Abstract
RNA editing is vast in some genetic systems, with up to thousands of targeted C-to-U and U-to-C substitutions in mitochondria and chloroplasts of certain plants. Efficient prognoses of RNA editing in organelle genomes will help to reveal overlooked cases of editing. We present PREPACT 2.0 (http://www.prepact.de) with numerous enhancements of our previously developed Plant RNA Editing Prediction & Analysis Computer Tool. Reference organelle transcriptomes for editing prediction have been extended and reorganized to include 19 curated mitochondrial and 13 chloroplast genomes, now allowing to distinguish RNA editing sites from “pre-edited” sites. Queries may be run against multiple references and a new “commons” function identifies and highlights orthologous candidate editing sites congruently predicted by multiple references. Enhancements to the BLASTX mode in PREPACT 2.0 allow querying of complete novel organelle genomes within a few minutes, identifying protein genes and candidate RNA editing sites simultaneously without prior user analyses.
Keywords: pyrimidine substitutions, RNA editing prediction, plants, protists, mitochondrial DNA, chloroplast DNA, BLASTX
Introduction
The term RNA editing has originally been coined for the surprising discovery that translatable reading frames are created by targeted uridine insertions in mitochondrial pre-mRNAs in the trypanosomes.1 After this exciting original finding numerous other processes of RNA editing, adding twists to the known pathways of gene expression, have been discovered in many evolutionary disparate groups of life.2 Like in trypanosomes, RNA editing is particularly abundant in the mitochondrial genetic systems of diverse eukaryotes. A prominent example along those lines is the slime mold Physarum polycephalum, which features hundreds of cytidine insertions as the dominant type of RNA editing.3 Similar in abundance is RNA editing in certain land plants, for which the lycophytes, which represent the most ancient surviving branch of vascular plant evolution, reveal particular astonishing examples. More than 2,000 events of mitochondrial C-to-U RNA editing have been identified in the club moss Selaginella moellendorffii.4 The sister lycophyte Isoetes engelmannii shows only slightly fewer editing events, but in this case they occur in both directions of pyrimidine exchange.5 Such “reverse” U-to-C editing events are also particularly abundant in ferns and hornworts. A transcriptome analysis of the hornwort Anthoceros formosae chloroplast, for example, has revealed nearly 1,000 C-to-U and U-to-C conversions.6
Pyrimidine-exchange editing in organelles of photosynthetic organisms has previously been regarded as a gain in early land plant evolution, given that no events were reported for ancestral green algae. However, two events of C-to-U RNA editing have recently been discovered in the mitochondrial transcriptome of the protist Naegleria gruberi7 and it is very likely that these RNA editing events are executed by the same nuclear factors as in plants, the so-called “DYW-type” pentatricopeptide repeat (PPR) proteins.8 This discovery opens up the possibility that many more instances of RNA editing may remain undiscovered in organelle genome sequences.
The plant-type of organelle RNA editing can be reasonably predicted given that the pyrimidine transitions mostly serve to re-establish evolutionary conserved codons in genes of the core standard set of chloroplast and mitochondrial gene complements. Particular obvious examples are the occasional creation of methionine start (AUG) or stop (UAR, UGA) codons by C-to-U editing from threonine (ACG) and from glutamine (CAR) or arginine (CGA) codons, respectively. Alternative examples include the removal of in-frame stop codons by reverse U-to-C editing. Aside from start and stop codon conversions, however, pyrimidine-exchange RNA editing can result in 22 other possible switches in meaning between codons for alanine and valine, arginine and cysteine, histidine and tyrosine, threonine and isoleucine or methionine and in any direction among the leucine, phenylalanine, proline and serine YYN codons. Predicting, comparing and annotating these RNA editing events is cumbersome, notably when they appear in large numbers.
The prognosis of mRNA editing events based on comparison with orthologous genes from non-editing taxa or from confirmed cDNA data of taxa utilizing RNA editing becomes more reliable with an increasing number of diverse reference sequences. Here we present a significant extension in functionality of our WWW server PREPACT, the “Plant RNA Editing Prediction & Analysis Computer Tool”.9 The PREPACT reference database has been entirely reshaped and extended and now relies on coding sequences directly translated from revised GenBank entries after taking RNA editing events into account, which have been manually curated. The latter step is necessary given that no standard annotation for RNA editing sites has hitherto been introduced for primary database accessions. We introduce a novel feature “RNA_editing” for annotation and internal representation of editing sites in the PREPACT reference database, which may be adapted in the future for primary database entries. The PREPACT 2.0 update now allows users to freely select from multiple organellar reference genomes for prediction of editing in their query sequences. In its BLASTX mode, PREPACT 2.0 even allows for comfortable simultaneous prediction of coding regions and RNA editing sites for entire organelle genome sequence queries within a few minutes.
Material and Methods
PREPACT core functionality
Like the initial version,9 the core of PREPACT 2.0 is written in the PHP hypertext preprocessor scripting language (http://php.net) using a MySQL (http://mysql.com) database backend and running on an Apache web server (http://httpd.apache.org). The graphical interface uses JavaScript elements (http://jquery.com, http://jqueryui.com, http://github.com/carhartl/jquery-cookie, http://github.com/flaviusmatis/flexibleArea.js, http://laktek.com/2008/10/27/really-simple-color-picker-in-jquery) for comfortable usage. Raster graphics output is generated by PHP scripts using ImageMagick (http://imagemagick.org). The PREPACT core functionality has been partly re-implemented, but still relies on the comparison of edited “reference” sequences aligned to unedited “test” sequence queries. Essentially, PREPACT suggests pyrimidine transitions (C-to-U, optionally U-to-C) in codons to result in improved amino acid matches in the reference sequences. Alignments for analysis can be provided directly by the user in FASTA format or can be generated by the BLASTX program,10,11 provided by the NCBI (http://blast.ncbi.nlm.nih.gov). The latter is enhanced with new functionality in PREPACT2 allowing simultaneous identification of protein coding genes and RNA editing sites without prior user analyses in non-annotated sequences.
Revision of the reference structure and database generation for BLASTX mode
The protein reference sequence database was entirely revised for PREPACT 2.0 and now relies on translated coding sequences of full organelle genomes taking known editing events into account. Original GenBank accessions (Table 1) were retrieved from NCBI, split into their various elements such as header, feature list, qualifiers and sequence origin and saved into the internal MySQL database after format checking. The fully-functional hierarchical tree of feature and qualifier objects within sequence objects is retained. A complete set of associated methods for position calculation, information retrieval and manipulation within PREPACT 2.0 makes it possible to check for potentially erroneous feature locations, translational mismatches and CDS naming issues during subsequent revision where necessary. Flexible regular expression-based search and replace classes scanning the sequence entries for necessary modifications are stored on a per-accession basis to curate the available organellar genomes. When present, annotated editing sites were parsed from the different formats currently present in primary accessions (Fig. 1) into a new PREPACT-internal “RNA_editing” feature (Fig. 2). This process simultaneously checked for consistency and more common mistakes (eg, annotation of the wrong DNA strand), which were resolved automatically, and remaining annotation errors (such as obvious mislabeling of editing positions or misannotation of splicing) were corrected manually. Where no editing was annotated at all (eg, most angiosperm chloroplast [cp] DNA entries) RNA editing annotation was introduced manually into the same modifications database.
Table 1.
Reference organelle genome and transcriptome data as incorporated in PREPACT 2.0.
| Genome/species | Accession | RNA editing events appl. | Reference on RNA editing |
|---|---|---|---|
| Mitochondrial genomes | |||
| Arabidopsis thaliana (thale cress) | NC_001284 | 430 | (38) |
| Beta vulgaris (sugarbeet) | NC_002511 | 359 | (39) |
| Brassica napus (rapeseed) | NC_008285 | 423 | (40) |
| Chaetosphaeridium globosum (green alga) | NC_004118 | 0 | – |
| Chara vulgaris (green alga, “stonewort”) | NC_005255 | 0 | – |
| Citrullus lanatus (watermelon) | NC_014043 | 456 | (41) |
| Cucurbita pepo (zucchini) | NC_014050 | 448 | (41) |
| Isoetes engelmannii (lycophyte, “quillwort”) | FJ010859+a | 3738 | (5) |
| Lotus japonicus (legume) | NC_016743 | 528 | Accession onlye |
| Marchantia polymorpha (liverwort) | NC_001660 | 0 | – |
| Millettia pinnata (pongam oiltree) | NC_016742 | 510 | Accession onlyf |
| Naegleria gruberi (heterolobosean protist) | NC_002573 | 2 | (7) |
| Nicotiana tabacum (tobacco) | BA000042c | 536 | Accession onlyg |
| Oryza sativa (rice) | NC_011033 | 481 | (42) |
| Physcomitrella patens (moss) | NC_007945 | 11 | (12) |
| Reclinomonas americana (jakobid protist) | NC_001823 | 0 | – |
| Selaginella moellendorffii (lycophyte, “spikemoss”) | JF338143+b | 2139 | (4) |
| Silene latifolia (white campion) | NC_014487 | 286 | (43) |
| Vitis vinifera (grapevine) | NC_012119 | 411 | (44) |
| Chloroplast genomes | |||
| Adiantum capillus veneris (fern) | AY178864d | 343 | (45) |
| Anthoceros formosae (hornwort) | NC_004543 | 970 | (6) |
| Arabidopsis thaliana (thale cress) | NC_000932 | 41 | (46) |
| Atropa belladonna (deadly nightshade) | NC_004561 | 40 | (37) |
| Chaetosphaeridium globosum (alga) | NC_004115 | 0 | – |
| Chara vulgaris (alga, “stonewort”) | NC_008097 | 0 | – |
| Hevea brasiliensis (rubber tree) | NC_015308 | 56 | (47) |
| Marchantia polymorpha (liverwort) | NC_001319 | 0 | – |
| Nicotiana tabacum (tobacco) | NC_001879 | 43 | (37;48;49) |
| Oryza sativa (rice) | NC_001320 | 35 | (50;51) |
| Physcomitrella patens (moss) | NC_005087 | 1 | (22;23) |
| Pisum sativum (pea) | NC_014057 | 26 | (50) |
| Zea mays (maize) | NC_016666 | 32 | (52;53) |
Notes: Annotation of RNA editing relied mostly on the reference genome accessions (NC_x) revised by the NCBI. Exceptions are the complex network-like recombining mtDNAs of the lycophytes aIsoetes engelmannii (FJ010859, FJ176330, FJ390841, FJ536259 and FJ628360) and bSelaginella moellendorffii (JF338143, JF338144, JF338145, JF338146 and JF338147) which at present rely on five separate original sequence accessions each and the cNicotiana tabacum mtDNA and the dAdiantum capillus-veneris cpDNA given that RNA editing information is absent in the corresponding organelle genome entries NC_006581 and NC_004766, respectively. No formal publications are presently available on the mitochondrial editomes of eLotus japonicus, fMillettia pinnata and gNicotiana tabacum. Dashes (–) indicate that RNA editing is hitherto not identified and assumed to be entirely absent in algae, marchantiid liverworts and protists like Reclinomonas americana. Note that due to gene duplications (eg, due to IRs in cpDNAs or redundancy in the recombining mtDNAs) the numbers of “applied RNA editings” in the reference sequences may be significantly larger than the number of actual C/U and U/C differences as reported in the literature.
Figure 1.
Four examples (A–D) of different free-text annotations of RNA editing in primary database plant organelle genome entries.
If indicated at all, RNA editing sites are mostly annotated using the miscellaneous feature (“misc_feature”) with additional information given in various ways in the “note” qualifier, which necessitates a parsing into a common standard for bioinformatic processing.
Figure 2.
Suggested new feature “RNA_editing” for annotation of editing events. The mandatory “type” qualifier distinguishes the different known cases of RNA editing by (A) substitution, (B) insertion or (C) deletion. A second obligatory qualifier “replace” gives the string of nucleotides to be replaced at the sequence location indicated for the RNA_editing feature. Its value is necessarily empty only for the deletion type of editing (C) but gives individual or stretches of nucleotides for the other two types of editing (A and B). As for the respective affected genetic locus, the “complement” function is to be used (A) where necessary to correctly refer to the DNA strand that is co-linear with the respective transcript. The “gene” qualifier is mandatory in case of overlapping CDS features, otherwise its use is encouraged for clarification.
Additional qualifiers may be optionally used (A, examples shown in italics) to introduce RNA editing site labels (“label”), indicate partial editing (“frequency”) or to convey additional biological information (“note”).
An auto-annotation module was created to process organelle genome entries without annotated RNA editing sites, but for which complete sets of cDNA are available; for example, the complex mitochondrial (mt) DNAs of lycophytes Isoetes engelmannii and Selaginella moellendorffii for which cDNAs exist as primary database entries, or Vitis vinifera, where editing information has been stored in REDIdb. The auto-annotation script aligns cDNA sequences to the corresponding CDS feature(s) in the organelle genome entries and automatically creates new “RNA_editing” features for these.
Out of a finally-curated organelle genome all CDS features are extracted, translated into proteins taking all corresponding RNA editing into account and stored as a BLAST database, which can be used for analysis. For genomes not being represented by a single accession (eg, the lycophytes mtDNA mentioned above), various accessions can be combined to a single BLAST database.
Generating BLAST results and “commons”
The nucleotide query sequence is run in all six reading frames by BLASTX against all selected reference databases individually. Results are optionally filtered for only the best local identity to the query. Accordingly, one BLAST hit can rule out another in the same reference database if it has a higher identity to the query in the part overlapping between these two. If at least one of the hits fulfils the limit given by the “filter threshold”, the hit that is longer and has higher identity to the reference within a range of same query positions is kept. The remaining (protein) alignments are loaded as edited nucleotide sequences from the internal database and analyzed for RNA editing before the results are displayed in individual tabs. For the calculation of “commons”, all predicted editing sites are grouped in rows by their position in the query sequence and labeled according to the chosen numbering scheme. If no editing is present in a reference database, it is checked whether there is actually no alignment covering this single position (‘−’) or if the alignment does not predict an editing event (‘0’). If an editing prediction matches a position in the reference database that has been edited for generation of the protein BLAST database, these sites are highlighted in red.
Results
RNA editing site annotation
The bioinformatic processing of RNA editing is currently impeded by lack of a standard for annotation of editing sites in the primary database sequence accessions. When RNA editing sites are indicated at all, their annotation so far relies on the multi-purpose “miscellaneous” feature (“misc_feature”) and variable free-text comments in “note” qualifiers in sequence accessions. Some examples in current GenBank entries are shown in Figure 1. Occasionally, such annotations are strangely ambiguous and particularly difficult for bioinformatic processing, for example, when the caret (^), normally used as the standard “in between” location indicator for database entries, is used to designate substitutional editing of individual nucleotides (Fig. 1C). Moreover, it frequently remains ambiguous whether protein translations provided by the submitter are based on DNA or on edited RNA sequences. Alternatively, in many cases, these translations are entirely hypothetical or a mixture of the two, which only took into account the necessary removals of stop codons and the introductions of start or stop codons to create intact open reading frames.
We here suggest a dedicated novel sequence feature “RNA_editing” that is compatible with the grammar of the GenBank flat file format and might hence be considered for future adaptations by the primary sequence databases (Fig. 2). For the purposes of PREPACT 2.0, the differently-formatted information on editing sites and de novo annotation of RNA editing sites in organelle genome sequences (Table 1) were parsed into this new standard. We suggest two mandatory qualifiers “type” and “replace” for the new RNA_editing feature to clearly describe the respective type of editing taking place and the resulting sequence changes introduced in the respective mature RNA (Fig. 2). The obligatory “type” qualifier has one of three alternative qualities: “substitution”, “insertion” or “deletion”. The second mandatory qualifier “replace” indicates the string of RNA nucleotides that are newly created by the editing event. The “replace” value will indicate the nucleotides newly introduced by an editing event or will be empty (“ ”) for the deletion- type of editing (Fig. 2C). The locations of the RNA_editing feature are given following database standards as individual positions or nucleotide ranges and are indicated as “complement()” when the affected transcript runs opposite to the sequence orientation of the sequence accession (Fig. 2A). The “in between” annotation using the caret (^) will only be used for the insertion-type of editing (Fig. 2B). As the RNA_editing annotations are not directly associated with a reading frame, the use of a “gene” qualifier to assign a certain editing event to a CDS with the same name is encouraged or even mandatory when there are overlapping reading frames present.
Further to the two mandatory qualifiers “type” and “replace” we suggest optional additional qualifiers to convey useful supplementary information (Fig. 2A). Several RNA editing events, for example in plant mitochondria, are realized only partially in a steady-state transcript population. We propose use of the already available qualifier “frequency” to characterize such partial editing events with a decimal value between 0 and 1 to reflect the fraction of editing revealed in a cDNA population. The qualifier “label” is suggested to indicate an informative label for the respective editing site. We have recently suggested such a nomenclature for substitutional editing events,9,12 which we also use to clearly identify editing sites in comparative studies and for analysis and output in PREPACT. Briefly, this label for RNA editing events consists of the respective gene name, followed by an ‘e’, the RNA nucleotide identity after editing (ie, U or C in plant organelle editing), the affected nucleotide position in the mature reading frame counting from position 1 of the start codon and the resulting amino acid change (see Fig. 2A).
Restructuring and extending the PREPACT 2.0 reference database
For the automatic ab initio prediction of coding sequences and RNA editing sites in a nucleotide query, PREPACT originally relied on BLASTX hits in manually-assembled collections of reference protein sequences.9 The reference data set has been completely reorganized and largely extended for PREPACT 2.0. Coding sequences (CDS) are now dynamically extracted from reference organelle genome entries with RNA editing sites automatically taken into account for protein translations, where applicable. To this end, RNA editing as currently documented in its variable ways (Fig. 1) or extracted from the literature or occasionally from other sources13 was parsed into the standard described above (Fig. 2). Numerous obvious errors in gene annotations (eg, CDS extensions, splice sites, strand complementarity), which occur to variable degrees in most database accessions, were corrected for internal representation in PREPACT.
Organelle gene naming issues
Our manual curation of reference genomes also included de novo annotation of previously non- annotated coding sequences (eg, chlN in the Marchantia polymorpha cpDNA) and the replacement of non-informative old gene labels with consensus names (eg, “orfB” with “atp8” or “orf25” with “atp4”). In other cases we used internal synonymising for different names that are currently in parallel use for orthologous genes, eg, mttB and tatC for the twin-arginine translocase subunit or ccb, yej and ccm for the suite of cytochrome c maturation genes in mitochondria to appropriately identify orthologous editing sites, as we will exemplarily demonstrate below. Such synonymising can also be advantageous for entirely biological reasons. A prominent example is the mitochondrial ccmF locus which in the course of plant evolution got disrupted differently into separate reading frames that were labeled in numerous different ways.14
The reference data
Altogether, PREPACT 2.0 currently offers 19 mitochondrial and 13 chloroplast genome sequences as references for comparison. We included several exhaustively analyzed transcriptomes and organelle genomes supposedly devoid of RNA editing as summarized in Table 1. The latter group also includes the mitochondrial genome of the jakobid protist Reclinomonas americana, which has a particularly rich gene complement.15 To avoid misleading results, however, we did not include those organelle genomes where transcriptome analyses are clearly only partial so far, such as for the chloroplast genome of Pinus thunbergii16 or the mitochondrial genomes of Cycas taitungensis17 and Phoenix dactylifera.18 Likewise, we did not include the maize (Zea mays) chondrome with ca. 100 RNA editing events annotated in database entry NC_007982 since it is likely that at least 300 additional ones were missing according to our estimate.
Querying PREPACT 2.0 in BLASTX mode: a complete mtDNA example
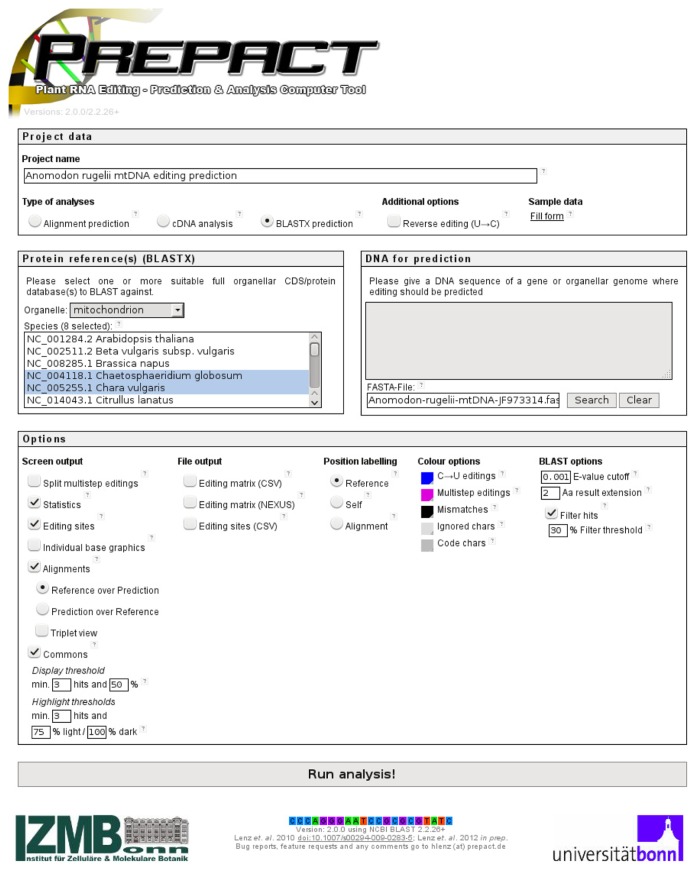
The “commons” function is a new core feature of PREPACT 2.0. It allows scanning for candidate RNA editing sites against multiple references simultaneously and prepares a comparative output for user evaluation. In describing the new features of PREPACT 2.0 we will here focus on the major enhancements for its BLASTX mode (Fig. 3), which essentially allows analysis of organelle sequences, largely without prior analyses by the user. In fact, the PREPACT 2.0 program code and server setup has been adapted to even allow querying of entire organelle genome sequences with hundreds of kilobases against multiple reference targets within minutes. A status window reflects progress of the analysis (Supplemental Fig. 1). Very long query sequences and simultaneous selection of very many references and a particularly detailed output including full alignments may require computation times of several minutes and slow down local browser display because of high memory consumption or even exceed server resource limits (approximately with queries > 500 kbp and >10 references). The recently determined complete mtDNA (104,239 bp) of the moss Anomodon rugelii19 is used here to exemplarily demonstrate the extended PREPACT 2.0 functionality, its query interface (Fig. 3) and the output (Figs. 4 and 5) and to discuss results and important considerations.
Figure 3.
The extended PREPACT 2.0 user interface.
Exemplarily shown is a query in BLASTX mode to scan for C-to-U editing candidate sites in the complete mtDNA of the moss Anomodon rugelii (104,239 bp, database accession JF973314). Multiple reference databases may be selected with the usual Ctrl-/Shift functionality. Selecting more than one reference automatically opens the “commons” options (bottom left) allowing to set thresholds selecting for editing sites congruently predicted from given fractions of the reference set for listing (preset to 50%) and two levels of highlighting (preset to 75% and 100% reference support) by light and dark shading. Additional integer thresholds may be set for display (preset ≥3) and highlighting (preset to ≥3) of hits present in only few or individual taxa. The labeling of editing sites may be chosen according to the proposed standard nomenclature for references (preset) or alternatively for query or alignment sequence positions when appropriate. BLAST search sensitivity may be adjusted (bottom right) and a newly introduced filter function allows for exclusion of secondary hits in protein paralogs according to length and similarity for overlapping hits (with “0%” excluding and “100%” allowing all overlaps). A number of codons (preset = 2) may be given by which alignments may be extended to possibly identify editing of start or stop codons when these are not included in BLASTX hits against the reference(s).
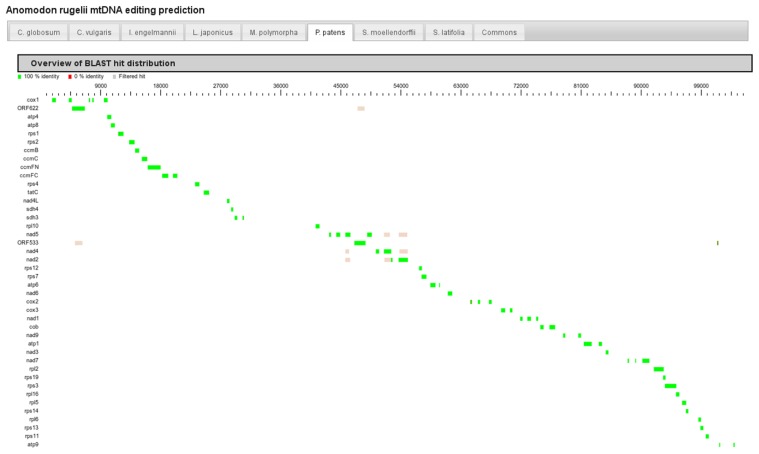
Figure 4.
PREPACT output in BLASTX mode: overview on hits in individual references.
The output is organized into tabs with results for the individual references that can be selected for display in the WWW browser. BLAST hits are displayed graphically on top of each reference tab, as exemplarily shown for the Physcomitrella patens reference. A very high mitochondrial gene synteny between the two moss mtDNAs is obvious for the Anomodon rugelii mtDNA query. Disrupted homologies along the horizontal lines suggest the presence of introns. Individual hits give short information on mouse-over and are directly linked to the respective analysis details included further down in the output as optionally selected in the query form (see Fig. 3), eg, of sequence alignments (see Fig. 6) or of statistics for sequences and BLAST hits, lists of editing sites and individual editing site graphics (not shown). BLAST hits are colored with decreasing similarities from bright green to dark red. The filtering of overlapping BLAST hits for exclusion in editing predictions is visualized by secondary hits in faded coloring, as here becomes obvious for the distantly related complex I paralogs nad2, nad4 and nad5 and the intron maturases orf533 and orf622.
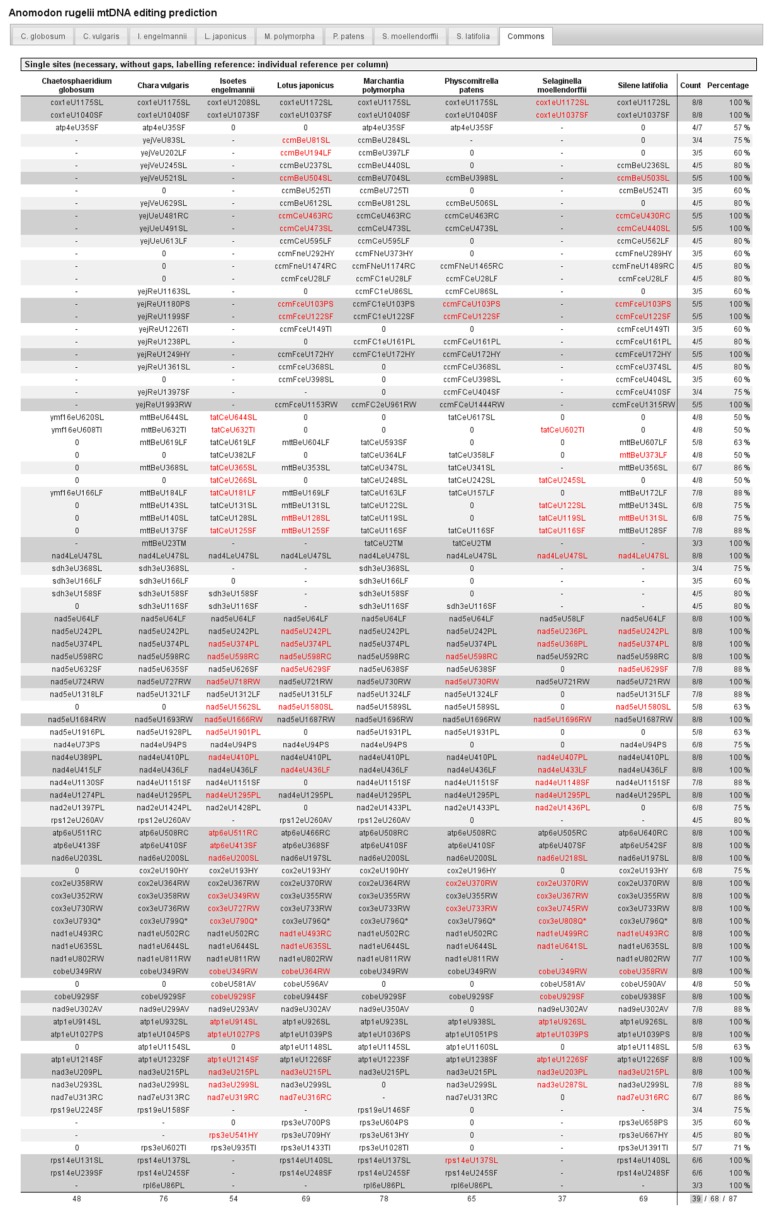
Figure 5.
The “commons” output in PREPACT 2.0 BLASTX mode.
The “commons” tab summarizes candidate RNA editing sites congruently predicted from the fractions of references as selected according to the thresholds chosen in the query form (see Fig. 3). The full list includes only sites fulfilling the display thresholds (here: minimally 50% and identification in ≥3 references). Light and dark gray shading highlights edits predicted for the indicated fractions of references (here: 75% and 100%, respectively) in an indicated minimum total number of references (here: 3). RNA editing events identified in the references are indicated in red. Symbols ‘-’ and ‘0’ indicate missing BLASTX homologies for a given region or no prediction for editing, respectively.
Improving BLASTX hit analyses: Filtering paralogs
A major earlier problem for BLASTX-based gene identification has been multiple hits due to similarities among the paralogous core subunits of chloroplast thylakoid membrane subunits (atpA-atpB, ndhB-ndhD-ndhF, psbA-psbD, psbB-psbC) and among mitochondrial respiratory chain complex I subunits (nad5-nad2-nad4). This issue has been successfully solved with a new filtering function that excludes secondary hits depending on extension and similarity of the overlap relative to a better primary hit according to an adjustable threshold preset to 30% (Fig. 3). Decreasing the filtering threshold may rarely be necessary to suppress identification of paralogs identified across large phylogenetic distances (‘0%’ would exclude all weaker hits conflicting with the top-scoring hit). Conversely, increasing its value (‘100%’ would suppress secondary hits only in case of complete overlapping with the top-scoring hit) promotes display of additional secondary hits, which may be interesting in exceptional cases (such as the multiple group II intron maturase paralogs in different organelle introns).
References and thresholds in the new “commons” function
Multiple references for identification of genes and editing sites may be selected from the choice of reference genomes/transcriptomes implemented in PREPACT (Table 1). In our example, we included the two green algae related to the plant lineage and the liverwort Marchantia polymorpha in all of which no RNA editing has been identified. Additionally, we here selected five highly diverse plant reference taxa with analyzed mitochondrial transcriptomes: the moss Physcomitrella patens with very rare RNA editing, the lycophytes Isoetes engelmannii and Selaginella moellendorffii with thousands of documented RNA editing sites as well as the “low-editing” Silene vulgaris and the comparatively “high-editing” Lotus japonicus flowering plant mtDNA references (Table 1).
The “commons” option allows restricting of the listing of RNA editing candidate sites to those that are congruently predicted from a given fraction of references (display threshold, preset to 50%). Two further thresholds (preset to 75% and 100%, respectively) may be adjusted for selective highlighting (Fig. 3). Additional filters are available to restrict candidate sites in “rare” genes (eg, maturases, ribosomal proteins) according to minimum numbers of parallel hits in the set of references for display or highlighting (both preset to ≥3) in the output.
The PREPACT 2.0 output for BLASTX mode
The PREPACT 2.0 output using the above example settings is shown in Figures 4 and 5. Results on gene identification and the prognoses for RNA editing are available as individual tabs for each reference selected in the query form (Figs. 4 and 5). Users may thus jump between reference tabs to inspect outputs in detail (eg, individual sequence alignments when needed, not shown) as selected in the query options (Fig. 3). A graphic overview of BLASTX hits along the query is displayed on top for each individual reference, as exemplarily displayed here for the top part of the output in the Physcomitrella patens reference tab (Fig. 4). Exon-intron gene structures in the query become immediately apparent with horizontally-arranged separate exon hits against the continuous protein reference targets in the BLASTX searches. Moreover, the sorting of matches between the query (horizontally) against the reference (vertically) also allows immediate recognition of gene syntenies in the organelle DNAs, as becomes very obvious for the two moss chondromes in our example (Fig. 4). Hits are colored from bright green to dark red according to increased expectancy for chance similarities. Secondary hits overlapping others with higher and/or more extended similarity that have been excluded for editing prognosis according to the filter settings are displayed with light shading. In the example shown this becomes clearly apparent for the nad5-nad4-nad2 cluster where similarities with the respective paralogs are appropriately excluded (Fig. 4). For convenient navigation in the output, BLASTX hits are actively linked to individual hit statistics, lists of editing sites and/or the individual alignments optionally displayed further down in the output as selected in the query form.
An additional tab “commons” (Fig. 5) summarizes the editing site predictions from the individual tabs according to the threshold settings in the query form. PREPACT 2.0 now distinguishes the absence of an editing prediction (0) from the absence of a BLASTX hit for a given position (-). This distinction becomes highly useful for genes that are absent (mostly due to nuclear gene transfer) in some of the reference genomes, thus allowing a smaller number of references to be taken into account for the threshold calculations. The absence of the entire suite of ccm genes (ccmB, ccmC and ccmF) from the chondromes of Chaetosphaeridium globosum, Isoetes engelmannii and Selaginella moellendorffii is an obvious bona fide case along those lines in the example output (Fig. 5). At the same time the ccm gene example also reflects the new gene synonymising functionality. The orthologous candidate RNA editing sites referring to identical query positions are given on the same line in spite of different gene labels in the reference database entries. Using the reference numbering for RNA editing site labeling, as chosen in our query example (Fig. 3), immediately allows recognition of amino-terminal length variation or indels in the homologous genes.
An advantage of the new reference data setup relying on edited coding sequences is that actual editing events previously identified in reference transcriptomes can be distinguished from “pre-edited” sites, ie, the edited versions of the affected pyrimidine, on the DNA level in others. We use red font for true editing events identified in a reference versus pre-edited sites on the DNA level in others given in black.
In our example querying the Anomodon rugelii mtDNA against the eight selected references using the “commons” default threshold settings as given in Figure 3, a total of 39 sites are consistently predicted against all reference genomes (100%, with minimally 3 hits total) as summarized in the bottom line of the output (Fig. 5). Most of these sites (27) are predicted by all 8 of 8 references, whereas others are also unequivocally predicted, but homologues for the respective loci are lacking in some references (2× 6/6 in rps14, 7× 5/5 in the suite of ccm genes, 2× 3/3 in rpl6 and for the start codon editing prediction in mttB/tatC and 7/7 for one site in nad1 lacking homology in Selaginella).
Additional support for the very strongly predicted editing sites as well as for the extended set included within the 75% threshold comes from the fact that the overwhelming majority of these sites are identified as being RNA editing positions (red) in at least one or more of the reference taxa. In fact, nad5eU64LF is the only example of a candidate site in the 8/8 subset that is consistently pre-edited in all reference taxa (Fig. 5). Similarly, RNA editing sites are also identified in the references for several other candidate sites that are included among the lower thresholds. Highly interesting cases are five candidate-editing sites in the tatC (or mttB) gene, all of which are independently corroborated by homologous editing in the reference taxa. However, two of them that would comparably be predicted for Physcomitrella patens have not been confirmed in cDNA analysis of that moss (“0”). In our example using the primary display threshold of ≥3 hits, a few candidate sites were not included in the output list since they were identified in only two homologues among the references (eight sites in maturase-like ORFs and one site each in rpl2 and rps7). No editing events have previously been documented for those positions in any of the references.
Note that our editing site nomenclature allows for quick identification of certain types of editing events also in long outputs (eg, by searching for a particular gene or for changes converting two particular codon identities) using the Ctrl-F search functionality of the WWW browser.
Re-checking the references
No RNA editing has so far been described for the organelle genomes of green algae like Chara vulgaris and Chaetosphaeridium globosum and the liverwort Marchantia polymorpha, which were accordingly here included as mtDNA and cpDNA references. The absence of RNA editing in the latter is most likely due to a loss of RNA editing in the entire clade of marchantiid, ie, complex-thalloid liverworts,20 recently further supported by an extended taxon sampling for selected mitochondrial loci.21 On the other hand, the recent discovery of rare RNA editing in the mtDNA of the protist Naegleria gruberi7 suggests that one should be careful about assuming that there is a general absence of RNA editing in certain organelle genomes.
We re-inspected the algal and Marchantia plastome and chondrome sequences using the new PREPACT 2.0 possibilities. To this end we replaced them individually using the monocot Oryza sativa as a reference but otherwise used the same stringent parameters as in our above example scan for candidate RNA editing sites in the Anomodon rugelii mtDNA. This search strategy indeed revealed no strongly-predicted mitochondrial RNA editing in Chaetosphaeridium globosum. However, two sites of mitochondrial RNA editing were consistently predicted for Chara vulgaris and four were consistently predicted for Marchantia polymorpha, all of which are edited in at least one of the reference taxa. This finding strongly warrants eventual re-inspecting of at least those loci on the cDNA level in the alga and in the liverwort.
Along similar lines we investigated the chloroplast reference set and likewise found that checking on cDNA level of some candidate sites may be warranted for the algae (eg, rps2eU463HY, psaAeU244LF, ndhFeU503SF and ndhDeU1450PS in Chaetosphaeri dium globosum). Notably, only one codon-changing edit was reported for the Physcomitrella patens cpDNA22,23 but one particular striking candidate RNA editing site was additionally suggested in our PREPACT 2.0 analyses: atpBeU1322SF. Indeed, this position turned out to be a sequence error in the database accession to be corrected into a genomic thymidine soon (Dr. M. Sugita, Nagoya, personal communication).
Improving start and stop codon editing predictions in both directions
A previous limitation of the BLASTX approach implemented in PREPACT had been that start and stop codons, which would need to be established by C-to-U editing, would be missed because T versus M and Stop versus Q or R mismatches were not included at the ends of BLASTX hits. This issue is now successfully addressed in PREPACT 2.0: An adjustable number of codons (preset to 2, see Fig. 3) flanking the BLASTX hits is re-investigated a posteriori to scan for additional possibilities of improving similarities towards the ends of the reference protein sequences.
Conversely, removal of in-frame stop codons has previously only been suggested when arginine or glutamine codons conserved in the reference could be reconstituted by candidate reverse editing. A novel “always edit stop” option may be activated when U-to-C prediction of editing is switched on (see Supplemental Fig. 2), which always suggests the conversion of in-frame stop codons, even when other codon identities are present in the references.
Here we demonstrate the capabilities of these new features using the mitochondrial atp6 gene of Isoetes engelmannii since this extremely edited locus combines the requirements of introducing start and stop codons with multiple stop removals of in-frame stops.5 The RNA editing prediction is exemplarily shown in an alignment with the Marchantia polymorpha reference (Fig. 6). The introduction of start and stop codons by C-to-U editing is now reliably predicted, given the codon extension option. Conversely, the “always edit stop” option suggests editing events eC145*R and eC415*R alongside three others removing stops, although a leucine and a glutamine codon are placed opposite the arginines that can be introduced here by reverse U-to-C editing.
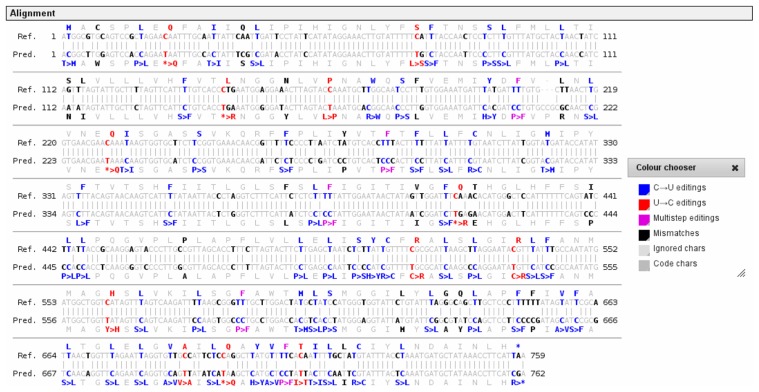
Figure 6.
Start and stop codon conversion in an extreme example of RNA editing.
The example shows the RNA editing prediction for the Isoetes engelmannii atp6 coding sequence (bottom), here exemplarily aligned with the Marchantia polymorpha reference (top). The extended capabilities of PREPACT 2.0 allow identification of C-to-U start and stop codon editing possibilities not included in the original BLASTX hits. In addition, they allow for the removal of two stop codons alongside three others by editing events atp6eC145*R and atp6eC415*R even though they are juxtaposed with a leucine and a glutamine, respectively, in the reference when the “always edit stop option” is activated. All 74 codon changes shown were confirmed on cDNA level. An updated version of the floating color picker, here shown next to the alignment, allows a yet more comfortable change of differentially coloring alignment positions in the WWW output.
In fact, all 74 codon edits as predicted from the alignment with Marchantia (and most of the other references) were indeed confirmed by cDNA analysis. Four additional sites of RNA editing detected in Isoetes atp6 cDNA were not predicted with Marchantia. Editing atp6eU430HY is an unexpected event with a histidine consistently conserved in all references and editing event atp6eU212PL is unpredictable, as it affects the inserted codon exclusive to Isoetes (Fig. 6). The two remaining editing events are moderately suggested from four in eight of the previous set of references (here again Oryza replacing Isoetes): atp6eUU280PF and atp6eU653PL.
Testing the predictions of RNA editing in entire organelle genomes
To obtain reasonable estimates for the success rates of C-to-U RNA editing prediction with the new PREPACT commons function, we ran the complete mtDNAs of the high editor Lotus japonicus and the low editor Silene vulgaris under the same conditions (ie, 75% threshold rate and a minimum of 3 parallel hits) as used above for the Anomodon rugelii mtDNA example with unknown editing. Since both chondromes are included in our references (Table 1), we selected Oryza sativa as an alternative additional reference, for comparability. These predictions based on the taxonomically wide sampling as used in the Anomodon example revealed sensitivities of 84% and 81%, respectively, to correctly identify the verified codon changes introduced by editing in Silene latifolia (total 267) and Lotus japonicus (total 524) at a rate of 3.7% and 4.2% false positives, respectively. To test how taxonomic proximity influences these results with respect to sensitivity and specificity, we replaced the phylogenetic wide sampling including the algae and non-seed plants with the full selection of available angiosperm chondrome references. In these runs, sensitivities rose to 90% and 92%, respectively, at the cost of only slightly decreased specificities with false positives still remaining below 5%.
We finally tested the functional innovations in PREPACT2 for predicting RNA editing in chloroplast DNA to its full extent, and to this end chose the Anthoceros formosae plastome, which is characterized as the most complex chloroplast editome yet published. More than 500 C-to-U RNA editing events and more than 400 U-to-C events have been identified in A. formosae chloroplasts, which altogether create 891 codon changes in chloroplast mRNAs.6 We performed test runs including prognosis of U-to-C editing with liberal stop codon removal, again using the previous 75% commons threshold settings (Supplemental Fig. 2). With this setup run against the set of other plastome references, we faithfully identified 89% of the verified codon-changing events at the cost of including 17% false positives. Hence, we consider the current preset threshold values to identify candidate edits (Fig. 3) a useful balance for sensitivity versus specificity. This is also the case for highly complex organelle transcriptomes showing pyrimidine exchanges in both directions, which include numerous stop codon removals and start and stop codon introductions by RNA editings.
User alignments with multiple references
The BLASTX mode of PREPACT 2.0 as described above will likely be of use to most users wishing to analyze new organelle sequence data. For comparative studies focusing on a single gene, however, the user may wish to provide sequence alignments containing multiple query sequences to be tested against multiple self-defined references. An example is a recent study investigating variable RNA editing frequencies among a wide sampling of liverworts and mosses in two selected mitochondrial loci.21 Such datasets may be used in the “alignment prediction” mode with reference sequences placed first in the alignment and their number indicated (Supplemental Fig. 3). In addition to the new analogous “commons” functionality as described above for the BLASTX mode, the alignment prediction mode of analysis also allows increased display functionality for highlighting and grouping of sites, as previously described for the differential cDNA analysis mode.9
Discussion
A simple hepta-peptide such as “PREPACT” may be encoded in 6,144 different ways by just considering the codon ambiguities of the universal genetic code. Allowing for pyrimidine editing, this number rises to an astounding 1,310,720 possibilities for alternative 21-nucleotide sequences on DNA level. The ambiguities of oligonucleotide primer design for cDNA amplification in heavily editing taxa are an obvious methodological consequence in the laboratory, while another major issue is the bioinformatic processing of editing.
No obvious evolutionary advantage has been associated with the abundant plant organelle RNA editing and as yet no reasonable explanation exists why hundreds or even thousands of edited RNA positions couldn’t be encoded as such in the DNA. Indeed, plant organelle RNA editing is currently best explained by a composite neutral evolution (CNE) model, which posits that an ancestral editing activity, functionally extended to operate on polyribonucleotide stretches allows the encoding genes to mutate more freely.24 Simple causes of genome evolution such as genomic GC or AT drifts may favor certain RNA editing mechanisms to affect more and more sites until they get evolutionarily “locked in” and can hardly be lost again. Frequent independent gains and losses of RNA editing positions in related taxa, mostly with corresponding pyrimidine transitions in the DNA, support the above hypothesis. The wide variability of RNA editing frequency ranging from apparently none in marchantiid liverworts,21 few sites in mosses, to thousands in lycophytes,4,5 support the idea of a freely evolving selfish mechanism.
Selfish as organelle RNA editing may seem, the functional implications in the cell are fascinating. The editing machinery has to select its RNA targets very specifically. Particular pentatricopeptide repeat (PPR) RNA-binding proteins are the obvious key players in recognizing the appropriate transcript sequences.25–27 Impressive steps forward have recently been made to decipher the code for sequence-specific RNA- binding encoded by the array of PPR repeats.28 Any such predictions of an RNA-binding code will, however, need testing against similar sites in an organelle transcriptome. Conversely, the identification of editing sites will prompt the quest for the appropriate RNA-binding protein factors. Along those lines, we hope that PREPACT may be of help to address this and similar issues, and possibly also reveal more cases of as yet hidden, unexpected RNA editing events like the ones recently elucidated for Naegleria gruberi.7
Here we have discussed the issues of proper annotation of RNA editing in primary database sequence accessions and have suggested a standard annotation that could be adapted in the primary databases in the future. Such a standard would significantly help with information exchange between the primary sequence repositories and efforts dedicated to the storage and analysis of data on RNA editing, such as PREPACT or other initiatives elsewhere.13,29–36 RNA editing database initiatives such as REDIdb include data on the investigation of single loci in taxa for which no extensive organelle transcriptome analyses have been performed. Aside from its RNA editing prognosis features, in the future PREPACT may hopefully concomitantly also serve as a database for curated full “editomes”, or “editotypes”,37 ie, the full inventories of editing sites for a given organelle genome. Querying PREPACT 2.0 with an organelle genome against its reference transcriptome will obviously reveal all RNA editing sites causing codon changes, and at the same time will allow for comparisons to be made to the editomes of other references.
For PREPACT 2.0 we have currently processed 19 mitochondrial and 13 chloroplast genomes as references in order to comfortably predict coding sequences and RNA editing in organelle sequence data. The major improvements of the new PREPACT 2.0 WWW server are (i) the exclusion of secondary BLASTX hits by filtering to avoid erroneous prediction of RNA editing from paralogs, (ii) the distinction of a lack of RNA editing prognosis from a simple lack of homology in individual references, (iii) the distinction of editing events from pre-edited sites in references through their re-organization as translated coding sequences from edited nucleotide sequences, (iv) the option to scan for potential introduction of start and stop codons by C-to-U editing beyond homologies identified as BLASTX hits, (v) the option of general stop-codon removal to create continuous reading frames irrespective of conserved arginine or glutamine codons in references, and finally (vi) the “commons” function relying on all of the above to summarize RNA editing predictions for fractions of the multiple-reference set according to threshold settings. Test runs with extensively analyzed mitochondrial transcriptomes show that success rates for faithful prognosis of mRNA editing may even exceed 90% at false positive rates below 5% when using the 75% commons threshold, which we currently recommend. Although prognoses benefit from taxonomically closer references, even a phylogenetically wide sampling including algae and non-seed land plants nevertheless reliably detects more than 80% of mRNA editing. Beyond the BLASTX-based functionality, PREPACT users may provide sequence alignments with multiple queries including multiple self-chosen references for predicting editing in individual loci using the same “commons” functionality. We hope to be able to provide timely updates of PREPACT 2.0 with the addition of novel organelle (transcriptome) reference data in the future, and that the PREPACT software will be of use to comfortably catalogue and predict RNA editing events in upcoming organelle sequences.
Supplementary Figures
The request status display indicating progress of an analysis after submission of a query.
Extensive requests employing large sequences and multiple output options may need several minutes for completion of the run and preparation of the output (see Figs. 4 and 5) which is indicated in several steps.
The PREPACT 2.0 query form exemplarily shown for the analysis of the Anthoceros formosae plastome.
All available chloroplast references are selected and the “always edit stop” option is included for analysis, which allows for proposal of reverse U-to-C editing to remove stop codons also where no arginine or glutamine codons are conserved in references. Additionally shown is the new quick choice colour selection option in the colour picker popup window for differential colouring of alignment positions (see Fig. 6).
The “Alignment prediction” mode for user-provided alignments of homologous organelle coding sequences in FASTA format.
Alignments may as respective first entries on top include any number (to be indicated, here: 3) of self-defined references. Like the differential cDNA analysis mode, the Alignment prediction mode also includes further options for comparative graphic display (bottom left).
Acknowledgements
We are grateful to our colleagues, Drs. A.B. Cremers and R. Manthey as well as M. Follwerk at the Fachgruppe Informatik of our faculty for providing the server environment for PREPACT, to members of our group for testing the program and to the countless people on the internet providing nifty code snippets for improvement of the world’s programs.
Footnotes
Author contributions
HL and VK designed the program features. HL did the programming and debugging. Both authors revised database entries, extracted RNA editing data from the literature and did test runs of the software. VK wrote the manuscript and both authors revised and approved the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
Author(s) disclose no funding sources.
References
- 1.Benne R, Van Den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46(6):819–26. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Knoop V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 2011;68(4):567–86. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bundschuh R, Altmüller J, Becker C, Nürnberg P, Gott JM. Complete characterization of the edited transcriptome of the mitochondrion of Physarum polycephalum using deep sequencing of RNA. Nucleic Acids Res. 2011;39(14):6044–55. doi: 10.1093/nar/gkr180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–58. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grewe F, Herres S, Viehöver P, Polsakiewicz M, Weisshaar B, Knoop V. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucl Acids Res. 2011;39(7):2890–902. doi: 10.1093/nar/gkq1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucl Acids Res. 2003;31(9):2417–23. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rüdinger M, Fritz-Laylin L, Polsakiewicz M, Knoop V. Plant-type mitochondrial RNA editing in the protist Naegleria gruberi. RNA. 2011;17(12):2058–62. doi: 10.1261/rna.02962911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knoop V, Rüdinger M. DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 2010;584(20):4287–91. doi: 10.1016/j.febslet.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Lenz H, Rüdinger M, Volkmar U, et al. Introducing the plant RNA editing prediction and analysis computer tool PREPACT and an update on RNA editing site nomenclature. Curr Genet. 2010;56(2):189–201. doi: 10.1007/s00294-009-0283-5. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V. RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genom. 2009;281(5):473–81. doi: 10.1007/s00438-009-0424-z. [DOI] [PubMed] [Google Scholar]
- 13.Picardi E, Regina TM, Verbitskiy D, Brennicke A, Quagliariello C. REDIdb: an upgraded bioinformatics resource for organellar RNA editing sites. Mitochondrion. 2011;11(2):360–5. doi: 10.1016/j.mito.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46(3):123–39. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 15.Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387(6632):493–7. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 16.Wakasugi T, Hirose T, Horihata M, Tsudzuki T, Kössel H, Sugiura M. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc Natl Acad Sci U S A. 1996;93(16):8766–70. doi: 10.1073/pnas.93.16.8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmans ML, Chaw SM, Lin CP, Shih AC, Wu YW, Mulligan RM. Editing site analysis in a gymnosperm mitochondrial genome reveals similarities with angiosperm mitochondrial genomes. Curr Genet. 2010;56(5):439–46. doi: 10.1007/s00294-010-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y, Wu H, Zhang T, et al. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PLoS One. 2012;7(5):e37164. doi: 10.1371/journal.pone.0037164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Xue JY, Wang B, Li L, Qiu YL. The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: dynamic and conservative evolution. PLoS One. 2011;6(10):e25836. doi: 10.1371/journal.pone.0025836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groth-Malonek M, Wahrmund U, Polsakiewicz M, Knoop V. Evolution of a pseudogene: exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in Marchantiidae. Mol Biol Evol. 2007;24(4):1068–74. doi: 10.1093/molbev/msm026. [DOI] [PubMed] [Google Scholar]
- 21.Rüdinger M, Volkmar U, Lenz H, Groth-Malonek M, Knoop V. Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria. J Mol Evol. 2012;74(1–2):37–51. doi: 10.1007/s00239-012-9486-3. [DOI] [PubMed] [Google Scholar]
- 22.Miyata Y, Sugiura C, Kobayashi Y, Hagiwara M, Sugita M. Chloroplast ribosomal S14 protein transcript is edited to create a translation initiation codon in the moss Physcomitrella patens. Biochim Biophys Acta. 2002;1576(3):346–9. doi: 10.1016/s0167-4781(02)00346-9. [DOI] [PubMed] [Google Scholar]
- 23.Miyata Y, Sugita M. Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J Plant Physiol. 2004;161(1):113–5. doi: 10.1078/0176-1617-01220. [DOI] [PubMed] [Google Scholar]
- 24.Gray MW. Evolutionary origin of RNA editing. Biochemistry. 2012;51(26):5235–42. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- 25.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433(7023):326–30. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 26.Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21(2):558–67. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salone V, Rüdinger M, Polsakiewicz M, et al. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581(22):4132–8. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 28.Barkan A, Rojas M, Fujii S, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8(8):e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mower JP. PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Jia L, Li Y. CURE-Chloroplast: a chloroplast C-to-U RNA editing predictor for seed plants. BMC Bioinformatics. 2009;10:135. doi: 10.1186/1471-2105-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J, Gopal S. Genetic algorithm learning as a robust approach to RNA editing site prediction. BMC Bioinformatics. 2006;7:145. doi: 10.1186/1471-2105-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du P, He T, Li Y. Prediction of C-to-U RNA editing sites in higher plant mitochondria using only nucleotide sequence features. Biochem Biophys Res Commun. 2007;358(1):336–41. doi: 10.1016/j.bbrc.2007.04.130. [DOI] [PubMed] [Google Scholar]
- 33.Du P, Li Y. Prediction of C-to-U RNA editing sites in plant mitochondria using both biochemical and evolutionary information. J Theor Biol. 2008;253(3):579–86. doi: 10.1016/j.jtbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Cummings MP, Myers DS. Simple statistical models predict C-to-U edited sites in plant mitochondrial RNA. BMC Bioinformatics. 2004;5:132. doi: 10.1186/1471-2105-5-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picardi E, Regina TM, Brennicke A, Quagliariello C. REDIdb: the RNA editing database. Nucl Acids Res. 2007;35(Database issue):D173–D7. doi: 10.1093/nar/gkl793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mower JP. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucl Acids Res. 2009;37(Web Server issue):W253–W9. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol. 2002;19(9):1602–12. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- 38.Giegé P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA. 1999;96(26):15324–9. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mower JP, Palmer JD. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Genet Genom. 2006;276(3):285–93. doi: 10.1007/s00438-006-0139-3. [DOI] [PubMed] [Google Scholar]
- 40.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucl Acids Res. 2003;31(20):5907–16. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol Biol Evol. 2010;27(6):1436–48. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genom. 2002;268(4):434–45. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 43.Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. Extensive loss of RNA editing sites in rapidly evolving silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics. 2010;185(4):1369–80. doi: 10.1534/genetics.110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, Pesole G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucl Acids Res. 2010;38(14):4755–67. doi: 10.1093/nar/gkq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf PG, Rowe CA, Hasebe M. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 46.Tillich M, Funk HT, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, et al. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 2005;43(5):708–15. doi: 10.1111/j.1365-313X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- 47.Tangphatsornruang S, Uthaipaisanwong P, Sangsrakru D, Chanprasert J, Yoocha T, Jomchai N, et al. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene. 2011;475(2):104–12. doi: 10.1016/j.gene.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 48.Sasaki T, Yukawa Y, Miyamoto T, Obokata J, Sugiura M. Identification of RNA editing sites in chloroplast transcripts from the maternal and paternal progenitors of tobacco (Nicotiana tabacum): comparative analysis shows the involvement of distinct trans-factors for ndhB editing. Mol Biol Evol. 2003;20(7):1028–35. doi: 10.1093/molbev/msg098. [DOI] [PubMed] [Google Scholar]
- 49.Hirose T, Kusumegi T, Tsudzuki T, Sugiura M. RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol Gen Genet. 1999;262(3):462–7. doi: 10.1007/s004380051106. [DOI] [PubMed] [Google Scholar]
- 50.Inada M, Sasaki T, Yukawa M, Tsudzuki T, Sugiura M. A systematic search for RNA editing sites in pea chloroplasts: an editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol. 2004;45(11):1615–22. doi: 10.1093/pcp/pch191. [DOI] [PubMed] [Google Scholar]
- 51.Corneille S, Lutz K, Maliga P. Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Mol Gen Genet. 2000;264(4):419–24. doi: 10.1007/s004380000295. [DOI] [PubMed] [Google Scholar]
- 52.Tsudzuki T, Wakasugi T, Sugiura M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J Mol Evol. 2001;53(4–5):327–32. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- 53.Maier RM, Zeltz P, Kössel H, Bonnard G, Gualberto JM, Grienenberger JM. RNA editing in plant mitochondria and chloroplasts. Plant Mol Biol. 1996;32(1–2):343–65. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The request status display indicating progress of an analysis after submission of a query.
Extensive requests employing large sequences and multiple output options may need several minutes for completion of the run and preparation of the output (see Figs. 4 and 5) which is indicated in several steps.
The PREPACT 2.0 query form exemplarily shown for the analysis of the Anthoceros formosae plastome.
All available chloroplast references are selected and the “always edit stop” option is included for analysis, which allows for proposal of reverse U-to-C editing to remove stop codons also where no arginine or glutamine codons are conserved in references. Additionally shown is the new quick choice colour selection option in the colour picker popup window for differential colouring of alignment positions (see Fig. 6).
The “Alignment prediction” mode for user-provided alignments of homologous organelle coding sequences in FASTA format.
Alignments may as respective first entries on top include any number (to be indicated, here: 3) of self-defined references. Like the differential cDNA analysis mode, the Alignment prediction mode also includes further options for comparative graphic display (bottom left).