic eminence; Cx: cortex; e: ectopia in region of the ventral striatum and central nucleus of the amygdala; GP: globus pallidus; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; MZ: mantle zone; Str: striatum; SVZ: subventricular zone; VPd: ventral pallidum; VZ: ventricular zone. Scale bar equals 500μm (A).
EGFP and NKX2-1 protein expression in control brains co-localized in a subset of cells derived from the MGE. EGFP/NKX2-1+ cells were observed in the MGE VZ and SVZ progenitors and a subset of their derived neurons, including the globus pallidus, and striatal interneurons (Xu et al., 2008; Figure 1G solid arrowheads), while interneurons migrating to the cerebral cortex showed little to no NKX2-1 protein expression (Figure 1G open arrowheads). In mutant brains however, most if not all EGFP labeled cells had detectable levels of NKX2-1 protein, with many cells strongly co-expressing NKX2-1 and EGFP in the LGE MZ, and in a region lateral to the globus pallidus (Figure 1G’ solid arrowheads). Thus, Zfhx1b mutants had a defect in their ability to repress Nkx2-1 RNA and protein expression, concomitant with failure of MGE-derived migration to the cerebral cortex. While Zfhx1b was required to repress Nkx2-1 expression, we did not find evidence that Nkx2-1 regulated Zfhx1b expression; this conclusion was based on in situ hybridization analysis of Zfhx1b expression in mice lacking Nkx2-1 in newly born MGE neurons at E15.5 (Nkx2-1 conditional mutant with Dlx5/6-Cre)(Figure S6).
Lhx6 RNA is expressed in tangentially migrating cells that are immature cortical and striatal interneurons, as well as cell types that remain in the subpallium (Flandin et al., 2011; Lavdas et al., 1999; Liodis et al., 2007; Sussel et al., 1999; Zhao et al., 2008). In the Zfhx1b mutant, Lhx6+ cells failed to be detected in the pallium, whereas they continued to be densely located throughout the MGE, and as a scattered population in the LGE and CGE (Figures 1K-1M’). On the other hand, Lhx8 and Gbx2 RNA expression was not appreciably changed in the mutants (Figures S1H-S1J’). Thus, Zfhx1b mutants may have a selective defect in cells fated to become pallial interneurons, but not cholinergic striatal interneurons. To explore this hypothesis we studied the phenotype at later developmental stages.
By E15.5, the tangential migration of immature cortical interneurons can be readily visualized by expression of Lhx6, Somatostatin (Sst), and EGFP (in Nkx2.1-Cre;CAG-EGFP brains) (Figures 2, S2D-S2F). By contrast, in Zfhx1b mutants (Nkx2.1-Cre), pallial expression of Lhx6, Sst, and EGFP was strongly attenuated (Figures 2, S2D’-S2F’). On the other hand, subpallial expression of these markers was increased in two locations: the striatum (asterisks Figures 2A-2C’, 2G-2I’, and S2D-S2F’) and a region contiguous with the caudoventral striatum, which we believe corresponds to the anlage of the central nucleus of the amygdala (labeled e, for ectopia; Figures 2E’, 2H’, S2D’; note that Figure S2T shows Dlx5 expression labeling the central nucleus of the amygdala, CeA). The ectopia in these regions also contained increased expression of Nkx2-1 and Sox6 (Figures 2D-2F’ and 2J-2L’). These genes are normally expressed in the subpallial projection neurons such as the globus pallidus, striatal interneurons and cortical interneurons (Sox6 only) (Azim et al., 2009; Batista-Brito et al., 2009).
Next, we tested whether the mutant cells that failed to migrate to the pallium had features of the globus pallidus or striatal interneurons. We examined expression of several globus pallidus markers including Kcnmb4, Kctd12, Gbx2 and Lhx8. Unlike the abnormal expression of Lhx6, Sst, Nkx2-1 and Sox6, expression of Kcnmb4, Kctd12, Gbx2 and Lhx8 appeared normal in the Zfhx1b mutants (Figures 2M-2O’, 2S-2U’, and S2J-S2O’), providing evidence that the abnormal collections of cells correspond either to abnormally migrated cortical interneurons, or to striatal interneurons, and not globus pallidus neurons. Furthermore, as Gbx2 and Lhx8 expression and function are linked to the development of striatal cholinergic interneurons (Chen et al., 2010; Zhao et al., 2003), these results provided evidence that increased striatal Nkx2-1 expression did not correspond to cells destined to become striatal cholinergic interneurons.
To distinguish whether the abnormal collections of cells in the mutant striatum were cortical or striatal interneurons, we examined expression of Cxcr7 and NPY. At E15.5 Cxcr7 marked migrating cortical interneurons, and few cells in the striatum (Figures 7J,7K and 7L), suggesting that it is a relatively specific cortical interneuron marker (Wang et al., 2011). In the mutant, there was a robust reduction of Cxcr7 expression in the pattern of migrating cortical interneurons, without a substantive increase in striatal expression (Figures 7J’,7K’,7L’); a similar result was seen for Cux2 (not shown). On the other hand, at E15.5, NPY expression strongly marks scattered striatal cells (probably interneurons), and relatively few migrating cortical interneurons (note, most of the cortical expression at this age resembles that of immature projection neurons in the cortical plate). In the mutant, there was a robust increase in NPY expression in the striatum (Figures 2P-2R’), in a pattern closely resembling the pattern of ectopic Nkx2-1, Lhx6 and Sox6 (Figures 2D-2L’). Thus, we propose that the mutant cortical interneurons are transformed towards GABAergic striatal interneurons.
Figure 7.
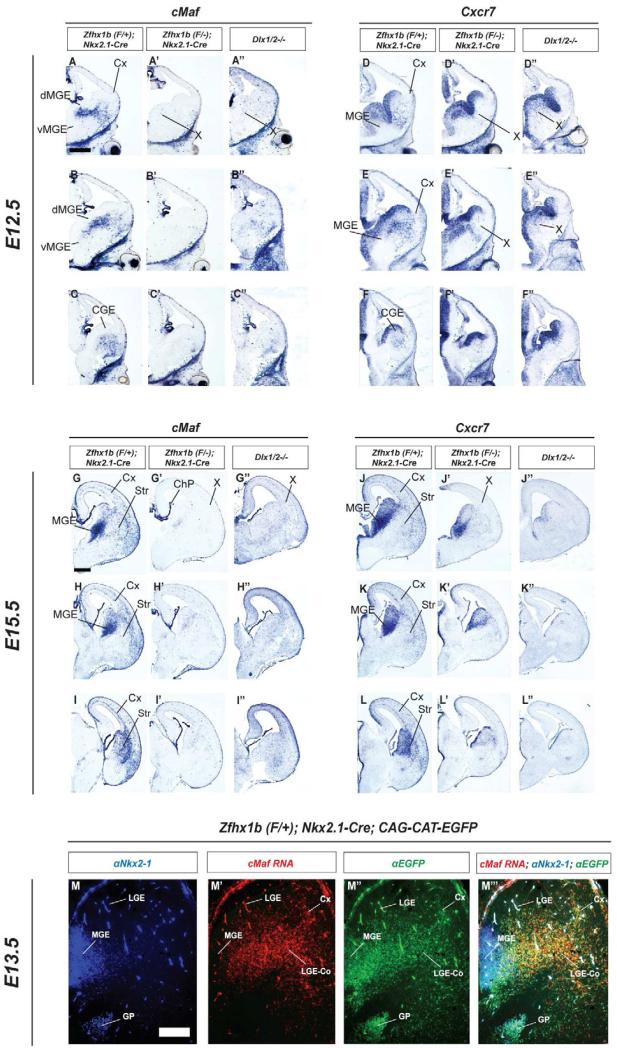
cMaf and CXCR7 are highly specific markers of the cortical interneuron lineage that are lost in Zfhx1b conditional mutants and Dlx1/2-/- mutants. Coronal hemisections of the E12.5 (A-F”) and E15.5 (G-L”) telencephalon comparing cMaf (A-C”; G-I”) and CXCR7 (D-F”; J-L”) RNA expression by in situ hybridization in three rostral-to-caudal planes of section in control (left panels), Zfhx1b;Nkx2.1-Cre conditional mutants (middle panels), and Dlx1/2-/- mutants (right panels). X shows reduced/absent cMaf+ or CXCR7+ cells in cortex or ganglionic eminences. Abbreviations: CGE: caudal ganglionic eminence; ChP: choroid plexus; Cx: cortex; LGE Co: LGE corridor; dMGE: dorsal medial ganglionic eminence; vMGE: ventral medial ganglionic eminence; Str: striatum. Scale bars are equal to 500μm (A) and 200uM (M).
Deleting Zfhx1b in SVZ of the MGE Using DlxI12b-Cre Phenocopies Loss of Zfhx1b function in the VZ (Nkx2.1-Cre)
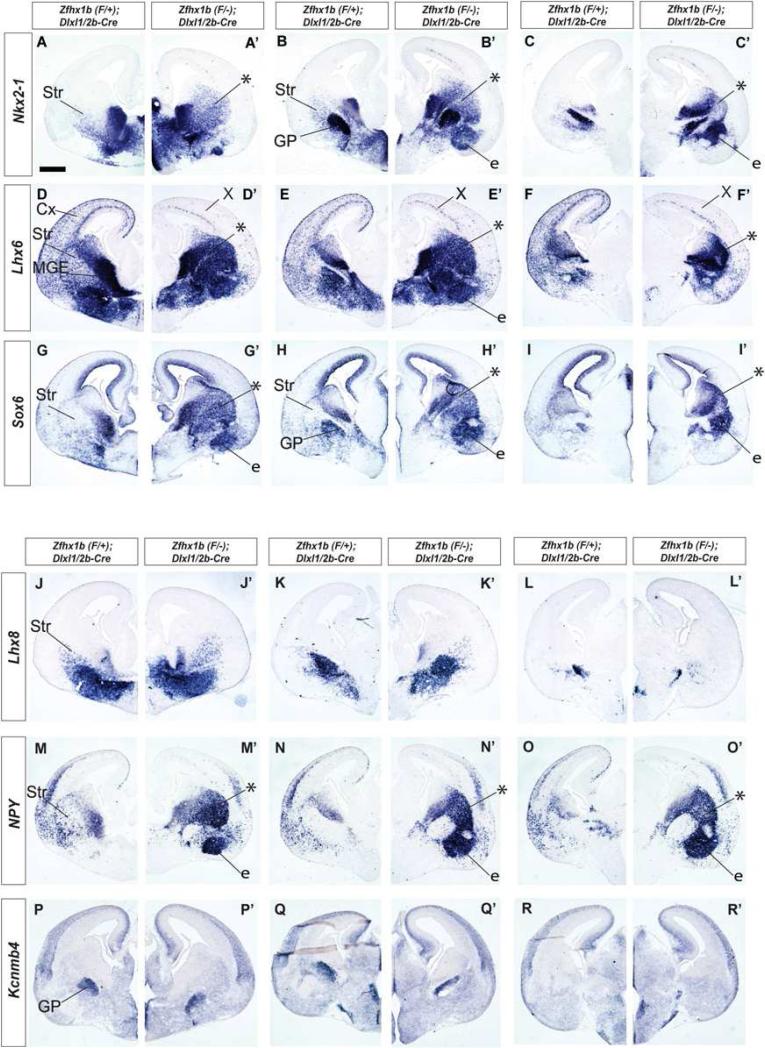
Towards defining the stage of differentiation when Zfhx1b is required for programming interneurons to migrate to the cortex, and not the striatum, we used the DlxI12b-Cre allele (Potter et al., 2008). DlxI1/2b-Cre expression begins in subpallial SVZ cells that express the mitotic marker Ki67 (Figures S1T-S1T”), suggesting that Cre recombination occurs in secondary progenitor cells that are mitotically active. Thus, DlxI12b-Cre induces recombination beginning in the SVZ of the entire subpallium, whereas Nkx2.1-Cre induces recombination in the VZ of the MGE and preoptic area (Figures 1D-1F’).
We analyzed the effect of deleting Zfhx1b using DlxI12b-Cre at E12.5 and E15.5. In general, all of the phenotypes of MGE-derived cells observed with the Nkx2.1-Cre were recapitulated with the DlxI12b-Cre (Figures 3, S1, S3), including the strong reduction of tangential migration to the cortex, indicated by analysis of Cre-dependent reporter EGFP expression, and Lhx6, Sst, and CXCR7 expression. Like Nkx2.1-Cre mutants, DlxI1/2b-Cre mutants showed increased numbers of striatal cells that expressed Lhx6, Nkx2-1, NPY, Sst and Sox6 (Figures 3A-3I’, 3M-3O’ and S3J-S3L’). Furthermore, these mutants did not show an increase in the number of cells that expressed markers of the globus pallius (Lhx8, Gbx2, Kcnmb4) or striatal cholinergic interneurons (Lhx8, Gbx2) (Figures 3J-3L’, 3P-3R’; and S1Q’-S1S’, and data not shown).
Figure 3.
Zfhx1b expression in the SVZ of the MGE is required for interneuron migration at E15.5. Coronal hemisections of the telencephalon comparing gene expression in three rostral-to-caudal planes of section in control (left side) and Zfhx1b DlxI12b-Cre conditional mutants (right side). In situ hybridization analysis of: Nkx2-1 (A-C’), Lhx6 (D-F’), Sox6 (G-I’), Lhx8 (P-R’), NPY (M-O’), Kcnmb4 (P-R’). Asterisks show increased numbers of labeled cells in the striatum. X shows reduced number of Lhx6+ cells in cortex. Abbreviations: CGE: caudal ganglionic eminence; Cx: cortex; e: ectopia in region of the ventral striatum and central nucleus of the amygdala; GP: globus pallidus; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; MZ: mantle zone; Str: striatum; SVZ: subventricular zone; VPd: ventral pallidum; VZ: ventricular zone. Scale bar equals 500μm (A).
Postnatal Analysis of cortical and striatal interneuron phenotypes in Nkx2.1-Cre;Zfhx1b mutants
Nkx2.1-Cre conditional mutants died between P17 and P21; at P15, mutants weighed ~30% less than their control littermates, a phenotype that was exacerbated by litter size. We did not observe seizures or other neurological/behavioral phenotypes.
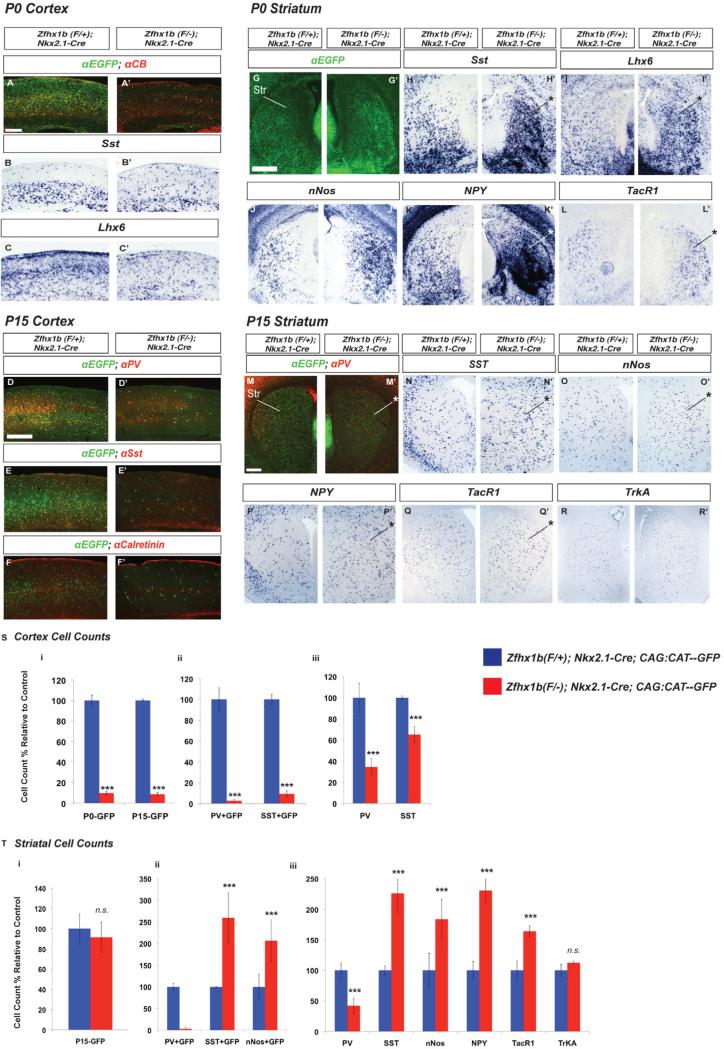
We analyzed postnatal day 0 (P0) and P15 Nkx2.1-Cre;Zfhx1bF/- conditional mutants to better understand the nature and extent of their cortical and striatal interneuron defects. In the P0 neocortex there was an ~90% reduction in the number of EGFP+ Cre-reporter marked cells, as well as a decrease in Calbindin (CB), Sst, and Lhx6 expressing interneurons (Figures 4A-4C’; 4S). Likewise, at P15 there was a >90% reduction in number of neocortical EGFP+ cells (Figure 4D-4F’; 4S). Next, we counted the number cortical interneurons in the Nkx2-1-Cre lineage that expressed Parvalbumin (PV) or Sst, which are the two main MGE-derived subtypes (Rudy et al., 2011). We saw a strong reduction in double labeled neurons, with the numbers of EGFP+ interneurons expressing Sst or PV reduced by >90% or more (Figure 4S). The expression of cortical Calretinin (CR), which predominantly marks CGE-derived cortical interneurons, showed little to no change in the Zfhx1b; Nkx2.1-Cre conditional mutant (Figure 4F-F’).
Figure 4.
Zfhx1b expression in the MGE regulates the numbers and fate of postnatal (P0 and P15) cortical and striatal interneurons. Coronal hemisections showing the neocortex (A-F) and the striatum (G-R), comparing gene expression in control (left side) and Zfhx1b;Nkx2.1-Cre conditional mutants (right side). (A, A’, D- F’ G,G’,M,M’) Two color immunofluorescence with anti-EGFP (Cre reporter; green) and interneuron markers (red); other panels show in situ hybridization results. (S) Cell counts, control relative to mutant, of: total Cre-reporter EGFP+ cell numbers in the P0 and P15 cortex (left); of the number of cells that had colocalization of the EGFP Cre-reporter with Sst or PV at P15 (middle); of the total levels of Sst and PV in the P15 cortex (right). (T) Cell counts, control relative to mutant, of: EGFP+ Cre-reporter cells in the P15 striatum (left); of EGFP-colocalization with PV, Sst and nNos in the P15 striatum (middle); of the total numbers of the markers PV, Sst, nNos, NPY, TacR1 and TrkA in the P15 striatum (right). Scale bar equals 500μm (A,D, G and M). *: p<.05; **: p<.01.
In the striatum at P0, as we saw at E15.5, there was an increase in the number of cells expressing EGFP (Cre reporter), Sst and Lhx6 (Figure 4G-4I’); consistent with the hypothesis that Zfhx1b mutant cells that were destined to go to the neocortex, instead migrated to the striatum. Additionally, there was a clear increase in the number of striatal cells expressing nNos, NPY and Nkx2-1 (Figures 4J-4K’, S4B and B’), while we observed no change in Lhx8, a marker for striatal cholinergic interneurons (Figures S4A and S4A’).
As NPY, Sst, and nNos are also expressed in subsets of cortical interneurons, their increased striatal expression does not provide unequivocal information about whether supernumerary cells correspond to cortical interneurons that failed to correctly migrate, or to interneurons that changed fate due to the mutation. To this end, we searched for a marker that is expressed in striatal, but not cortical interneurons. Substance P receptor (TacR1) is robustly expressed in striatal interneurons (Ardelt et al., 1996). We found that TacR1 is almost exclusively expressed in striatal and not cortical interneurons at E15.5, P0 and P15 (Figures 4L, 4L’, 4Q, 4Q’, S4D-S4F’, and data not shown). In Zfhx1b-Nkx2.1-Cre mutants at P0, there was increased striatal TacR1 expression (Figure 4Q-4Q’), supporting the idea that at least some of the mutant cells are adopting a striatal interneuron identity.
At P15 the number of mutant cells (EGFP+) was roughly the same as in controls, and they were evenly dispersed within the striatum, lacking the cell clusters and ectopia (striatal and caudal amygdala) that were apparent at younger ages (Figure 4M-4M’, 4T). The elimination of the excess mutant striatal cells appears to occur through apoptosis, which is robust at P0 (expression of activated cleaved-caspase 6), particularly in the ectopia (Figure S4C-S4C’).
Despite the cell death, Zfhx1b conditional mutants at P15 continued to have significantly increased numbers of striatal nNOS, NPY, Sst and TacR1 expressing cells (183%, 230%, 225%, and 164%; Figures 4N-4Q’, 4T). Importantly, total striatal PV+ cells were decreased by 58% (Figure 4S). Furthermore, there was no detectable change in TrkA expression (Figure 4R-4R’, 4T), which marks striatal cholinergic interneurons. We saw very few CR+ cells in the control striatum (1-3 cells per section), which did not noticeably change in the Zfhx1b conditional mutant (data not shown). Thus, Nkx2.1-Cre;Zfhx1b mutants have a selective increase in striatal interneurons expressing nNos, NPY, Sst and TacR1, but have reduced PV interneurons, and no change in cholinergic or CR interneurons.
We also analyzed the gross morphological properties of nNos/NPY/Sst striatal interneurons in the Zfhx1b mutant, and found that, like control brains, Zfhx1b conditional mutants had Sst processes restricted to the matrisomes (Chesselet and Graybiel, 1986), in a lateral to medial gradient (Figure S4G-S4I’- arrowheads mark CB-poor striosomes), suggesting that the overproduced nNos/NPY/Sst interneurons in the Zfhx1b conditional mutant share grossly similar morphological properties with wildtype striatal interneurons.
Zfhx1b Expression Is downstream of Dlx1/2 in the Developing Basal Ganglia
Dlx1 and Dlx2 are necessary for subpallial development, including interneuron migration to the cortex (Anderson et al., 1997a; Long et al., 2009a; Long et al., 2009b; Yun et al., 2002a); thus we examined Zfhx1b RNA expression in Dlx1/2 constitutive null mutants using in situ hybridization (Figures 5A-B’). In control brains at E12.5 and E15.5, Zfhx1b was expressed in the VZ and SVZ of the subpallium, in addition to its previous described expression in the cortical plate and SVZ (Miquelajauregui et al., 2007; Seuntjens et al., 2009). Zfhx1b expression in the subpallial MZ was restricted to dispersed cells in the LGE and to a nucleus forming near the ventral medial ganglionic eminence (MGE) (Figures 1A; 5A, 5B). In Dlx1/2-/- mutants, Zfhx1b expression was strongly and specifically decreased in the SVZ of the entire subpallium; expression in the subpallial VZ was maintained, albeit perhaps reduced (Figures 5A-B’).
Figure 5.
Dlx1&2 are required for Zfhx1b expression in the SVZ of the subpallium. (A-B’) Coronal hemisections of the telencephalon comparing Zfhx1b expression in Dlx1/+/- and Dlx1/2-/- at E12.5 (A-A’) and E15.5 (B,B’). Note the greatly reduced Zfhx1b expression in the SVZ of the LGE and MGE. X notes reduction in Zfhx1b expression in the SVZ. (C-K) Regulatory elements near Zfhx1b that drive subpallial expression are bound by DLX2 in vivo, and are positively regulated by DLX2. (C) Relative genomic position of two ultraconserved DNA elements near the Human Zfhx1b (Zeb2) locus, #675 (D) and #649 (E) (data from http://genome.ucsc.edu/). (D and E) Genomic alignment of enhancers #675 and #649; each contain a number of conserved consensus homeobox sites (asterisks). (F and G) Base-resolution view of regions with homeobox sites within #649 and #675, which are heavily conserved across vertebrate species and are similar to known DLX2 binding sites (Potter et al., 2008). (H and I) Whole mount E11.5 enhancer-lacZ transgenic mouse embryos that demonstrated lacZ expression (X-Gal staining) in the ganglionic eminences (subpallium) (H’ and I’). (J) Luciferase assay demonstrating DLX2-dependent transcriptional activation (pCAGGS vector) mediated by enhancers #649 and #675 upstream of luciferase (pGL4.23 vector). (K) Blue bars: DLX2 ChIP qPCR assay (N=3) demonstrates anti-DLX2 binding to chromatin from E13.5 ganglionic eminences to subdomains of enhancers #675 and #649 and the positive control (Dlx5/6 enhancer), and not to the negative control region (a non-conserved domain upstream of Dlx2). Red bars: Addition of a DLX2 peptide blocks the anti-DLX2 binding. Abbreviations: Cx: cortex; LGE: lateral ganglionic eminence; Luc. Luciferase; MGE: medial ganglionic eminence; MZ: mantle zone; SP: subpallium; SVZ: subventricular zone, VZ: ventricular zone. Scale bar equals 500μm (A and B). ***: p<.001; n.s. : not significant.
Towards defining the mechanisms that regulate Zfhx1b expression in the developing subpallium, we identified two regulatory elements near the Zfhx1b locus that drive expression in the developing subpallium. These enhancers, here named #649 and #675, were identified by virtue of their extremely strong evolutionary conservation (Figures 5C-G) and their reproducible enhancer activity in the forebrain of mouse embryos in transgenic experiments (Figures 5H-I) (Visel et al., 2007; Visel et al., 2008). The other genes in this region do not have known expression in the developing subpallium. Analysis of enhancer activity at E11.5 in transgenic whole mounts and sections showed that both enhancers drive LacZ expression in the subpallium, including the SVZ of the MGE (Figures 5H,I). The spatial overlap of enhancer activities and Zfhx1b mRNA expression suggests that these two elements are distant-acting transcriptional activators of Zfhx1b in the developing subpallium. Computational analysis identified multiple candidate homeobox binding sites (asterisks in Figures 5D,E and highlighted regions in 5F,G).
To test whether Dlx2 can regulate these candidate Zfhx1b enhancers we used a luciferase reporter assay. Co-transfection of a luciferase reporter construct containing enhancers #649 and #675 with a Dlx2 expression vector in P19 cells showed that DLX2 strongly activates luciferase transcription when these elements are present (Figure 5J).
To determine whether DLX2 directly regulates enhancers 649 or 675, we performed chromatin immunoprecipitation (ChIP)-qPCR of E13.5 basal ganglia using a DLX2 antibody. We found enrichment over several homeodomain-containing regions of enhancers 649 and 675, with a particular domain of #675 (region #3) showing the strongest enrichment as compared to control regions of the of the genome (Figure 5K). Also, the relative enrichment of the enhancer fragments was eliminated when a DLX2 polypeptide was included in the immunoreaction as a negative control (Figure 5K).
In summary, we have identified two candidate distant-acting gene regulatory elements whose activity patterns suggest that they contribute to Zfhx1b expression in the developing subpallium. These enhancer elements are activated by DLX2 in luciferase reporter assays, and are bound by DLX2 in vivo, providing strong evidence that subpallial Zfhx1b expression directly depends on Dlx1/2. Consistent with this, we observed marked alterations of Zfhx1b expression in the subpallial SVZ of Dlx1/2-deficient mice. Taken together, these results raise the possibility that the loss of Zfhx1b expression in the SVZ could contribute to the defects in differentiation and interneuron migration seen in Dlx1/2 mutants. To investigate this possibility, we compared the phenotypes of the conditional Zfhx1b and Dlx1/2-/- mutants.
Dlx1/2-/- Constitutive and Zfhx1b Conditional Mutants Have Similar Changes in Gene Expression Related to Their Defects in Interneuron Migration
As Zfhx1b expression in the subpallial SVZ was greatly reduced in the Dlx1/2 mutants (Figures 5A-B’), and because interneuron migration to the cortex was greatly reduced in both the Dlx1/2-/- and Zfhx1b mutants, we hypothesized that loss of Zfhx1b may underlie some of the interneuron migration phenotype of the Dlx1/2-/- mutants. To evaluate this idea, we compared gene expression phenotypes of the Dlx1/2-/- constitutive null mutant with the Zfhx1b conditional (Nkx2.1-Cre) mutant at E15.5 (Figure 6).
Figure 6.
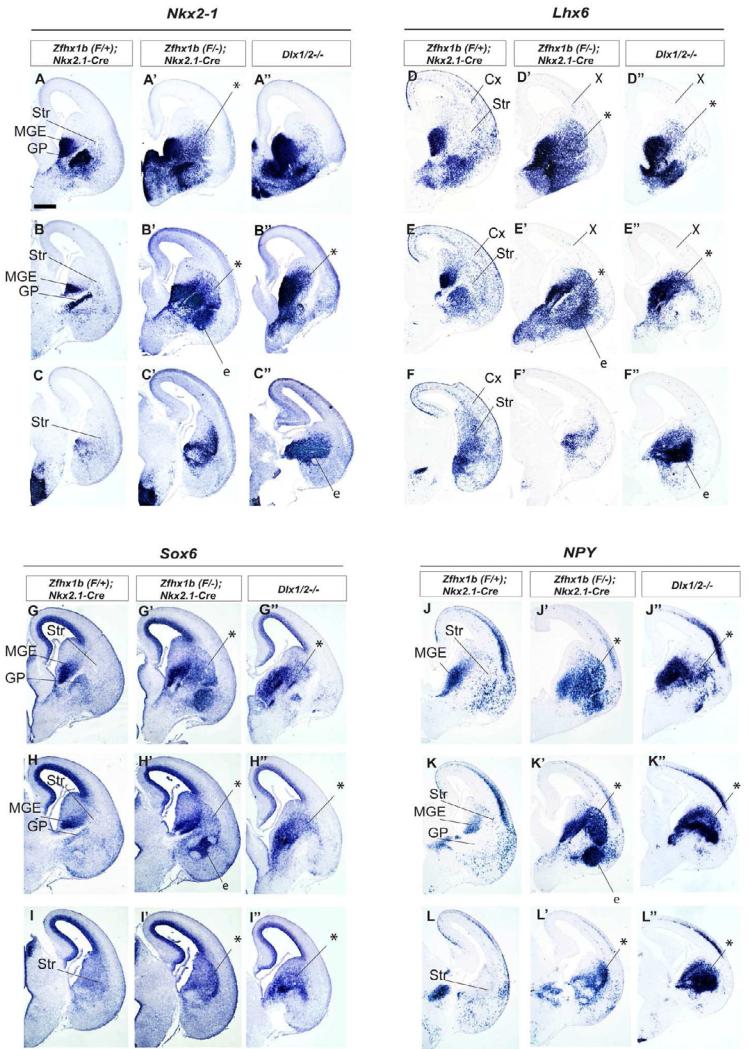
Zfhx1b;Nkx2.1-Cre and Dlx1/2-/- mutants both fail to repress Nkx2-1 and Sox6, lose cortical interneurons, and accumulate MGE cells in their striatum. Coronal hemisections of the E15.5 telencephalon comparing gene expression in three rostral-to-caudal planes of section in controls (left side) and mutants (right side). In situ hybridization analysis of Nkx2-1 (A-C”), Lhx6 (D-F”), Sox6 (G-I”), NPY (J-L”) expression was assessed for control, Zfhx1b;Nkx2.1-Cre mutants and Dlx1/2-/- mutants. Asterisks show increased numbers of labeled cells in the striatum. X shows reduced number of Lhx6+ cells in cortex. Abbreviations: CGE: caudal ganglionic eminence; Cx: cortex; e: ectopia in region of the ventral striatum and central nucleus of the amygdala; GP: globus pallidus; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; MZ: mantle zone; Str: striatum; SVZ: subventricular zone; VPd: ventral pallidum; VZ: ventricular zone. Scale bar equals 500μm (A).
Indeed, changes in Nkx2-1 and Sox6 expression were similar in the Dlx1/2-/- and Zfhx1b mutants. Nkx2-1 and Sox6 RNAs are normally expressed in similar patterns at E15.5 (Figures 6A,B,C,G,H and I); in wild type brains, both RNAs are maintained in the MGE, and in putative striatal interneurons and globus pallidus neurons, while they are downregulated in the MGE-derived cortical interneuron lineage (Nkx2.1-, Sox6low). Both Dlx1/2-/- and Zfhx1b mutants show increased numbers of Nkx2-1+ and Sox6+ cells in the striatum/LGE (asterisks in Figure 6; note: the Dlx1/2-/- mutant striatum is small due to Dlx-function in the LGE) (Anderson et al., 1997b; Yun et al., 2002b).
Lhx6+ cells are lacking throughout the cortex and increased in the striatum of both the Dlx1/2-/- and Zfhx1b mutants (Figures 6D-F”). Likewise, Sst+ and NPY+ cortical interneurons were lost, whereas their expression was increased in the striatum (Figures 6J-L”; Sst data not shown).
RNA Expression Array Analysis Identifies Candidate Mediators of Zfhx1b Function
Towards identifying the molecular mechanisms underlying the Zfhx1b mutant phenotype we used an RNA expression microarray analysis. We compared gene expression from the E12.5 MGE of Nkx2.1-Cre; Zfhx1bF/- mutants to that of Nkx2.1-Cre; Zfhx1bF/+ control littermates. Table S1 (see Table S3 for an extended version) lists the most highly up-regulated and down-regulated genes. Overall, a larger number of genes were found to be significantly up-regulated than down-regulated in Zfhx1b mutants, which may reflect Zfhx1b's function as a recruiter of repressive transcriptional complexes (van Grunsven et al., 2003; Verschueren et al., 1999; Verstappen et al., 2008). We verified the results for many of the genes by performing in situ RNA hybridization on E12.5 control and mutant brains (Figure S6 and data not shown).
We were most interested in genes that were altered in both the Nkx2.1-Cre and DlxI12b-Cre Zfhx1b mutants, given that both mutants showed altered interneuron migration and specification. Six genes fell into this category: cMaf, MafB, CXCR7, Dlk1, Cited1 andGpc4 (Figures 7, S6). Dlk1, Cited1 and Gpc4 were up-regulated in the MGE both mutants (Figure S6). cMaf, MafB, and CXCR7 were down-regulated in the MGE and migrating interneurons; later in the paper we focused more on these genes (Figures 7, S6); below we discuss the other genes.
Dlk1 expression was strongly increased in the VZ and SVZ of the MGE in the Nkx2.1-Cre mutant, and increased weakly only in the SVZ of the MGE in the DlxI12b-Cre mutant. (Figures S6J-S6L’ and S6HH-S6JJ”). Given that Zfhx1b's function was required in the SVZ of the MGE, the increase in Dlk1 expression could play a role in the phenotype. Dlk1 encodes a secreted delta-like ligand (Ferrón et al., 2011; Moon et al., 2002) that could alter Notch signaling. We used electroporation to increase Dlk1 expression in wild type MGE, but failed to identify a change in interneuron migration (data not shown).
Other genes related to Notch-signaling were also identified in the array analysis, including the Id2 and Id4 helix-loop-helix and Sox6 HMG-box transcription factors (Table S1). Expression of Id4 was increased in the VZ of the Nkx2.1-Cre mutant; however, no change in expression was detected in the DlxI12b-Cre mutant (Figures S6M-S6O’ and S6KK-S6MM’); this implies that Id4 does not contribute to the interneuron phenotype. Id2 expression showed a subtle expression increase in the Nkx2.1-Cre mutant (not shown); like Id4, we did not find a change in its expression in the DlxI1/2b-Cre mutant (not shown). Sox6 expression was also increased based on the array and an increase was seen in both Nkx2.1-Cre and DlxI1/2b-Cre mutants by in situ hybridization at E12.5 (Table S1 and Figures S1F-S1F’ and data not shown), which became more pronounced at E13.5 and E15.5 (Figures 2J-L’ and 3G-I’). Of note, Sox6 represses MGE expression of Ascl1 (Mash1) (Azim et al., 2009), a basic-helix-loop-helix transcription factor whose expression is promoted by Notch-signaling.
Expression of Cited1, a p300-binding transcriptional co-activator that promotes signaling in the TGF-beta pathway (Gerstner and Landry, 2007) was increased in the SVZ of the ventral MGE and POA in both the Nkx2.1-Cre and DlxI12b-Cre mutants (Figures S5S-S5U’ and S5QQ-S5SS’). This is of interest given that ZFHX1B acts as a SMAD-binding transcriptional co-repressor (Vandewalle et al., 2008). Thus, Cited1 and Zfhx1b may function antagonistically in MGE development. Gpc4 expression in the SVZ of the MGE was increased in both mutants (Figure S5V-S5X’ and S5TT-S5WW’). Glypicans (GPC) are extracellular matrix proteins that promote FGF-signaling (Jen et al., 2009).
Expression of genes related to oligodendrogenesis, including Olg1 and GPR17 (Chen et al., 2009; Lu et al., 2000) were down-regulated on the array in the Nkx2.1-Cre mutant (Table S1); we failed to detect GPR17 expression by in situ hybridization and Olg1 expression was weak. For this reason, we studied Olg2 expression; its expression was reduced in the SVZ of the MGE at E12.5 (Figures S5P-S5R’). The down-regulation of oligodendrocyte markers may be related to the increase in ID4 RNA; ID proteins can repress oligodendrogenesis (Wang et al., 2001). By E15.5, we did not detect a change in Olg2 expression (Figures S2P-S2R’). The DlxI12b-Cre mutant did not show changes in Olg2 expression (Figure S5NN-S5PP’) suggesting the Zfhx1b function in the VZ, and not SVZ, regulates oligodendrogenesis.
Zfhx1b Is Required for Expression of Genetic Markers of Cortical Interneurons: cMaf, MafB and Cxcr7
The gene expression array showed a ~3-fold reduction in the expression of cMaf (v-Maf), a leucine zipper-containing transcription factor (Table S1). cMaf, and its relative MafB, have been reported to be expressed in cortical interneurons (Cobos, 2006; Faux et al., 2009; Zhao et al., 2008). Likewise, the array identified reduced expression of CXCR7, whose expression and function are required during cortical interneuron migration (Sánchez-Alcañiz et al., 2011; Wang et al., 2011).
We compared cMaf, MafB and Cxcr7 RNA expression at E12.5 and E15.5 and identified some important features (Figures 7, S2A-S2C and S6D-S6F). cMaf, MafB and Cxcr7 RNAs were expressed in the SVZ of the dorsal MGE (and not the ventral MGE), and were maintained in cells migrating through the LGE and CGE and then into the cortex at E12.5 and E15.5 (Figure 7, S2A-S2C and S6D-S6F). cMaf, MafB and CXCR7 appear to be excellent markers of the cortical interneuron lineage, as we did not detect their expression in other MGE-derived structures, such as the ventral pallidum or globus pallidus at E12.5, E15.5 or P0 (Figures 7, S2A-S2C, S6D-S6F and data not shown). In the E15.5 and P0 striatum, these genes showed little expression (Figure 7), except for MafB and cMaf in a very small population of cells (data not shown). Thus, unlike other cortical interneuron markers that are also expressed in striatal interneurons (e.g. Dlx1, Lhx6, parvalbumin, Sst), cMaf, MafB and CXCR7 expression largely mark only cortical interneurons.
We then compared cMaf, MafB and Cxcr7 expression in E12.5 and E15.5 control (Zfhx1b heterozgyotes), Zfhx1b conditional mutants (Nkx2.1-Cre and DlxI12b-Cre) and the Dlx1/2-/- constitutive mutant (Figures 7, S2A-S2C”, S3A-S3I’). cMaf expression was nearly eliminated in all three mutants. Much of the remaining cMaf expression was in scattered blood cells and in the choroid plexus (Figure 7 H’ and H”). MafB and Cxcr7 were also greatly reduced in the SVZ of the ganglionic eminences, although they were not as strongly down-regulated as cMaf (Figures 7, S2A-S2C’, S3D-S3I’, S6D-S6I’ and S6BB-S6GG’). Therefore, Zfhx1b (and Dlx1&2) were required for cMaf, MafB and Cxcr7 expression, which are highly specific markers of immature migrating cortical interneurons (cMaf and MafB are specific for MGE-derived interneurons). Thus, the loss of cMaf, MafB and Cxcr7 expression in Zfhx1b conditional mutants (Nkx2.1-Cre and DlxI12b-Cre) provides additional evidence that cortical interneurons fail to be specified.
Next, we determined at a cellular resolution when cMaf expression begins in developing cortical interneurons (Figures 7M-M”’). Given that cMaf expression in the dorsal MGE and migrating cortical interneurons is dependent on Zfhx1b expression, and that repression of Nkx2-1 in these regions is also Zfhx1b dependent, we analyzed whether or not cMaf and Nkx2-1 are co-expressed in these cells. To this end, we performed a triple-labeling analysis: fluorescent in situ hybridization to detect cMaf in combination with immunofluorescence to label NKX2-1 and the EGFP+ Nkx2.1-Cre lineage (EGFP expression from CAG-CAT-EGFP, the Cre reporter allele). We found that cMaf RNA was expressed in EGFP+ cells derived from the MGE lineage that were migrating through the LGE corridor (LGE-Co) on route to the cortex (Figures 7M’, M” and M’”). These cells were NKX2-1-. On the other hand, NKX2-1 was expressed in cMaf- cells of the globus pallidus (Figures 7 M, M” and M”’). Thus, as NKX2-1 expression is repressed in immature cortical interneurons, cMaf expression begins.
Discussion
Herein we demonstrate that Zfhx1b subpallial expression is directly positively regulated by Dlx1&2, and is required in the MGE to generate cortical interneurons that express Cxcr7, MafB and cMaf. In its absence, Nkx2-1 expression is not repressed, and cells that ordinarily would become cortical interneurons are transformed towards the NPY/nNos/Sst subtype of striatal GABAeric interneuron. Furthermore, it is possible that the Zfhx1b-/- phenotype is also caused by defects in migration and differentiation that contribute to the formation of subpallial ectopia. However, below we largely concentrate on discussing the evidence that Zfhx1b regulates cell-type specification.
Zfhx1b regulates MGE cell-type generation
The MGE generates multiple cell types, including GABAergic interneurons of the cortex and striatum, GABAergic projection neurons of the basal ganglia (e.g. GP), cholinergic neurons of the striatum and basal telencephalon and oligodendrocytes (Flandin et al., 2010; Petryniak et al., 2007; Xu et al., 2008). The MGE generates roughly 60% of all GABAergic cortical interneurons; these express PV, Sst, NPY and nNos (Gelman and Marín, 2010; Rudy et al., 2011).
The MGE also generates striatal interneurons. There are three subtypes of GABAergic striatal interneurons: PV+, nNos/NPY/Sst+ and CR+ (Tepper et al., 2010). The striatum also has cholinergic interneurons. The cholinergic population is marked and regulated by Gbx2, Islet1, and Lhx8 (Chen et al., 2010; Fragkouli et al., 2009), and the neurotrophin receptor TrkA (Sanchez-Ortiz et al., 2012).
The ventral MGE is not a major source for cortical interneurons based on fate mapping using Shh-Cre (Flandin et al., 2010). Thus, the dorsal MGE must be the source of most MGE-derived cortical and striatal GABAergic interneurons. It is poorly understood whether these cell types are generated from distinct subregions, from distinct but intermixed progenitors, or from the same progenitors in a stochastic or temporally modulated program. While the same neuroepithelial progenitor can generate different types of cortical interneurons (PV and Sst)(Brown et al., 2011), it is not known whether cortical and striatal interneurons are derived from the same progenitor.
Zfhx1b was required to generate GABAergic cells that migrate to the cortex, and to repress the generation GABAergic cells that migrate to the striatum. We suggest that Zfhx1b promotes a fate switch between cortical interneurons and nNos/NPY/Sst striatal interneurons through repression of Nkx2-1 expression. Furthermore, Zfhx1b mutants have reduced striatal PV interneurons (Figure 4M,4M’); thus, Zfhx1b could also control this fate decision. Zfhx1b is required in the MGE SVZ, and not the VZ, to promote the specification of pallial interneurons, as we observed largely the same phenotype using DlxI12b-Cre (SVZ recombination, Figures 3, S1, S3) and Nkx2.1-Cre (VZ recombination; Figures 1, 2, S1, S2).
We identified perhaps the first specific early marker of dorsal MGE-derived cortical interneurons: cMaf (Figure 7). cMaf and Mafb expression are dependent on Zfhx1b and Dlx1/2 function (Figure 7, S2A-C’, S3D-F’, S6D-S6F’ and S6BB-S6DD’ (Cobos, 2006; Long et al., 2009a; Long et al., 2009b) Notably, neither cMaf, nor Mafb are strongly expressed prenatally in neurons of the striatum (interneurons and medium spiny neurons), suggesting that prenatally they may be specific markers of the cortical interneuron lineage. Currently, Maf function in the brain has only been studied in the hindbrain (Cordes and Barsh, 1994).
Zfhx1b connects the Nkx2-1 and Dlx transcription pathways
There is genetic evidence for at least three parallel (although interacting) transcriptional pathways in the MGE that are required for cortical interneuron development: the 1) Ascl1 (Mash1); 2) Dlx; and 3) Nkx2-1 pathways (Long et al., 2009a,b). The Nkx2-1 pathway is the core mediator of MGE regional and cell identity (Butt et al., 2007; Sussel et al., 1999); it functions through induction of Lhx6 and Lhx8 (Sussel et al., 1999). Lhx6 is essential for induction of Mafb and Shh in neurons, maintaince of Sox6 in interneurons, and the differentiation of Sst and Parvalbumin cortical interneurons (Lhx6) (Liodis et al., 2007; Zhao et al., 2008); Lhx8 is required in cholinergic striatal interneurons (Lhx8) (Fragkouli et al., 2009).
Only a subset of MGE neuronal derivatives maintain Nkx2-1 and Lhx8 expression, such as the globus pallidus and cholinergic striatal interneurons (Marin et al., 2000), whereas MGE-derived cortical interneurons suppress Nkx2-1 and Lhx8 expression (Nóbrega-Pereira et al., 2008). We propose that Dlx and Nkx2-1 pathways interact at this step. We demonstrated that Dlx1/2 were required for Zfhx1b expression in the subpallial SVZ (Figure 4), and that Zfhx1b was required for repression of Nkx2-1, but not of Lhx8 (Figures 2D-F’, 2M-O’, 3A-C’ and 3J-L’). Thus, in the absence of Zfhx1b, dorsal MGE-derived neurons continued to express Nkx2-1, Sox6 and Lhx6, and migrate into the striatum and not the cortex. These cells failed to express markers of cortical interneurons (Cxcr7, cMaf and MafB) (Figures 7 A-L”, S5A-S5I’), but highly expressed the striatal GABAergic subtype markers NPY, nNos and Sst. (Figures 2P-R’ and 3M-O’ (Tepper et al., 2010). Additionally, there was increased expression of TacR1, which is robustly expressed in Sst+ and ChAT+ striatal interneurons, and in very few cortical interneurons (Ardelt et al., 1996; Figure S4D-S4F’).
Finally, Zfhx1b mutants did not exhibit clear phenotypes of striatal cholinergic interneurons or the GP. Thus, we propose a distinct Zfhx1b-independent mechanism for the generation of the GP and cholinergic neurons; the latter depends on the maintenance of Lhx8, perhaps in combination with Islet1 and Gbx2 (Chen et al., 2010; Fragkouli et al., 2009).
Downstream of Zfhx1b in the MGE Cells
It is unclear whether Zfhx1b has a common molecular mechanism in all developing cells. Zfhx1b mediates some of its functions through interactions with SMAD proteins, and thus participates in TGF-beta signaling (Vandewalle et al., 2008). Expression of the SMAD-binding transcriptional co-activator Cited1 was increased in Zfhx1b mutants (Sup. Figure 6). The link to SMAD signaling is intriguing because SMAD dominant negative expression can inhibit interneuron tangential migration (Maira et al., 2010).
In the pallium, Zfhx1b functions in both progenitors and neurons. In hippocampal progenitors, it functions upstream of Wnt signaling to control development of the entire region (Miquelajauregui et al., 2007). In neocortical neurons Zfhx1b regulates neurotrophin-3 and Fgf9 expression, to control cortical progenitors (Seuntjens et al., 2009). We did not observe similar regulatory changes in the Zfhx1b mutant MGE.
While Zfhx1b in the MGE SVZ regulates the switch between cortical and striatal interneurons, Zfhx1b is also expressed in the VZ of the MGE (Figure 1A). Two genes related to Notch signaling were up-regulated in the mutant MGE VZ, including the secreted delta-like ligand Dlk1 (Ferrón et al., 2011; Moon et al., 2002) and the HLH transcription factor ID4 (Yun et al., 2004) (Figure S6J-S6O’). Dlk1 up-regulation in the VZ and SVZ could alter the balance of cell fate decisions.
Previous studies suggested that Nkx2-1 promotes interneuron integration into the striatum via repression of Npn2/Sema3-dependent repulsion (Marín et al., 2001; Nóbrega-Pereira et al., 2008). We did not detect a change in Npn2 and Npn1 RNA expression in migrating immature Zfhx1b mutant interneurons at E12.5. On the other hand, van den Berghe et al (in press) present evidence that Zfhx1b regulates interneuron migration through the netrin receptor Unc5b.
Zfhx1b and Human Disease
Mowat-Wilson syndrome (MWS) is caused by a heterozygous mutation or deletion of the Zfhx1b (ZEB2, SIP1), and is characterized by a distinctive facial appearance, intellectual disability, and variable other features including seizures, agenesis of the corpus callosum, and Hirschsprung disease (Mowat et al., 2003). Given Zfhx1b's critical role in cortical interneuron development, we propose that cortical interneuron defects contribute to the seizure phenotype of MWS. Furthermore, since Dlx1&2 regulate Zfhx1b expression in the subpallium, and Dlx1&2 also regulate craniofacial and enteric nervous system development (Qiu et al., 1995) it will be intriguing whether Zfhx1b is also downstream of Dlx function during development of these tissues.
Experimental Procedures
See Supplemental Experimental Procedures for detailed description of methods.
Mice
Zfhx1bF/F mice were genotyped according to (Miyoshi et al., 2006). CAG-CAT-eGFP mice were genotyped according to (Kawamoto et al., 2000). Zfhx1bF/F males were crossed to Beta-Actin Cre mice (Lewandoski et al., 1997) to generate the Zfhx1b null allele, which was followed by a cross to wildtype mice to eliminate the Beta-Actin Cre allele. Zfhx1b+/-mice were crossed with Nkx2.1-Cre I1/2b-Cre mice, and male Zfhx1b+/-; Cre+ mice were crossed with female Zfhx1bF/F mice with or without the CAG-CAT-EGFP allele to generate conditional mutant embryos.
Histochemistry
Embryonic and postnatal brains were prepared and immunostained (Flandin et al., 2010), or assayed by in situ hybridization (Jeong et al., 2008). Protocols can be found on our lab website http://physio.ucsf.edu/rubenstein/protocols/index.asp, with modifications for dual immuno/in situ fluorescence analysis described in the Supplementary Experimental Procedures.
Cell culture, transfections and luciferase assays
P19 cells were cultured as described in (Farah et al., 2000). Experimental conditions were tested in triplicate by transfection of cells in 12-well plates using Fugene 6 (Roche). Cotransfection of a Renilla luciferase expression construct was used as a normalization control for a dual-luciferase assay. The following amounts of DNA were used in each well: 80ng pGL4.73 (Renilla Luciferase, Promega), 240 ng pCAGGs-empty or pCAGGS-Dlx2, 240ng pGL4.23-empty (Luciferase, Promega) or pGL4.23-enhancer. Luciferase and Renilla Luciferase quantification was done using a Promega Dual-Lucifase Assay Kit and a microplate luminometer (Veritas). Chi-square test showed that the levels of activation were significant *: p<0.05.
Chromatin immunoprecipitation (ChIP)
ChIP was performed similar to a published method (McKenna et al., 2011) with modifications described in Supplementary Experimental Procedures.
Supplementary Material
Acknowledgements
This work was supported by the research grants to JLRR from: Nina Ireland, Weston Havens Foundation, Genentech, NIMH R37 MH049428 and R01 MH081880; to GM: Predoctoral Training in Neurobiology T32 GM007449; and to LAP and AV from: NINDS R01NS062859A and NHGRI R01HG003988.; to GLM from: Predoctoral Training in Neurobiology T32 GM007449. LAP and AV conducted research at the E.O. Lawrence Berkeley National Laboratory, performed under DOE DE-AC02-05CH11231, University of California. DH is funded by: Queen Elisabeth Medical Foundation (1113), Scientific Research-Flanders (G.0954.11N), Research Council Univ. of Leuven GOA, InfraMouse Hercules, and Belspo IUAP7/07. We thank Professor Melinda Duncan at the University of Delaware for providing Zfhx1bF/+ mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information for this article includes six Supplemental Figures, two Supplemental Tables and Supplemental Experimental Procedures.
References
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Ardelt AA, Karpitsky VV, Karause JE, Roth KA. The neostriatal mosaic:basis for the changing distribution of neurokinin-1 receptor immunoreactivity during development. J Comp Neurol. 1996;16:463–75. doi: 10.1002/(SICI)1096-9861(19961216)376:3<463::AID-CNE8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Machold R, Klein C, Fishell G. Gene expression in cortical interneuron precursors is prescient of their mature function. Cereb Cortex. 2008;18:2306–2317. doi: 10.1093/cercor/bhm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KN, Chen S, Han Z, Lu C-H, Tan X, Zhang X-J, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. Clonal Production and Organization of Inhibitory Interneurons in the Neocortex. Science. 2011:480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJB, Cobos I, Golden J, Kessaris N, Pachnis V, Anderson S. Transcriptional Regulation of Cortical Interneuron Development. Journal of Neuroscience. 2007;27:11847–11850. doi: 10.1523/JNEUROSCI.3525-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJB, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008:722–732. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chatterjee M, Li JYH. The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J Neurosci. 2010;30:14824–14834. doi: 10.1523/JNEUROSCI.3742-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu H, Wang S, Koito H, Li J, Ye F, Hoang J, Escobar SS, Gow A, Arnett HA, et al. The oligodendrocyte-specific G protein–coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 2009:1398–1406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I. Cellular Patterns of Transcription Factor Expression in Developing Cortical Interneurons. Cerebral Cortex. 2006;16:i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Faux C, Rakic S, Andrews W, Yanagawa Y, Obata K, Parnavelas JG. Differential gene expression in migrating cortical interneurons during mouse forebrain development. J Comp Neurol. 2009:NA–NA. doi: 10.1002/cne.22271. [DOI] [PubMed] [Google Scholar]
- Ferrón SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, Morante-Redolat JM, Laborda J, Guillemot F, Bauer SR, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011:381–385. doi: 10.1038/nature10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JLR. The Progenitor Zone of the Ventral Medial Ganglionic Eminence Requires Nkx2-1 to Generate Most of the Globus Pallidus But Few Neocortical Interneurons. Journal of Neuroscience. 2010;30:2812–2823. doi: 10.1523/JNEUROSCI.4228-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein, John LR. Lhx6 and Lhx8 Coordinately Induce Neuronal Expression of Shh that Controls the Generation of Interneuron Progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 2009:3841–3851. doi: 10.1242/dev.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Marín O. Generation of interneuron diversity in the mouse cerebral cortex. Eur J Neurosci. 2010:2136–2141. doi: 10.1111/j.1460-9568.2010.07267.x. [DOI] [PubMed] [Google Scholar]
- Gerstner JR, Landry CF. Expression of the Transcriptional Coactivator CITED1 in the Adult and Developing Murine Brain. Developmental Neuroscience. 2007;29:203–212. doi: 10.1159/000096389. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Maruhashi M, Nelles L, Van de Putte T, Verschueren K, Miyoshi T, Yoshimoto A, Kondoh H, Huylebroeck D. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis. 2002;32:82–84. doi: 10.1002/gene.10048. [DOI] [PubMed] [Google Scholar]
- Jen Y-HL, Musacchio M, Lander AD. Glypican-1 controls brain size through regulation of fibroblast growth factor signaling in early neurogenesis. Neural Dev. 2009:33. doi: 10.1186/1749-8104-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Li X, McEvilly RJ, Rosenfeld MG, Lufkin T, Rubenstein JLR. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135:2905–2916. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470:263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harbor Symposia on Quantitative Biology. 1997;62:159–168. [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 Activity Is Required for the Normal Migration and Specification of Cortical Interneuron Subtypes. Journal of Neuroscience. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JLR. Dlx1 and Mash1 Transcription Factors Control MGE and CGE Patterning and Differentiation through Parallel and Overlapping Pathways. Cerebral Cortex. 2009a;19:i96–i106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JLR. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009b;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Maira M, Long JE, Lee AY, Rubenstein JLR, Stifani S. Role for TGF-beta superfamily signaling in telencephalic GABAergic neuron development. J Neurodev Disord. 2010:48–60. doi: 10.1007/s11689-009-9035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001:872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JLR, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. Journal of Neuroscience. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquelajauregui A, Van De Putte T, Polyakov A, Nityanandam A, Boppana S, Seuntjens E, Karabinos A, Higashi Y, Huylebroeck D, Tarabykin V. Smad-interacting protein-1 (Zfhx1b) acts upstream of Wnt signaling in the mouse hippocampus and controls its formation. Proceedings of the National Academy of Sciences. 2007:12919. doi: 10.1073/pnas.0609863104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Maruhashi M, Van de Putte T, Kondoh H, Huylebroeck D, Higashi Y. Complementary expression pattern of Zfhx1 genes Sip1 and deltaEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev Dyn. 2006;235:1941–1952. doi: 10.1002/dvdy.20799. [DOI] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim K-H, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat DR, Wilson MJ, Goossens M. Mowat-Wilson syndrome. Journal of medical genetics. 2003;40:305–310. doi: 10.1136/jmg.40.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega-Pereira S, Gelman D, Bartolini G, Pla R, Pierani A, Marin O. Origin and Molecular Specification of Globus Pallidus Neurons. Journal of Neuroscience. 2010:2824–2834. doi: 10.1523/JNEUROSCI.4023-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marín O. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JLR. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, Mckinsey GL, Ekker M, Rubenstein JLR. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Molecular and Cellular Neuroscience. 2008:1–20. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes & Development. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Alcañiz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, López-Bendito G, Stumm R, Marín O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ortiz E, Yui D, Song D, Li Y, Rubenstein JL, Reichardt LF, Parada LF. TrkA gene ablation in basal forebrain results in dysfunction of the cholinergic circuitry. Journal of Neuroscience. 2012;32:4065–4079. doi: 10.1523/JNEUROSCI.6314-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave K-A, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2003:1–10. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave K-A, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Tecuapetla F, Koós T, Ibáñez-Sandoval O. Heterogeneity and Diversity of Striatal GABAergic Interneurons. Frontiers in Neuroanatomy. 2010;4 doi: 10.3389/fnana.2010.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grunsven LA, Michiels C, Van De Putte T, Nelles L, Wuytens G, Verschueren K, Huylebroeck D. Interaction between Smad-interacting protein-1 and the corepressor C-terminal binding protein is dispensable for transcriptional repression of E-cadherin. Journal of Biological Chemistry. 2003:26135. doi: 10.1074/jbc.M300597200. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cellular and Molecular Life Sciences. 2008;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. Journal of Biological Chemistry. 1999:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Verstappen G, Van Grunsven LA, Michiels C, Van De Putte T, Souopgui J, Van Damme J, Bellefroid E, Vandekerckhove J, Huylebroeck D. Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Human Molecular Genetics. 2008:1175–1183. doi: 10.1093/hmg/ddn007. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nature genetics. 2008:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JLR. CXCR4 and CXCR7 Have Distinct Functions in Regulating Interneuron Migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JLR. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002a;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JLR. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002b:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JLR. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marín O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JLR, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA. 2003;100:9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.