Abstract
Over 100 genotypes of human papillomaviruses (HPVs) have been identified as being responsible for unapparent infections or for lesions ranging from benign skin or genital warts to cancer. The pathogenesis of HPV results from complex relationships between viral and host factors, driven in particular by the interplay between the host proteome and the early viral proteins. The E2 protein regulates the transcription, the replication as well as the mitotic segregation of the viral genome through the recruitment of host cell factors to the HPV regulatory region. It is thereby a pivotal factor for the productive viral life cycle and for viral persistence, a major risk factor for cancer development. In addition, the E2 proteins have been shown to engage numerous interactions through which they play important roles in modulating the host cell. Such E2 activities are probably contributing to create cell conditions appropriate for the successive stages of the viral life cycle, and some of these activities have been demonstrated only for the oncogenic high-risk HPV. The recent mapping of E2-host protein-protein interactions with 12 genotypes representative of HPV diversity has shed some light on the large complexity of the host cell hijacking and on its diversity according to viral genotypes. This article reviews the functions of E2 as they emerge from the E2/host proteome interplay, taking into account the large-scale comparative interactomic study.
Keywords: HPV, E2, protein interactions, network, viral pathogenesis, productive life cycle, cervical cancer.
E2 PROTEIN IN THE HPV LIFE CYCLE AND PATHOGENESIS
The E2 protein is pivotal for both the productive cycle and persistent infections by human papillomaviruses (HPVs). The productive cycle causes benign hyperproliferative lesions (genital or cutaneous warts). However, some lesions have the potential to progress toward malignancies in case of unresolved infection with high-risk HPV (HR-HPV). Carcinogenic conversion is a complex process occurring over long periods of time, thus requiring long-lasting infections. Indeed, persistence of HR-HPV is considered as a major risk factor for cancer development.
The E2 proteins have been mostly studied for the genital α-type HPV, more particularly HR-HPV 16 and 18, which are associated with about 70% of HPV-induced genital cancers and therefore represent major health concerns. Less information is available about E2 of low-risk genital HPV (LR-HPV) or of cutaneous HPVs. Yet, the complete deciphering of E2 activities in the productive life cycle is still required to find a way to eradicate genital and cutaneous warts. In addition, decoding the roles of E2 in viral persistence is crucial to improve the comprehension of the HR-HPV associated carcinogenic conversion, which arises from persistently infected cells. It also may help to understand the pathology of some cutaneous HPVs which persist without generating lesions (asymptomatic infections) in the general population, and trigger productive infection only in immunocompromised individuals.
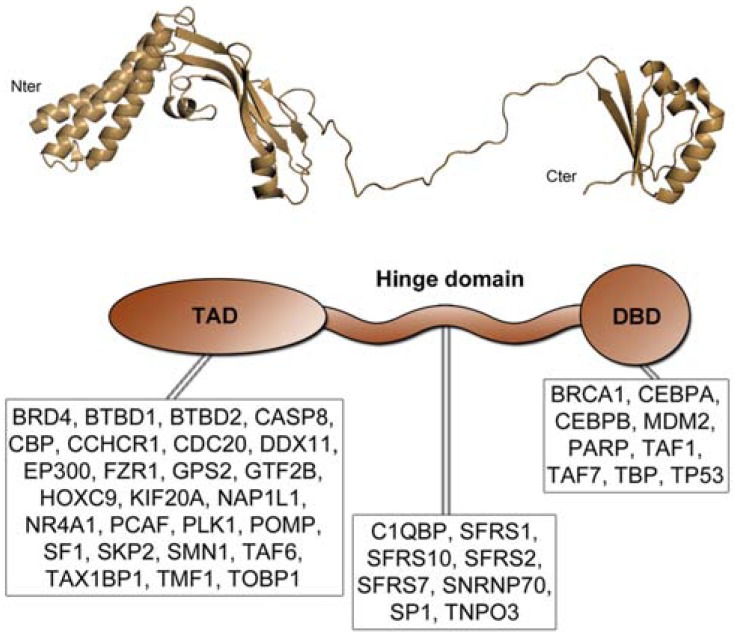
The E2 proteins contain 350 to 500 amino acids consisting of two conserved modular domains, a 200 amino acids N-terminal domain referred as TAD (TransActivation Domain), and a 100 amino acid C-terminal DNA Binding Domain (DBD). These two domains are separated by an unstructured Hinge region, not conserved and of variable length among the different HPV genotypes. The crystal structure of the two conserved domains has been determined for several genital α-HPV E2 proteins [1, 2], but no information is available concerning the structure of E2 from the cutaneous HPVs.
The DNA binding domain of E2 exhibits a typical three-dimensional structure, which has homology to other viral factors and is considered prototypic of some viral regulation factors [3]. This domain contains both a strong dimerization interface and amino acids making contacts with DNA located along an α-helix [1]. Indeed, E2 binds as a dimer to specific palindromic DNA sequences ACCGN4 CGGT designated E2 binding sites (E2BS). At least four of these E2BS are located within the regulatory region (LCR for Long Control Region) of all HPV genomes [4] (Fig. 1). Through the binding to the regulatory region, the E2 protein recruits at the viral genome a range of host cell factors necessary to support its transcription, replication, and mitotic segregation. By binding to the same E2BS, E2 ensures the coordinated regulation of viral DNA transcription and replication, thereby appearing as a pivotal factor of the early/late switch of the productive viral life cycle. An effect on mRNA splicing has been detected as well, suggesting that E2 controls viral gene expression through both transcription regulation and mRNA processing.
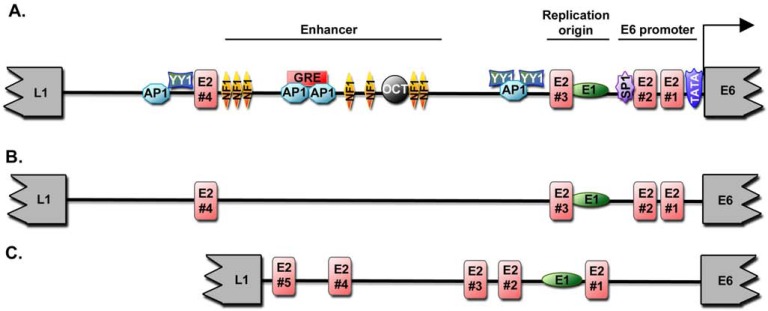
Fig. (1).
HPV Long Control Region. (A) Schematic map of the long control region of HPV16. The end of the L1 and beginning of the E6 ORF are indicated. The E2 binding sites (E2BS) are numbered from 1 to 4 and colored in red. (B) Schematic representation of a prototypic genital HPV LCR with the position of the E2 and E1 binding sites. (C) Schematic representation of a prototypic cutaneous HPV LCR with the position of the E2 and E1 binding sites.
Several groups have demonstrated a number of additional functions that are independent of E2 binding to the viral genome, such as NFκB activation [5], induction of apoptosis (for review [6]) or regulation of the host cell cycle [7]. These “autonomous” activities directly impact on the host cells and are likely to be required for the implementation of the productive viral life cycle within stratified epithelia. For example, the stimulation of TNF-induced NFκB activation by E2 was proposed to play a role in the commitment of infected keratinocytes to activation of the differentiation program, which is necessary for the productive phase of viral life cycle [5]. Some of these autonomous E2 functions are specific to HR-HPV, leading to the notion that the E2 proteins have specially developed activities in correlation with the oncogenic power of HPV. For example, a proapoptotic activity is specifically associated with the E2 proteins of HR-HPV and not with the LR-HPV [6, 8, 9]. It was hypothesized that the HR-HPV E2 proteins evolved apoptotic activities to counterbalance the transforming properties of the E6/E7 oncoproteins, which would otherwise be detrimental for progression of viral life cycle. In addition, the HR-HPV E2 proteins cause a G2/M cell cycle arrest, which can induce genomic instability, recognized as a precursor of carcinogenesis [7]. In the case of cutaneous HPV8, E2 was shown to be sufficient to induce the development of skin cancer in transgenic mice [10]. The correlation between E2 and modulation of mechanisms involved in cell transformation has raised the hypothesis that E2 may have a direct role in the early steps of carcinogenic conversion, and thus contribute to the oncogenic potential of HR-HPV in addition to E6 and E7. In late stages of carcinogenesis, the genital HPV genome is integrated into the host chromatin, with the preferential disruption of the E2 gene [11, 12]. The E6/E7 genes are subsequently upregulated and trigger key events for cell transformation. During the advanced steps of malignant progression, the pro-apoptotic activity of HR-HPV E2 proteins may favor the selective inactivation of E2-expressing cells, thereby promoting the clonal selection of E6/E7 over-expressing cells. The role of E2 in the different stages of HPV-associated carcinogenesis has been extensively discussed recently [13].
THE E2 INTERACTOME
Like other viral proteins, E2 exerts its functions by targeting different cell factors. Indeed, almost all of the activities associated with E2 have been linked to direct interaction with host cell factors, either to support viral genome expression, segregation and replication, or to provide conductive cell conditions.
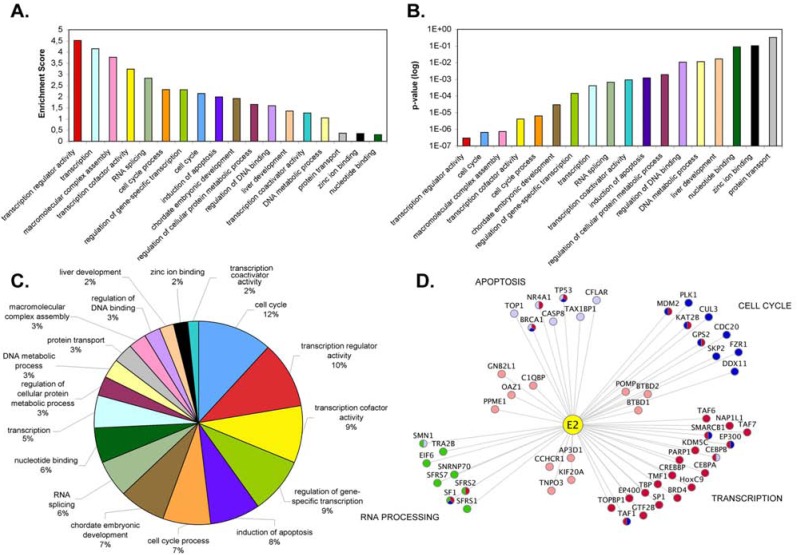
To date, most of the interaction data concerning the E2 proteins arose from separated studies of E2-associated functions. In total, we referenced 57 cellular proteins binding to one or several E2 proteins by database mining (VirHostNet, VirusMINT and PubMed; Table 1). Given the major public health concern of cervical cancers, prior studies have primarily focused on E2 proteins from the mucosal HR-HPV HPV16 and HPV18, creating a bias toward the most studied E2 proteins. Interactions were detected with the three domains of E2, although most of them concerned the E2 TAD (Fig. 2). To get further insights into the E2-host interplay, we merged all known E2 interactions to build a network, and conducted an analysis based on the GO (Gene Ontology) terms associated with the E2-targeted cellular proteins. For that aim, we used the DAVID database [14], which generates a functional annotation of the data and allows the clustering of similar annotations into functional groups. 18 clusters have been identified within the list of published E2 interacting partners. Fig. (3) represents the parameters of each cluster: enrichment score, p-value and prevalence (percentage of GO-terms included in the cluster). By combining these parameters, four main functional families emerged presenting overall the highest criteria (high enrichment score, low p-value and high prevalence): transcription regulation, RNA processing, apoptosis and modulation of the cell cycle (Fig. 3D). This analysis provides insight into the range of E2 functions arising from E2-host interplay, but because of the limited number of genotypes represented, it gives only restricted information regarding HPV pathogenesis.
Table 1.
Literature Curated Interactions. List of Interactions Found for the HPV E2 Proteins in the VirHostNet, virusMINT and PubMed Databases
| ENSembl ID | Official Gene Symbol | Name | Synonyms | HPV Type | Refs. |
|---|---|---|---|---|---|
| ENSG00000065000 | AP3D1 | adaptor-related protein complex 3, delta 1 subunit | ADTD | HPV16 | [123] |
| ENSG00000012048 | BRCA1 | breast cancer 1, early onset | BRCC1, PPP1R53, RNF53 | HPV18 | [124] |
| ENSG00000141867 | BRD4 | bromodomain containing 4 | CAP, HUNK1, HUNKI, MCAP | HPV16,18,31,11,6,8,1 | [55, 125, 126] |
| ENSG00000064726 | BTBD1 | BTB (POZ) domain containing 1 | HPV16 | [90] | |
| ENSG00000133243 | BTBD2 | BTB (POZ) domain containing 2 | HPV16 | [90] | |
| ENSG00000108561 | C1QBP | complement component 1, q subcomponent binding protein | gC1Q-R, gC1qR, HABP1, p32, SF2p32 | HPV5 | [60] |
| ENSG00000064012 | CASP8 | caspase 8, apoptosis-related cysteine peptidase | Casp-8, FLICE, MACH, MCH5 | HPV16,18 | [66] |
| ENSG00000206355 | CCHCR1 | coiled-coil alpha-helical rod protein 1 | C6orf18, HCR | HPV16 | [90] |
| ENSG00000117399 | CDC20 | cell division cycle 20 homolog | CDC20A, p55CDC | HPV16,18 | [7] |
| ENSG00000245848 | CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha | C/EBP-alpha, CEBP | HPV16,18,8 | [103] |
| ENSG00000172216 | CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | C/EBP-beta, CRP2, IL6DBP, LAP, NFIL6, TCF5 | HPV16,18,8 | [103] |
| ENSG00000003402 | CFLAR | CASP8 and FADD-like apoptosis regulator | c-FLIP, CASH, CASP8AP1, Casper, CLARP, FLAME, FLIP, I-FLICE, MRIT | HPV16 | [68] |
| ENSG00000160917 | CPSF4 | cleavage and polyadenylation specific factor 4 | CPSF30, NAR | HPV16 | [62] |
| ENSG00000005339 | CREBBP | CREB binding protein | CBP, KAT3A, RSTS, RTS | HPV18 | [31] |
| ENSG00000036257 | CUL3 | cullin 3 | HPV16 | [88] | |
| ENSG00000013573 | DDX11 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 | CHL1, CHLR1, KRG2 | HPV16,11 | [56] |
| ENSG00000242372 | EIF6 | eukaryotic translation initiation factor 6 | b(2)gcn, EIF3A, ITGB4BP, p27BBP | HPV16 | [123] |
| ENSG00000100393 | EP300 | E1A binding protein p300 | KAT3B, p300 | HPV18,5,8 | [32] |
| ENSG00000183495 | EP400 | E1A binding protein p400 | CAGH32, DKFZP434I225, KIAA1498, KIAA1818, P400, TNRC12 | HPV16 | [23] |
| ENSG00000105325 | FZR1 | fizzy/cell division cycle 20 related 1 | CDC20C, CDH1, FZR, FZR2, HCDH, HCDH1, KIAA1242 | HPV16,18 | [7] |
| ENSG00000204628 | GNB2L1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | Gnb2-rs1, H12.3, RACK1 | HPV16 | [123] |
| ENSG00000132522 | GPS2 | G protein pathway suppressor 2 | HPV16 | [127] | |
| ENSG00000137947 | GTF2B | general transcription factor IIB | TFIIB | HPV16 | [128] |
| ENSG00000180806 | HoxC9 | homeobox C9 | HOX3, HOX3B | HPV16 | [90] |
| ENSG00000114166 | KAT2B | K(lysine) acetyltransferase 2B | GCN5, GCN5L, P/CAF, PCAF | HPV16,18,11,6 | [30] |
| ENSG00000126012 | KDM5C | lysine (K)-specific demethylase 5C | DXS1272E, JARID1C, SMCX, XE169 | HPV16 | [23] |
| ENSG00000112984 | KIF20A | kinesin family member 20A | RAB6KIFL | HPV16,11 | [57] |
| ENSG00000135679 | MDM2 | Mdm2 p53 binding protein homolog | HDM2, HDMX, MGC5370 | HPV16 | [129] |
| ENSG00000187109 | NAP1L1 | nucleosome assembly protein 1-like 1 | MGC23410, MGC8688, NAP1, NAP1L, NRP | HPV18,5,8 | [34] |
| ENSG00000123358 | NR4A1 | nuclear receptor subfamily 4, group A, member 1 | GFRP1, HMR, N10, NAK-1, NGFIB, NUR77, TR3 | HPV16 | [70] |
| ENSG00000104904 | OAZ1 | ornithine decarboxylase antizyme 1 | AZI, MGC138338, OAZ | HPV16 | [123] |
| ENSG00000143799 | PARP1 | poly (ADP-ribose) polymerase 1 | ADPRT, PARP, PPOL | HPV18 | [130] |
| ENSG00000166851 | PLK1 | polo-like kinase 1 | PLK | HPV5 | [131] |
| ENSG00000132963 | POMP | proteasome maturation protein | C13orf12, HSPC014, UMP1 | HPV16 | [123] |
| ENSG00000214517 | PPME1 | protein phosphatase methylesterase 1 | PME-1 | HPV16 | [123] |
| ENSG00000168066 | SF1 | splicing factor 1 | ZFM1, ZNF162 | HPV16 | [90] |
| ENSG00000136450 | SFRS1 | serine/arginine-rich splicing factor 1 | ASF, MGC5228, SF2, SF2p33, SFRS1, SRp30a | HPV5 | [60] |
| ENSG00000161547 | SFRS2 | serine/arginine-rich splicing factor 2 | PR264, SC-35, SC35, SFRS2, SFRS2A | HPV5 | [60] |
| ENSG00000115875 | SFRS7 | serine/arginine-rich splicing factor 7 | 9G8, AAG3, HSSG1, RBM37, SFRS7, ZCCHC20, ZCRB2 | HPV5 | [60] |
| ENSG00000145604 | SKP2 | S-phase kinase-associated protein 2 (p45) | FBL1, FBXL1 | HPV18 | [85] |
| ENSG00000099956 | SMARCB1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | BAF47, hSNFS, Ini1, RDT, Sfh1p, SNF5L1, Snr1 | HPV18 | [33] |
| ENSG00000172062 | SMN1 | survival of motor neuron 1, telomeric | BCD541, SMA, SMA1, SMA2, SMA3, SMA@, SMNT | HPV16,18,11 | [64] |
| ENSG00000104852 | SNRNP70 | small nuclear ribonucleoprotein 70kDa (U1) | RNPU1Z, RPU1, Snp1, SNRP70, U1-70K | HPV5 | [58] |
| ENSG00000185591 | SP1 | Sp1 transcription factor | HPV18,8 | [39. 132] | |
| ENSG00000147133 | TAF1 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | BA2R, CCG1, CCGS, DYT3, DYT3/TAF1, KAT4, NSCL2, TAF2A, TAFII250 | HPV16,18 | [133] |
| ENSG00000106290 | TAF6 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | MGC:8964, TAF2E, TAFII70, TAFII80, TAFII85 | HPV18 | [134] |
| ENSG00000178913 | TAF7 | TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa | TAF2F, TAFII55 | HPV5 | [135] |
| ENSG00000106052 | TAX1BP1 | Tax1 (human T-cell leukemia virus type I) binding protein 1 | CALCOCO3, TXBP151 | HPV16,18 | [91] |
| ENSG00000112592 | TBP | TATA box binding protein | GTF2D1, SCA17, TFIID | HPV16,8 | [135] |
| ENSG00000144747 | TMF1 | TATA element modulatory factor 1 | ARA160, TMF | HPV16 | [90] |
| ENSG00000064419 | TNPO3 | transportin 3 | IPO12, MTR10A, TRN-SR, TRN-SR2 | HPV5 | [60] |
| ENSG00000198900 | TOP1 | topoisomerase (DNA) I | HPV16 | [78] | |
| ENSG00000163781 | TOPBP1 | topoisomerase (DNA) II binding protein 1 | KIAA0259, TOP2BP1 | HPV16 | [136] |
| ENSG00000141510 | TP53 | tumor protein p53 | LFS1, p53 | HPV16,18 | [9,137] |
| ENSG00000136527 | TRA2B | transformer 2 beta homolog | Htra2-beta, SFRS10 | HPV5 | [60] |
| ENSG00000082512 | TRAF5 | TNF receptor-associated factor 5 | RNF84 | HPV1,3,5,6,8,9,11,16,18,33,39 | [5] |
| ENSG00000175104 | TRAF6 | TNF receptor-associated factor 6, E3 ubiquitin protein ligase | RNF85 | HPV1,3,5,6,8,9,11,16,18,33,39 | [5] |
Fig. (2).
E2 proteins structure and interactions. HPV16 E2 threedimensional structure representing the three domains: Transactivation domain in the N-terminal part (TAD); Hinge unstructured domain in the centre, and DNA binding domain in the C-terminal part (DBD). A schematic representation of E2 is depicted below the 3D structure, with E2-interacting cellular proteins assigned to the interaction domain when it was determined.
Fig. (3).
Functional gathering of E2 interacting partners. DAVID analysis was performed on the E2 known interacting partners based on their associated GO (Gene Ontology) terms. Several parameters were taken into account to extract the four main categories: high enrichment score (A), low p-value (B), and high prevalence (C). (D) Representation of the four main functional families targeted by E2. Cellular proteins shared by different categories are multi-colored. The network representation was generated by Cytoscape [122].
In order to have a more global outlook of E2 interactions, we recently performed a comparative interactomic assay with the E2 proteins from 12 genotypes representative of HPV diversity in terms of tropism (cutaneous and mucosal) and pathogenic potential (high risk or low risk) and spanning over three HPV clades (α, α and μ HPV) [15]. It consisted of a large-scale identification of E2-interaction partners by yeast two-hybrid, followed by validation of the interactions in mammalian cells with the 12 E2 proteins. 98 cellular proteins were found to interact with at least one of the E2 proteins, of which 76 were new cellular E2 partners. This approach provided the first overview of E2 activities across multiple genotypes. We will discuss below the E2 functions in relation to E2-host protein-protein interactions, in light with the recently performed large-scale interactomic study.
E2 and Transcription Regulation
The most represented category of E2 partners emerging from known interactions is Transcription, which appears in several clusters, involving both many GO term associations and numerous E2 partners (Fig. 3). Not surprisingly, such prevailing targeting of cellular transcriptional machinery was reproduced in the interactome with the 12 E2 proteins, substantiating the prominent role of E2 as a transcriptional regulator.
E2 regulates HPV gene transcription either positively or negatively. The regulation of the early promoter transcription depends on the relative position of E2BS along the regulatory region. Within the genital HPV group, two E2BS overlap sequences of the early promoter. In such configuration, E2 acts as a repressor: by binding to these sites, E2 interferes with the binding of the cellular transcription factors TBP and SP1 by steric hindrance [16-18]. Conversely, in the LCR of cutaneous HPVs the E2BS are located farther from the early transcription initiation start, which leads to an overall transcriptional activation by E2 [19]. In addition, the stability of E2/DNA complexes differ according to the sequence of E2BS [16, 20, 21], and this has been proposed to modulate the effect of E2 on early promoter transcription [22].
The early promoter repression is not only due to steric hindrance, but is also the outcome of E2 binding to cellular proteins involved in the recruitment of repressive complexes on the LCR. For example, interaction of E2 with transcriptional regulatory proteins as the BRD4 protein, the histone demethylase SMCX, and the EP400 component of histone acetyltransferase complex have been shown to be required for efficient repression of the HPV18 LCR [23]. In line with these observations, we found in the comparative E2 interactome analysis a high proportion of targeted factors categorized as transcription repressors (Fig. 4B), highlighting the importance of repression in E2-mediated transcriptional regulation [15].
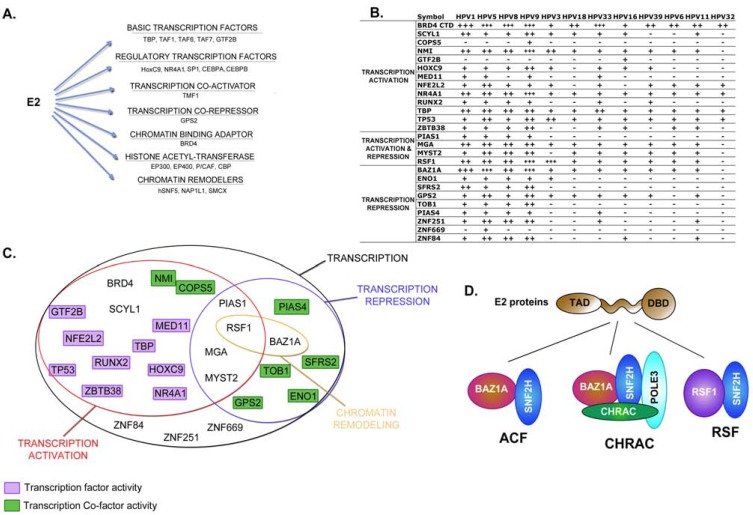
Fig. (4).
Complex regulation of transcriptional mechanisms. (A) Known E2 interacting partners of the transcription family and their associated function either as transcription regulators or chromatin factors. (B) Summary of the interaction strength detected between 12 E2 proteins and cellular proteins involved in regulation of transcription. The symbol “-” stands for a lack of interaction and from “+” to “+++”, an increasing strength of interaction as measured in [15] (C) Gathering of E2 targets involved in transcription regulation by main function. (D) Representation of E2 association with the SNF2H-related chromatin remodeling complexes.
In addition, the E2 proteins are assumed to activate transcription from the late HPV promoter, which depends on epithelial differentiation and is not easily accessible experimentally. Consequently, little is known about the regulation of late genes by E2 in the context of viral life cycle. The transcriptional activation capacities of E2 were mostly studied using synthetic promoters containing multimerized E2BS upstream of a minimal promoter, or in in vitro transcription assays. It was determined that the HPV E2 proteins can activate promoters over large distances [24], and exhibit different transcriptional activation capacities. Notably the high risk E2 proteins were found to be intrinsically more potent transcriptional activators than the low risk E2 proteins [25]. These differences might result from different interplays between the E2 proteins and the host cell machinery.
Interactions detected with E2 over a wide series of studies identified a large spectrum of transcriptional targets, ranging from basic factors to chromatin-related factors (Fig. 4A), suggesting that the E2 proteins are able to interfere at multiple levels of transcriptional processes.
Proteins with transcriptional activation properties have been shown to act, at least partially, by manipulating stability and assembly of preinitiation complexes [26]. TFIID is a pivotal component of the cellular transcription initiation complex, and is composed of TBP and TBP-associated factors (TAF) [27]. Regulatory, sequence-specific transcription factors bound in promoter regions allow efficient recruitment of the preinitiation complex through co-activators, mediating protein-protein interactions between regulatory transcription factors and the basal transcription machinery. As depicted in Fig. (4A), several interactions have been detected between E2 and both basal (TAF, TBP, GTF2B) or regulatory (SP1, HOXC9, NR4A1, C/EBP) transcription factors, as well as with co-activators (TMF), indicating that transcriptional properties rely, at least partly, on modulation of preinitiation complex formation on specific promoters.
Other targets of E2 act at the chromatin level to regulate transcription. Indeed, the best-characterized partner necessary for E2-dependent transcription is BRD4, which binds to acetylated histones and stimulates RNA polymerase II-dependent transcription by recruiting distinct transcriptional regulators [28]. Mutation of the amino acids responsible for the interaction with BRD4 (notably I73 and R37 for HPV16 E2 [29]) strongly affects the transcriptional capacities of E2, making BRD4 an essential mediator of E2’s transcriptional properties [28].
In addition, the E2 proteins are able to modulate transcription through modification of the histone code, by targeting histone-modification factors. Notably, several histone acetyltransferases PCAF, EP300, EP400 or CBP were shown to interact with E2 [30-32]. Histone acetyltransferases function within macromolecular complexes, recruited to their target promoters by interacting with sequence specific transcription factors. Histone acetylation both loosens nucleosome-DNA interactions and promotes interaction of modified histones with other transcription activators proteins.
Lastly, the HPV E2 proteins interact with members of chromatin remodeling complexes, involved in the deposition or displacement of nucleosomes. Two members of chromatin remodeling complexes were known E2 targets, NAP1L1 and hSNF5 [33, 34]. NAP1L1 is primarily involved in replication-coupled nucleosome assembly by mediating the incorporation of histones H2A-H2B dimers in nucleosomes. Its direct binding to E2 from genotypes 5, 8 and 18 has been shown to enhance their transcriptional activation capacities [34], though this interaction might primarily impact on the replication activating functions of E2. On the other hand, the SWI/SNF chromatin remodeling complex was recently shown to enhance HPV18 E2-dependant transcription through direct binding of the core component hSNF5 to E2 [33]. Such interactions might counteract the repressive effect of chromatin through the recruitment of chromatin remodeling complexes to the LCR, in order to regulate transcription.
Numerous interactions thus emerged from studies performed over time, giving clues to some characteristics of E2 transcriptional functions, particularly the broad range of targeted transcriptional processes. However, as discussed earlier, it is difficult to determine to what extent these characteristics are conserved, given that they were described only with a restricted number of HPV genotypes.
The recent comparative mapping of the E2-host protein-protein interactions allowed the identification of 26 transcription regulators targeted by E2, showing a partial overlap with known E2 partners consisting of 8 proteins. It provides an experimental assessment of the interplay between E2 and the transcriptional machinery across multiple genotypes. We were surprised that in fact, if consistently predominant for all genotypes tested, the targeting of cellular transcription factors was quite heterogeneous among different E2 proteins (Fig. 4B, C) [15]. Panel B in Fig. (4) shows the positive interactions engaged by the different E2 proteins and transcription factors, in correlation with the associated regulatory functions provided by their associated GO terms. Overall, our study corroborated the targeting of a large spectrum of transcription processes by all E2 proteins, which therefore likely represents a common trait of E2 transcriptional function. The range of E2 transcriptional targets encompasses transcription factors and cofactors, associated with transcriptional activation, repression, or possibly with both depending on the promoter context. This analysis highlights a prominent targeting of activators, representing more than half of the partners for all E2 proteins. As noted earlier, E2 also interacted with cellular proteins dedicated to transcriptional repression, and this targeting was more pronounced for the cutaneous β-type E2 proteins than for the α-type E2 proteins.
10 transcription factors are targeted by almost all E2 proteins, and BRD4 was consistently found among the strongest interactors [15]. These common targets may represent a “transcription core”, essential for the transcriptional functions of all E2 proteins. Of the common targets, only the basic transcription factor TBP binds to the HPV regulatory region. The other sequence-specific regulatory transcription factors of this core are TP53, MGA and NR4A1. TP53 primarily activates the transcription of cell cycle arrest or cell death genes [35]. MGA functions either as a repressor or as an activator of MYC-MAX target genes through binding to core E-box DNA sequences upon heterodimerization with the MAX transcription factor [36]. NRA41 (NUR77) is an Orphan nuclear receptor that may act by binding to NGFI-B response element in promoters of cellular genes [37]. Given that none of these factors harbor specific binding sites in HPV LCR, their interactions with the E2 proteins may primarily impact on the transcription of cellular genes rather than of viral genes. It thus emerges that part of E2 transcriptional functions might be involved in the modulation of expression of a common set of cellular genes. Several E2 proteins were indeed shown to affect the transcription of cellular genes [38-42], which could contribute to create cell conditions appropriate for the successive stages of the viral life cycle.
Other factors of the transcription core are co-activators such as MYST2, NMI or the GPS2 co-repressor. GPS2 is a subunit of the NCoR co-repressor complex [43] and its binding to E2 may be involved in the repression of HPV LCR, or affect the expression of NCoR1-repressed genes. NMI increases coactivator protein recruitment to sequence-specific transcription factors [44] and thus may be involved in E2-dependent transcriptional activation. MYST2 belongs to the histone H4-specific acetyltransferase complex HBO1, which acts as a coactivator of TP53-dependent transcription but has also been shown to specifically repress Androgen-Receptor (AR)-mediated transcription [45]. It is thought to regulate DNA replication through chromatin acetylation, and therefore its interaction with E2 may be involved in viral DNA replication, in addition to regulation of transcription.
The two remaining commonly targeted factors are the RSF1 and BAZ1A components of chromatin-remodeling complexes. Interestingly, both factors are part of chromatin remodeling complexes containing the SNF2H helicase. SNF2H is the catalytic subunit of four chromatin-remodeling complexes: CHRAC, RSF, ACF/WCRF, and WICH, which all contain other subunits regulating the specificity or catalytic activity of SNF2H. BAZ1A and RSF1 are accessory, non-catalytic subunits of three of the SNF2H-related complexes: ACF, CHRAC and RSF (Fig. 4D). The RSF chromatin remodeling complex is primarily involved in transcription regulation by mediating nucleosome deposition and positioning along regular arrays. It can either facilitate transcription or direct transcriptional repression by interacting with transcription factors, as was shown for the HBV pX protein [46]. Its interaction with a wide range of E2 proteins suggests that it may have a general role for E2 transcription functions, potentially both in repression and activation. RSF possesses not only chromatin remodeling activity but also chromatin assembly activity in vitro, and it has been proposed that RSF plays a role in silent chromatin formation [47]. Its recruitment by E2 could thus be involved in long-term silencing of HPV genome associated with viral persistence. Targeting of BAZ1A could have implications in E2-mediated transcriptional repression as well. Indeed, BAZ1A has been shown to interact with NCoR1 [48, 49]. E2 binding to both GPS2 and BAZ1A thus highlights a double targeting of the NCoR complex, which underlines the importance of NCoR-mediated repression for E2 transcriptional functions during infection. In addition, SNF2H forms the ACF complex with BAZ1A, which generates evenly spaced nucleosomes, creating a repressive state of chromatin [50, 51]. It is also involved in DNA replication, in particular it facilitates replication of heterochromatin regions [49]. Together with POLE3 and CHRAC subunits, BAZ1A and SNF2H assemble in the CHRAC complex, which also sets up repressive chromatin, and is involved in nucleosome assembly upon DNA replication [52, 53].
Overall, we propose that factors of the transcription core are instrumental for the transcriptional functions of all E2 proteins, and essential for common E2-dependent transcription processes as the timely regulation of viral gene expression in the course of the viral life cycle.
The remaining non-shared interactions are likely to reflect a degree of specificity in E2 intrinsic transcriptional properties, and may point to mechanistic variability in E2 transcriptional functions. For example, the comparative interactomic study reveals that 16E2 is the only one to bind the general transcription factor TFIIB (GTF2B), while it binds less efficiently to BRD4 when compared to the other E2 proteins [15] (Fig. 4B). The 16E2 protein has been shown in in-vitro studies to exhibit especially potent transcriptional activation properties, and this was proposed to rely on a more efficient recruitment of basal transcriptional factors than other E2 proteins [22]. The comparative interaction mapping substantiates this hypothesis and suggests it could be due to GTF2B binding. By contrast, BRD4 would not be expected to significantly contribute to the strength of HPV16 E2 transcriptional activity.
All other targeting specificity is associated with cutaneous HPV. Interestingly, most of the cutaneous-specific factors (ENO2, SFRS2, TOB) mediate transcription repression. Their preferred interaction with cutaneous HPV E2 proteins possibly underlies the decreased efficiency of E2-dependent transcription that was observed with synthetic promoters [15]. Conversely, the low level of transactivation is not related to reduced binding to BRD4, since the β-type cutaneous E2 proteins bind to high levels to BRD4. The activation properties of the cutaneous E2 proteins may thus result from a combined recruitment of repressors together with BRD4 and other activators.
The variety of cellular factors targeted by E2 provides an experimental appraisal of the mechanistic diversity of E2 transcriptional activity. It highlights an intricate interplay between the E2 proteins and the host cell transcriptional machinery, and allows the identification of a core set of common targets.
E2 and Viral DNA Replication
By binding to the LCR, E2 also activates the replication of viral DNA. Indeed, the viral origin of replication lies within the promoter-proximal region and contains E2BS as well as a binding site for the viral helicase E1. E2 both binds at high affinity to its cognate sites and to E1 through the TAD, consequently promoting the loading of E1 to the origin of replication ([54] for review). E2 also acts on viral DNA replication by recruiting the host cell replication factors TOP1 or RPA through direct interaction. Only few additional replication factors were identified in the comparative interactome, like POLDIP2, interacting with DNA polymerase δ and PCNA, or ORC3L, a protein involved in the assembly of the pre-replication complex.
E2 and Segregation of the Viral Genome
E2 is also critical for viral genome partitioning [4]. As such, E2 is required for the maintenance of viral episome in the basal dividing cells of the epithelium in the early steps of the viral life cycle. It is also essential to maintain a reservoir of HPV genomes during long-term infections, and is thus a crucial determinant of viral persistence. E2 acts as a bridge between viral episomes and host chromosomes, through binding to viral genome and protein-protein interactions with different factors of the mitotic apparatus. Initially, the targeting of E2 to mitotic chromatin has been demonstrated to rely on the binding of E2 TAD with the double bromodomain protein BRD4 [55]. It was later shown that, while the binding to BRD4 is conserved for all HPV E2 proteins and is consistently required for their transcriptional activation capacities, its role in the targeting of E2 to mitotic chromatin is variable [4]. In fact, several other factors of the mitotic apparatus have since been proposed as relevant for E2-mediated HPV genome partitioning, as the ChLR1 helicase or MKlp2 kinesin [4, 56, 57]. We could confirm the binding of MKlp2 to almost all the tested E2 proteins, and also identified the mitotic spindle-associated factor CEP350, which could be relevant for partitioning of the viral genome.
E2 and RNA Processing
It has been also shown that E2 could assist the splicing of mRNA. This is achieved for the cutaneous E2 proteins through interactions with SR factors, known to be key regulators of RNA processing mechanisms [58]. The mRNA processing machinery functions along with transcription, and some transcriptional activators are also able to interact with essential splicing factors [59]. RNA processing is a highly complex process, involving the formation of the spliceosome composed of mRNA substrates, snRNA (small nuclear RNA), snRNP (small ribonucleoproteins), and associated with splicing regulatory proteins, notably SR proteins.
The relation between E2 and RNA processing, contrary to other E2 functions, has first been uncovered with cutaneous HPVs. β-type E2 proteins harbor a long hinge enriched in arginine, serine and glycine residues. Arginine/serine (RS) dipeptide repeats are the characteristics of the SR superfamily of proteins, primarily involved in processing of mRNA. These observations led to the hypothesis that E2 proteins could share similar functions as cellular SR proteins or interact with them [58]. Accordingly, the HPV5 E2 protein was found to interact with a number of SR proteins [58, 60]. Among others, 5E2 interacts with SFRS1, best known as ASF/SF2 [60], an essential sequence specific splicing factor, as well as with SFSR2 (SC35) and SFRS7 (9G8). By binding to both its cognate DNA elements and to SR proteins, HPV5 E2 was shown to regulate the splicing of transcripts near the promoter, probably by increasing the local concentration of SR factors [58]. The mucosal α-type HPV E2 proteins appear to modulate mRNA splicing as well, in another way since 16E2 protein activates the expression of splicing factors such as ASF/SF2 (SFRS2) [40]. This function appears to be critical given that viral genomes expressing an E2 protein defective for SFRS1 upregulation show reduce levels of viral RNA [40] (reviewed in [61]). Also, 16E2 was recently shown to inhibit the polyadenylation of viral mRNA [62]; further substantiating the participation of E2 to viral mRNA processing.
Regulation of RNA by E2 might be even broader, since E2 interacts with cellular factors regulating RNA processing other than SR proteins. Indeed, SMN1, an RNA binding protein involved in the assembly of snRNP [63], interacts with mucosal HPVE2 proteins and enhances E2-mediated transcriptional activation [64]. For the cutaneous HPV5, E2 binds TNPO3 [60], a protein that mediates the nuclear import of the splicing factors SFRS1 and SFRS2 by recognizing phosphorylated RS domains [65]. These interactions suggest that the regulation of RNA processing by the E2 proteins is extensive and thus must be pivotal for regulation of HPV gene expression.
In the comparative interactomic study conducted with the 12 E2 proteins, we corroborated interactions of E2 proteins with SFRS1, 2 and 7 proteins of the SR family [15]. The interaction data set provides evidence that the targeting of SR splicing factors is not restricted to the β-type E2 proteins, but also extends to the α-type E2 proteins, and therefore is conserved among all HPV genotypes. Interaction with SFRS7 was the strongest for both the cutaneous and the mucosal E2 protein, while SFRS2 only interacted with the E2 proteins from β/μ cutaneous HPV, suggesting that variations may occur in the regulation of mRNA splicing according to HPV tropism. The α-type HPV E2 proteins exhibit greatly reduced interaction levels, in accordance with the presence of short R-alternating sequences in their hinge regions. This indicates that E2 proteins encoded by the genital α-HPVs are able to interact with SR proteins but less efficiently than those from the cutaneous HPVs, which might explain why it had not been detected. A conserved role of E2 in the regulation of RNA splicing through interaction with SR-splicing factors nevertheless emerges, probably involved in the synthesis of viral mRNA, as initially proposed for HPV5 E2 [58]. It substantiates a pivotal role of E2 in the timely regulation of mRNA production through alternative splicing, enabling the sequential expression of viral proteins along the viral life cycle [40]. SRFS1 and 7 are the strongest of all the interactors of cutaneous E2 proteins, suggesting that binding to splicing factors is prevailing for the viral gene expression by these E2. For the α-type E2 proteins, the interaction with splicing factors is not predominant and might thus be less important. Altogether, the E2 proteins nevertheless emerge as primary factors to orchestrate viral regulation, both at the level of transcription and of mRNA splicing.
E2 and Apoptosis
The pro-apoptotic activity of E2 is one of the first described E2 functions independent of its binding to the viral genome (reviewed in [6]). A striking aspect of the involvement of E2 in apoptosis is that it was demonstrated to be specific for the HR-HPV E2 proteins [8, 9], first supporting the idea that E2 proteins might have developed specific activities that correlate with the HPV oncogenic power. The two main pro-apoptotic pathways are dependent on caspase activation. On one hand, the intrinsic pathway can be induced by p53 and involves mitochondrial dysfunctions leading to the activation of caspase 9. On the other hand, the extrinsic pathway is dependent on death receptors signaling and activation of caspase 8. Both pathways eventually merge in the activation of effector caspases, which leads to subsequent cell death. Several interactions between HR-HPV E2 and effectors of both the extrinsic and the intrinsic pathways have been identified over the years, notably with p53, CASP8 and C-FLAR [66-68]. Overall, the interplay between HR-HPV E2 proteins and the apoptotic machinery is complex, as reflected by the variety of interactions identified (see [13]). Other known interactions may have implications for E2-induced apoptosis. For example, SMN1 was shown by Strasswimmer and colleagues to interact with E2 [64]. Mutation in this protein results in a neurodegenerative disease called SMA (Spinal Muscular Atrophy) and recent evidence demonstrated that knock down of SMN1 increases neuronal cell death which might be counteracted by over-expression of the anti-apoptotic Bcl-xL [69]. We speculate that binding of SMN1 to E2 might thus favor apoptosis. A yeast-two hybrid screen also identified NR4A1 as an interacting partner of HPV16 E2 [70]. This protein is a member of the nuclear-receptor superfamily and plays a role in regulating cell growth and apoptosis [71]. NR4A1 triggers cytochrome c release from the mitochondria [72] both in the context of cancer cells and of virus-induced apoptosis [73, 74] by interacting with Bcl2 [75]. This interaction could thus modulate the intrinsic apoptotic pathway and this reinforces the notion that E2 uses various redundant approaches to regulate cell death. One of the final steps of apoptosis following caspase activation, is the alteration of DNA structures (strand breaks, oxidized bases). TOP1 is known to be involved in modification of the topology of supercoiled DNA by decreasing the torsional stress on DNA [76]. TOP1 has recently been linked to apoptosis since down regulation of its expression by RNA interference reduces apoptotic DNA fragmentation [77]. The interaction with E2 [78] might stabilize TOP1 and favor apoptosis completion, suggesting that E2 could impact on multiple steps of apoptosis.
In the large-scale study conducted in our lab, regulation of apoptosis emerged as a functional family targeted by E2, which indicates that the 12 studied E2 proteins have an intrinsic capacity to interact with apoptosis regulators. Thus, the regulation of cell death and survival pathways is not restricted to the mucosal HR-HPV but extends to the LR-mucosal as well as to the cutaneous HPVs. Our data highlight the targeting of both positive and negative regulators of apoptosis, suggesting that the E2 proteins may interfere oppositely with cell death pathways at different steps of the viral life cycle. However, for some of the targets, the effect on cell death can depend on the context (level of damage for TP53) or the protein isoform (BCL2L1), so that the final outcome of the interaction may vary according to cellular conditions, and is difficult to anticipate from this complex picture.
Among the 12 proteins of this family, three were bound by all E2 proteins (CASP8, TAX1BP1 and TP53). For TP53 and CASP8, the binding of E2 may not have similar functional consequences according to HPV genotype. The detection of an interaction between p53 and all the E2 proteins was unexpected based upon previous results describing an interaction of p53 only with the HR-mucosal E2 proteins [9]. From our recent data, we infer that E2 binding to p53 is actually conserved among HPV genotypes, but might have a different impact on p53 function and induce apoptosis only in the case of HR-HPV E2 proteins. For the LR-HPV E2 proteins, binding to p53 may alter other activities, perhaps the induction of a cell cycle arrest. This notion is further supported by the detection of interactions between Caspase 8 and all the E2 proteins tested. Yet, it was shown that among the mucosal HPV, only HR-HPVE2 proteins are able to induce apoptosis through direct binding to caspase 8 [8, 66]. E2-mediated caspase 8 activation is linked to the accumulation of E2 protein in the cytoplasm due to an active nucleo-cytoplasmic shuttling, which does not occur in the case of LR-mucosal HPV [8]. However, the comparative interaction mapping provides evidence for a conserved E2 binding to caspase 8 across all HPV genotypes, suggesting that the outcome of the interaction between the LR-HPV E2 proteins and caspase 8 might be different from that of HR-HPV E2 proteins. It could notably play a role in keratinocyte differentiation, since a role of caspase 8 in skin homeostasis has been recently recognized [79]. For both p53 and caspase 8, the functional impact of interactions with the non-apoptotic E2 proteins could be the opposite of interactions with the proapoptotic E2 proteins, i.e. interfering with death induction. This binding could thus be differentially involved in viral pathogenesis according to HPV oncogenicity. Another hypothesis is that these interactions take place at different times of the HPV life cycle. It can be envisioned that the LR-HPV E2 induces apoptosis only at late stages of the viral life cycle in the upper layers of infected epithelium, in which case it would have escaped detection in cell culture conditions. Indeed, the LR-HPV E2 proteins could trigger cell death only after nuclear membrane breakdown which occurs during keratinocyte differentiation, and would allow the accumulation of E2-Caspase 8 complexes in the cytoplasm enabling caspase activation. Such delayed apoptotic induction could be involved in common viral processes such as viral particles release.
Overall, the functional targeting of apoptosis regulatory factors shows that the E2 proteins actively take part in the manipulation of cell death or survival pathways, tightly orchestrated by the virus throughout infection.
E2 and the Proteasome Ubiquitin System
Controlling or rewiring the ubiquitin-proteasome pathway by viral proteins is likely to have a great impact on the host cell. The consequences could be multiple: degradation of antiviral molecules, and diversion of ubiquitin ligase substrate specificity. There are two types of E3 ubiquitin ligases, the HECT (Homologous to E6AP C-terminus) proteins with an intrinsic catalytic activity, and the Cullin-based complexes composed of a cullin, a RING finger protein, and one or more substrate specificity adapters [80-82].
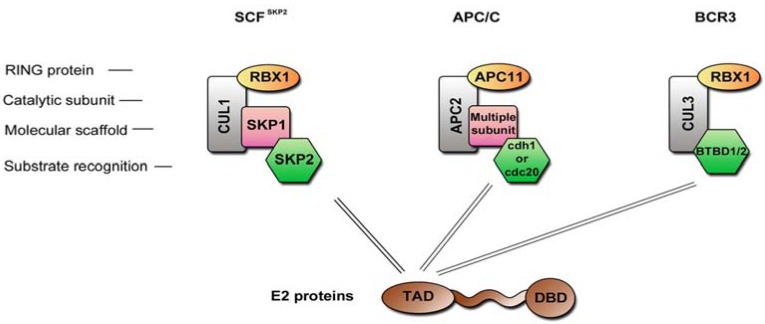
Several interactions have been shown between the E2 proteins and elements of the ubiquitin proteasome system over the past years. E2 proteins from HR-HPV16 and 18 interact with CDC20 and FZR1 (also known as cdh1), two substrate recognition subunits of the ubiquitin ligase complex APC/C (Anaphase Promoting Complex, Fig. 5), while in contrast the LR-HPV6 and 11 E2 proteins are not able to engage in such interaction [7]. The APC/C complex is responsible for anaphase transition through the degradation of specific substrates such as cyclin B. The binding of HR-HPVE2 proteins to CDC20 and FZR1 has been shown to inhibit APC/C-mediated cyclin B degradation, which leads to a metaphase arrest and genomic instability [7]. Another APC/C substrate is SKP2 [83], a substrate recognition subunit of the ubiquitin ligase complex SCF (Fig. 5), and its stabilization by E2 activates this E3 ligase complex. Since SCF is responsible for the degradation of the cell cycle negative regulators p21 and p27, this activation results in an enhanced G1 to S transition [84]. Therefore it appears that identified interactions between the HR-HPVE2 proteins and APC/C ubiquitin ligase complex strongly impact on cell cycle. In addition it has been demonstrated by Bellanger et al. that the complex SCF SKP2 can degrade HPV18 E2 protein itself, suggesting a complex and controlled feedback mechanism [85].
Fig. (5).
Cullin-based E3 ubiquitin ligase complexes. Schematic representation of three E3 ubiquitin ligase complexes that have been shown to be regulated by the HPV E2 proteins.
Few interactions with proteins involved in ubiquitination mechanisms have also been shown to be involved in the regulation of E2 stability itself. The E2 proteins have short half-lives, and this rapid turnover has been shown to be the result of ubiquitin-mediated proteasome degradation targeting of the N-terminal domain [8, 86-88]. Identification of the ubiquitin ligases responsible for the degradation of some E2 emerged only recently: HPV18 E2 can be degraded by the ubiquitin ligase complex SCFSKP2, and this degradation is mediated by direct interaction with the adaptor SKP2 [85] while for HPV16, it was demonstrated that a cullin 3-based complex (BRC3) mediates ubiquitination and subsequent degradation [88]. It was hypothesized that 16E2 is recruited to this complex through BTB domain proteins, which are substrate adaptors of cullin 3-based complexes (Fig. 5) [88, 89]. In line with this hypothesis, two BTB domain–containing proteins, BTBD1 and BTBD2, were identified as partners of HPV16E2 protein in a yeast two-hybrid screen [90]. The process of E2 protein degradation is even more complex, since several cellular proteins have been shown to modulate in E2 stability as TAX1BP1 or BRD4 [87, 91, 92]. Moreover, since HPVs infect different tissue niches, it is unlikely that all E2 proteins are degraded in the same way, and to our knowledge, there are no data available on the stability of E2 proteins from cutaneous β- or μ-HPV types.
Previous studies thus converge on the targeting by the HR-HPV E2 of cullin-based E3 ligase complexes mediated by interactions with substrate adaptors that ensure the specificity of the protein to be degraded. The comparative interaction mapping reinforces the notion of a preferred targeting of E3 ubiquitin ligases by the E2 proteins. Indeed, of 16 factors of the ubiquitin-proteasome pathway found to interact with E2, eight are part of E3 ubiquitin ligase complexes. In line with previously published works, most of them correspond to substrate-adaptors of cullin-based complexes, of which BTBD1 and SPOP are BTB-proteins forming complexes with cullin3, FBX022 is a F-box protein subunit of cullin1-based complex, CDC20 and CDH1 are part of the APC/C. The HR-specificity of E2 binding to CDC20 was recovered among the mucosal HPV, but this binding was also detected with cutaneous E2 proteins, suggesting that the involvement of this interaction in HPV pathogenesis differs according to HPV tropism. Of note, the SPOP adapter binds all E2 proteins, with reduced efficiency for HPV16 E2. Conversely, the HPV16 E2 protein was the only one to bind another adapter of cullin3-complexes, BTBD1, suggesting that the targeting of E2 to cullin3-based complex is conserved but could be mediated through diverse interactions with adaptors [15].
In addition, two HECT domain family proteins HUWE1 and WWP2 were identified as novel E2 partners, even further broadening the potential impact of E2 through interaction with ubiquitin ligases. This is compatible with the notion that E2 could not only modify the action of ubiquitin ligases toward their natural substrates, as was shown for cyclin B or SKP2, but also could divert their substrate specificity toward new substrates, the E2 proteins then acting as novel substrate adaptors. In that case, the targeting of ubiquitin ligase complexes by E2 would have a similar outcome than the binding of E6 to E6AP, which induces the hijacking of E6AP activity toward ubiquitylation of p53 [93]. Such potential E2-dependent substrates are expected to bind to E2 only when in complex with the Ubiquitin ligase and are therefore not accessible through binary protein-protein interaction mapping.
Intracellular Transport
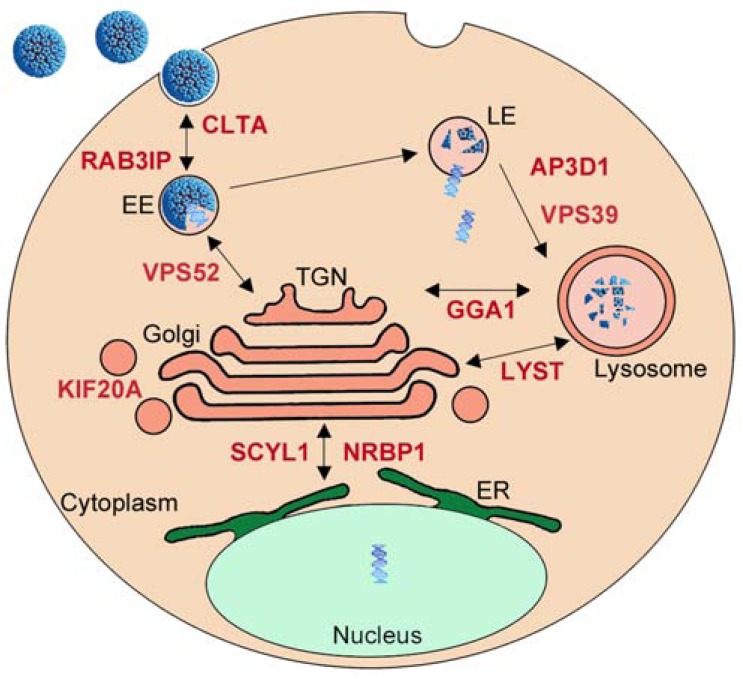
Among known E2-interacting proteins, several are assigned with GO-terms related to intracellular protein transport, such as KIF20A, AP3D1, TNPO3 or TMF1. AP3D1 is part of the complex AP-3, which regulates sorting of membrane proteins [94], and KIF20A acts as a motor for the transport of vesicle and has been shown to associate with a protein from the Rab family, RAB6A [95]. TMF1, an interactor of HPV16 E2, was also demonstrated to be involved in Rab-6 dependent transport processes [96]. Other members of the Rab family have been implicated in the entry pathway of certain types of HPV such HPV31 [97]. Additionally, E2 was shown to interact with TNPO3, involved in the entry steps of other viruses such as HIV-1 [98]. However, the targeted factors are not numerous enough to represent an enriched functional family based on the criteria of GO-term analysis (Fig. 1). The large-scale interactomic strategy performed with the 12 E2 proteins identified additional proteins involved in intracellular trafficking, which now emerges as a functional family targeted by E2 [15]. Indeed, 15 cellular E2 targets were classified in intracellular transport family, among which 13 are newly identified E2 partners [15]. Notably, the targeted proteins are more concentrated on vesicular transport, affecting dynamics and maintenance of intracellular membranous organelles (Clathrin, Rab-family protein, lysosomal transport, vesicle transport to and from the golgi apparatus). Even if interactions of the E2 proteins with such factors is surprising when considering that E2 is primarily a nuclear protein, it might be hypothesized that they play a role in the translocation of HPV episomes to the nucleus. Indeed, the regulation of vesicle transport is important for entry processes during HPV infection, and we noticed that most of E2 targets overlap with the pathways of HPV entry in the cell as depicted in Fig. (6). E2 is able to link the viral DNA and it interacts with the minor capsid protein L2 [99], which is pivotal for the trafficking of viral genomes from the capsid to the nucleus [100]. It is thus tempting to speculate that E2 participates in the delivery and transport of viral genomes through the cytoplasm by interacting with the identified proteins, and in collaboration with L2. This would imply that E2 is encapsidated in the virion bound to the viral genome, and that it plays a role at very early steps of viral infection. Pseudovirions systems have been developed and they indicate that E2 is not necessary for proper delivery of DNA materials to the nucleus, in contrast to L2 [101]. However, only sparse information is available on the involvement of E2 in pseudovirions infectivity, and these studies were only done with BPV1 pseudvirions. In the BPV1 system, a study reported that E2 enhanced encapsidation of full-length viral DNA and may be packaged with the pseudovirion [102]. These findings, however, were not corroborated in another study showing that E2 expression does not alter BPV1 pseudovirions production and infectivity [101]. In light of the emergence of the functional targeting of intracellular transport pathways, we feel that this issue should be re-evaluated with HPV pseudovirions. We wonder whether the use of over-optimized systems for pseudovirion production (codon optimized ORF) could hide a potential involvement of E2 under normal conditions. Also, it should be taken into consideration that the pseudovirion systems allow the efficient delivery only for small DNA molecule. A potential role of E2 could be uncovered with viral genomic DNA containing E2 binding sites, thereby mimicking authentic HPV genome encapsidation and translocation.
Fig. (6).
Intracellular transport and virus entry. Overlap between E2 targets involved in intracellular transport and the pathways of virus entry. EE: early endosome; LE: late endosome; ER: endoplasmic reticulum.
Overall, this compilation of observations raises the intriguing possibility that E2 could take an active part in very early steps of HPV infection, which we feel is worth investigating.
E2 and Keratinocyte Migration/Differentiation
Several lines of evidence point to a modulation of cellular genes involved in keratinocyte migration and differentiation by the E2 proteins. Indeed, E2 represses the transcription of ITGB4, a major integrin for keratinocyte attachment to the matrix, and leads to the detachment of keratinocyte from the underlying matrix [38]. E2 also activates MMP9, a protein involved in cell migration, invasion and metastasis [41]. In addition, E2 was shown to alter the transcription of differentiation markers through cooperation with cellular transcription factors such as C/EBP [103]. Accordingly, in a microarray study, E2 was shown to modify the expression profile of cellular genes, among which many were involved in cell differentiation [104]. It was also observed that E2 induces phenotypical changes typical of terminally differentiated cells, with increased expression of differentiation markers [105]. E2 therefore promotes differentiation by acting as a transcription factor which modulates the expression of a set of cellular genes.
More surprisingly, the regulation of cell differentiation by E2 might be mediated through direct protein-protein interaction. Indeed, it was recently published that E2 stimulates TNF-induced NFκB activation through direct interaction with TRAF5, an intermediate of the NFκB signaling pathway [5]. This could be involved in cell commitment to differentiation, since activation of NFκB by TNF induces an epithelial differentiation program [106]. Other known cellular proteins interacting with E2 have been implicated in cell differentiation or migration, such as CCHCR1, which affects the balance between proliferation and differentiation in keratinocytes [107, 108], or GNB2L1 reported to be involved in migration of carcinoma cells [109]. The interaction of these proteins with E2 in the context of infection might impact on their stability, localization and function, and thus promote the detachment of keratinocytes from the basal lamina and further induction of differentiation.
In the comparative E2 interactomics study, we detected a targeting concentrated around proteins involved in cellular adhesion. Interactions were identified with PVRL1, a protein involved in cell-to-cell junctions [110], PTK2B, a member of the focal adhesion kinase family [111] or FN1, the fibronectin responsible for the attachment to the extracellular matrix [112]. Overall, this reinforces the idea that E2 might impact on the cohesion of the infected epithelium.
We also uncovered a direct interaction of all E2 proteins with ITGB4, which as discussed before, is also transcriptionally repressed by E2 [38]. Such redundant targeting may highlight the importance of inactivating ITGB4 for progression of the viral life cycle. It might provide an early signal to induce differentiation, which requires migration of keratinocyte from basal to upper layers of the epithelia. Interestingly, ITGB4 interaction with E2 is conserved across all HPV genotypes tested, and ITGB4 was also identified as a target of E7 [138]. Theses observations reinforce the notion that the targeting of ITGB4 might be pivotal, and that its sequential regulation by E2 and E7 potentially essential for progression of the viral life cycle.
Overall, these observations point to a potential redundant effect of E2 on keratinocyte differentiation and migration through modulation of gene expression as well as direct targeting of cell-cell and cell-matrix contacts.
CONCLUDING REMARKS
Viral proteins are platforms of interaction that are known to orchestrate host proteins localization and degradation, control their expression levels and post-translational modifications, and rewire signal transduction in the infected cell [113, 114]. Recently, through improvement of detection technologies [115, 116], significant efforts have been made to generate comprehensive protein-protein interaction network maps in order to improve the understanding of virus-host relationship [117-119]. Additional steps should be focused on the development of integrative comparative interactomic approaches in order to decipher the interaction profiles of viral proteins from different pathogenic viruses. It should give insight into how genomic variations drive phenotypic divergence, but also on how viral proteins diversely hijack host cellular structures and pathways.
In the case of HPVs, a great divergence exists between infections with the different viral strains, ranging from variations in the tropisms (oral, genital, cutaneous epithelia) to variations in the pathogenic potential. The variability of HPV-associated lesions likely results from a diverse interplay between viral and host proteins. However, only recent studies have started to encompass large-scale analyses, which will reveal their wide impact on the comprehension of HPV pathogenesis.
Functional analysis of the E2 interactome provides a complete picture of the range of E2 functions, viewed through its interactions with the host proteome. It highlights a prevailing impact of E2 on the cellular processes of transcription, RNA processing, apoptosis, ubiquitination, and intracellular transport. The emergence of this latter unexpected functional family raises the provocative hypothesis that E2 may be involved in early steps of viral infection. In addition, a constant targeting of positive and negative regulators of the same cell processes emerges, indicating a duality in E2 functions, potentially driving opposing activities during the different steps of the viral life cycle. We did not identify any specific cellular functional targeting according to tropism or pathogenic power, suggesting that modulation of these cell processes takes part in the general regulation of the viral life cycle of all HPVs. However, this common functional targeting proved to be achieved through diversified patterns of interactions. Such variations in the interactions profiles most probably drive the contribution of E2 proteins to HPV pathogenesis.
Furthermore, the assessment of previously known interactions with the 12 E2 indicates that interaction data cannot be generalized from functional studies addressing only a subset of E2 proteins. For example, the binding of GTF2B had been detected for both BPV1 E2 and 16 E2, and was assumed to be conserved and involved in the activity of all E2 proteins. However, binding to this factor turned out to be specific for 16E2 and as such may account for some particular aspect of its transcriptional function as discussed earlier. By contrast, other interactions, as with TP53 or caspase 8, had been detected in connection to E2 functions specific for a subset of genotypes, but arise in the comparative interaction mapping as conserved across all HPV genotypes. These observations underline the importance of combining functional studies and interaction mapping to decipher E2 activities in relation with HPV pathogenesis.
Studying viral-host interactions has become an important challenge since targeting protein-protein interaction is now considered as an attractive approach for the development of new therapeutic strategies [120]. The use of small peptide based strategies to counteract specific interaction is studied for long [121]. But the ultimate option to develop an efficient treatment would be to identify a pan-HPV inhibitor. To that end, efforts should be put into the expansion of systematic and comparative approaches to identify protein-protein interactions. The targeting of E2 interactions would be a valuable therapeutic strategy, since E2 is an essential factor of the life cycle of all HPVs, is expressed early in the infection process, and is required for viral persistence.
ACKNOWLEDGEMENTS
This work was supported by funding from the Pasteur Institute. MM is the recipient of a ministerial doctoral fellowship.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1. Hegde RS, Androphy EJ. Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: implications for its DNA binding-site selection mechanism. J Mol Biol. 1998;284:1479–89. doi: 10.1006/jmbi.1998.2260. [DOI] [PubMed] [Google Scholar]
- 2. Antson AA, Burns JE, Moroz OV, et al. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature. 2000;403(6771):805–9. doi: 10.1038/35001638. [DOI] [PubMed] [Google Scholar]
- 3. Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 4. McBride AA, Oliveira JG, McPhillips MG. Partitioning viral genomes in mitosis: same idea, different targets. Cell Cycle. 2006;5:1499–502. doi: 10.4161/cc.5.14.3094. [DOI] [PubMed] [Google Scholar]
- 5. Boulabiar M, Boubaker S, Favre M, Demeret C. Keratinocyte sensitization to tumour necrosis factor-induced nuclear factor kappa B activation by the E2 regulatory protein of human papillomaviruses. J Gen Virol. 2011;92:2422–7. doi: 10.1099/vir.0.032466-0. [DOI] [PubMed] [Google Scholar]
- 6. Blachon S, Demeret C. The regulatory E2 proteins of human genital papillomaviruses are pro-apoptotic. Biochimie. 2003;85:813–9. doi: 10.1016/j.biochi.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 7. Bellanger S, Blachon S, Mechali F, Bonne-Andrea C, Thierry F. High-risk but not low-risk HPV E2 proteins bind to the APC activators Cdh1 and Cdc20 and cause genomic instability. Cell Cycle. 2005;4:1608–15. doi: 10.4161/cc.4.11.2123. [DOI] [PubMed] [Google Scholar]
- 8. Blachon S, Bellanger S, Demeret C, Thierry F. Nucleocytoplasmic shuttling of high risk human Papillomavirus E2 proteins induces apoptosis. J Biol Chem. 2005;280:36088–98. doi: 10.1074/jbc.M505138200. [DOI] [PubMed] [Google Scholar]
- 9. Parish JL, Kowalczyk A, Chen HT, et al. E2 proteins from high- and low-risk human papillomavirus types differ in their ability to bind p53 and induce apoptotic cell death. J Virol. 2006;80:4580–90. doi: 10.1128/JVI.80.9.4580-4590.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfefferle R, Marcuzzi GP, Akgul B, et al. The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J Invest Dermatol. 2008;128:2310–5. doi: 10.1038/jid.2008.73. [DOI] [PubMed] [Google Scholar]
- 11. Choo KB, Pan CC, Han SH. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology. 1987;161:259–61. doi: 10.1016/0042-6822(87)90195-4. [DOI] [PubMed] [Google Scholar]
- 12. Corden SA, Sant-Cassia LJ, Easton AJ, Morris AG. The integration of HPV-18 DNA in cervical carcinoma. Mol Pathol. 1999;52:275–82. doi: 10.1136/mp.52.5.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bellanger S, Tan CL, Xue YZ, Teissier S, Thierry F. Tumor suppressor or oncogene?. A critical role of the human papillomavirus (HPV) E2 protein in cervical cancer progression. Am J Cancer Res. 2011;1:373–89. [PMC free article] [PubMed] [Google Scholar]
- 14. Huang da W, Sherman BT, Zheng X, et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi1311s27. Chapter 13: Unit 13 11. [DOI] [PubMed] [Google Scholar]
- 15. Muller M, Jacob Y, Jones L, et al. Large scale genotype comparison of human papillomavirus e2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog. 2012;8:e1002761. doi: 10.1371/journal.ppat.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demeret C, Desaintes C, Yaniv M, Thierry F. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J Virol. 1997;71:9343–9. doi: 10.1128/jvi.71.12.9343-9349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dong G, Broker TR, Chow LT. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–27. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dostatni N, Lambert PF, Sousa R, Ham J, Howley PM, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–71. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 19. Guido MC, Zamorano R, Garrido-Guerrero E, Gariglio P, Garcia-Carranca A. Early promoters of genital and cutaneous human papillomaviruses are differentially regulated by the bovine papillomavirus type 1 E2 gene product. J Gen Virol. 1992;73(Pt 6):1395–400. doi: 10.1099/0022-1317-73-6-1395. [DOI] [PubMed] [Google Scholar]
- 20. Sanchez IE, Dellarole M, Gaston K, de Prat Gay G. Comprehensive comparison of the interaction of the E2 master regulator with its cognate target DNA sites in 73 human papillomavirus types by sequence statistics. Nucleic Acids Res. 2008;36:756–69. doi: 10.1093/nar/gkm1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stubenrauch F, Pfister H. Low-affinity E2-binding site mediates downmodulation of E2 transactivation of the human papillomavirus type 8 late promoter. J Virol. 1994;68:6959–66. doi: 10.1128/jvi.68.11.6959-6966.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hou SY, Wu SY, Chiang CM. Transcriptional activity among high and low risk human papillomavirus E2 proteins correlates with E2 DNA binding. J Biol Chem. 2002;277:45619–29. doi: 10.1074/jbc.M206829200. [DOI] [PubMed] [Google Scholar]
- 23. Smith JA, White EA, Sowa ME, et al. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc Natl Acad Sci USA. 2010;107:3752–57. doi: 10.1073/pnas.0914818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thierry F, Dostatni N, Arnos F, Yaniv M. Cooperative activation of transcription by bovine papillomavirus type 1 E2 can occur over a large distance. Mol Cell Biol. 1990;10:4431–7. doi: 10.1128/mcb.10.8.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kovelman R, Bilter GK, Glezer E, Tsou AY, Barbosa MS. Enhanced transcriptional activation by E2 proteins from the oncogenic human papillomaviruses. J Virol. 1996;70:7549–60. doi: 10.1128/jvi.70.11.7549-7560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–77. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 27. Goodrich JA, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–9. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 28. Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 29. Abbate EA, Voitenleitner C, Botchan MR. Structure of the papillomavirus DNA-tethering complex E2: Brd4 and a peptide that ablates HPV chromosomal association. Mol Cell. 2006;24:877–89. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 30. Lee D, Hwang SG, Kim J, Choe J. Functional interaction between p/CAF and human papillomavirus E2 protein. J Biol Chem. 2002;277:6483–9. doi: 10.1074/jbc.M105085200. [DOI] [PubMed] [Google Scholar]
- 31. Lee D, Lee B, Kim J, Kim DW, Choe J. cAMP response elementbinding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J Biol Chem. 2000;275:7045–51. doi: 10.1074/jbc.275.10.7045. [DOI] [PubMed] [Google Scholar]
- 32. Muller A, Ritzkowsky A, Steger G. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J Virol. 2002;76:11042–53. doi: 10.1128/JVI.76.21.11042-11053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cha S, Seo T. hSNF5 is required for human papillomavirus E2-driven transcriptional activation and DNA replication. Intervirology. 2011;54:66–77. doi: 10.1159/000318871. [DOI] [PubMed] [Google Scholar]
- 34. Rehtanz M, Schmidt HM, Warthorst U, Steger G. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol Cell Biol. 2004;24:2153–68. doi: 10.1128/MCB.24.5.2153-2168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bieging KT, Attardi LD. Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 2012;22:97–106. doi: 10.1016/j.tcb.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–74. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 37. Zhao Y, Bruemmer D. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol. 2010;30:1535–41. doi: 10.1161/ATVBAHA.109.191163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oldak M, Smola H, Aumailley M, Rivero F, Pfister H, Smola-Hess S. The human papillomavirus type 8 E2 protein suppresses beta4-integrin expression in primary human keratinocytes. J Virol. 2004;78:10738–46. doi: 10.1128/JVI.78.19.10738-10746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steger G, Schnabel C, Schmidt HM. The hinge region of the human papillomavirus type 8 E2 protein activates the human p21(WAF1/CIP1) promoter via interaction with Sp1. J Gen Virol. 2002;83:503–10. doi: 10.1099/0022-1317-83-3-503. [DOI] [PubMed] [Google Scholar]
- 40. Mole S, Milligan SG, Graham SV. Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J Virol. 2009;83:357–67. doi: 10.1128/JVI.01414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akgul B, Garcia-Escudero R, Ekechi C, et al. The E2 protein of human papillomavirus type 8 increases the expression of matrix metalloproteinase-9 in human keratinocytes and organotypic skin cultures. Med Microbiol Immunol. 2011;200:127–35. doi: 10.1007/s00430-011-0183-4. [DOI] [PubMed] [Google Scholar]
- 42. Lee D, Kim HZ, Jeong KW, et al. Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J Biol Chem. 2002;277:27748–56. doi: 10.1074/jbc.M203706200. [DOI] [PubMed] [Google Scholar]
- 43. Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–23. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 44. Zhu M, John S, Berg M, Leonard WJ. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell. 1999;96:121–30. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 45. Sharma M, Zarnegar M, Li X, Lim B, Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J Biol Chem. 2000;275:35200–8. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- 46. Shamay M, Barak O, Doitsh G, Ben-Dor I, Shaul Y. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J Biol Chem. 2002;277:9982–8. doi: 10.1074/jbc.M111354200. [DOI] [PubMed] [Google Scholar]
- 47. Hanai K, Furuhashi H, Yamamoto T, Akasaka K, Hirose S. RSF governs silent chromatin formation via histone H2Av replacement. PLoS Genet. 2008;4:e1000011. doi: 10.1371/journal.pgen.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ewing AK, Attner M, Chakravarti D. Novel regulatory role for human Acf1 in transcriptional repression of vitamin D3 receptor-regulated genes. Mol Endocrinol. 2007;21:1791–6. doi: 10.1210/me.2007-0095. [DOI] [PubMed] [Google Scholar]
- 49. Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–32. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- 50. Racki LR, Yang JG, Naber N, et al. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–21. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–83. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J Mol Biol. 1999;293:187–98. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 53. Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 54. Desaintes C, Demeret C. Control of papillomavirus DNA replication and transcription. Semin Cancer Biol. 1996;7:339–47. doi: 10.1006/scbi.1996.0043. [DOI] [PubMed] [Google Scholar]
- 55. You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–60. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 56. Parish JL, Bean AM, Park RB, Androphy EJ. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol Cell. 2006;24:867–76. doi: 10.1016/j.molcel.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 57. Yu T, Peng YC, Androphy EJ. Mitotic kinesin-like protein 2 binds and colocalizes with papillomavirus E2 during mitosis. J Virol. 2007;81:1736–45. doi: 10.1128/JVI.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lai MC, The BH, Tarn WY. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J Biol Chem. 1999;274:11832–41. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- 59. Ge H, Si Y, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell. 1998;2:751–9. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- 60. Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importinbeta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 61. McFarlane M, Graham SV. Human papillomavirus regulation of SR proteins. Biochem Soc Trans. 2010;38:1116–21. doi: 10.1042/BST0381116. [DOI] [PubMed] [Google Scholar]
- 62. Johansson C, Somberg M, Li X, et al. HPV-16 E2 contributes to induction of HPV-16 late gene expression by inhibiting early polyadenylation. EMBO J. 2012;31(14):3212–27. doi: 10.1038/emboj.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–9. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 64. Strasswimmer J, Lorson CL, Breiding DE, et al. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum Mol Genet. 1999;8:1219–26. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- 65. Allemand E, Dokudovskaya S, Bordonne R, Tazi J. A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1. Mol Biol Cell. 2002;13:2436–47. doi: 10.1091/mbc.E02-02-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thierry F, Demeret C. Direct activation of caspase 8 by the proapoptotic E2 protein of HPV18 independent of adaptor proteins. Cell Death Differ. 2008;15:1356–63. doi: 10.1038/cdd.2008.53. [DOI] [PubMed] [Google Scholar]
- 67. Webster K, Parish J, Pandya M, Stern PL, Clarke AR, Gaston K. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J Biol Chem. 2000;275:87–94. doi: 10.1074/jbc.275.1.87. [DOI] [PubMed] [Google Scholar]
- 68. Wang W, Fang Y, Sima N, et al. Triggering of death receptor apoptotic signaling by human papillomavirus 16 E2 protein in cervical cancer cell lines is mediated by interaction with c-FLIP. Apoptosis. 2011;16:55–66. doi: 10.1007/s10495-010-0543-3. [DOI] [PubMed] [Google Scholar]
- 69. Garcera A, Mincheva S, Gou-Fabregas M, et al. A new model to study spinal muscular atrophy: neurite degeneration and cell death is counteracted by BCL-X(L) Overexpression in motoneurons. Neurobiol Dis. 2011;42:415–26. doi: 10.1016/j.nbd.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 70. Olejnik-Schmidt AK, Schmidt MT, Gozdzicka-Jozefiak A. Orphan nuclear hormone receptor NR4A1 interacts with HPV16 E2 regulatory protein. Cell Mol Biol Lett. 2006;11:102–8. doi: 10.2478/s11658-006-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367:277–81. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 72. Li H, Kolluri SK, Gu J, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–64. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 73. Holmes WF, Soprano DR, Soprano KJ. Comparison of the mechanism of induction of apoptosis in ovarian carcinoma cells by the conformationally restricted synthetic retinoids CD437 and 4-HPR. J Cell Biochem. 2003;89:262–78. doi: 10.1002/jcb.10505. [DOI] [PubMed] [Google Scholar]
- 74. Lee JM, Lee KH, Weidner M, Osborne BA, Hayward SD. Epstein-Barr virus EBNA2 blocks Nur77- mediated apoptosis. Proc Natl Acad Sci USA. 2002;99:11878–83. doi: 10.1073/pnas.182552499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lin B, Kolluri SK, Lin F, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–40. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 76. Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–13. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 77. Sordet O, Goldman A, Pommier Y. Topoisomerase II and tubulin inhibitors both induce the formation of apoptotic topoisomerase I cleavage complexes. Mol Cancer Ther. 2006;5:3139–44. doi: 10.1158/1535-7163.MCT-06-0463. [DOI] [PubMed] [Google Scholar]
- 78. Clower RV, Hu Y, Melendy T. Papillomavirus E2 protein interacts with and stimulates human topoisomerase I. Virology. 2006;348:13–18. doi: 10.1016/j.virol.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 79. Lee P, Lee DJ, Chan C, Chen SW, Ch'en I, Jamora C. Dynamic expression of epidermal caspase 8 simulates a wound healing response. Nature. 2009;458:519–23. doi: 10.1038/nature07687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 81. Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 82. Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–8. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 84. Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–7. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 85. Bellanger S, Tan CL, Nei W, He PP, Thierry F. The human papillomavirus type 18 E2 protein is a cell cycle-dependent target of the SCFSkp2 ubiquitin ligase. J Virol. 2010;84:437–44. doi: 10.1128/JVI.01162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bellanger S, Demeret C, Goyat S, Thierry F. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J Virol. 2001;75:7244–51. doi: 10.1128/JVI.75.16.7244-7251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gagnon D, Joubert S, Senechal H, Fradet-Turcotte A, Torre S, Archambault J. Proteasomal degradation of the papillomavirus E2 protein is inhibited by overexpression of bromodomain-containing protein 4. J Virol. 2009;83:4127–39. doi: 10.1128/JVI.02468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zheng G, Schweiger MR, Martinez-Noel G, et al. Brd4 regulation of papillomavirus protein E2 stability. J Virol. 2009;83:8683–92. doi: 10.1128/JVI.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–7. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 90. Olejnik-Schmidt AK, Schmidt MT, Kedzia W, Gozdzicka-Jozefiak A. Search for cellular partners of human papillomavirus type 16 E2 protein. Arch Virol. 2008;153:983–90. doi: 10.1007/s00705-008-0061-6. [DOI] [PubMed] [Google Scholar]
- 91. Wang X, Naidu SR, Sverdrup F, Androphy EJ. Tax1BP1 interacts with papillomavirus E2 and regulates E2-dependent transcription and stability. J Virol. 2009;83:2274–84. doi: 10.1128/JVI.01791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lee AY, Chiang CM. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J Biol Chem. 2009;284:2778–86. doi: 10.1074/jbc.M805835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Beaudenon S, Huibregtse JM. HPV E6, E6AP and cervical cancer. BMC Biochem. 2008;9(Suppl 1):S4. doi: 10.1186/1471-2091-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Styers ML, Kowalczyk AP, Faundez V. Intermediate filaments and vesicular membrane traffic: the odd couple's first dance? Traffic. 2005;6:359–65. doi: 10.1111/j.1600-0854.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 95. Echard A, Jollivet F, Martinez O, et al. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279:580–5. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 96. Yamane J, Kubo A, Nakayama K, Yuba-Kubo A, Katsuno T, Tsukita S. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp Cell Res. 2007;313:3472–85. doi: 10.1016/j.yexcr.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 97. Smith JL, Campos SK, Wandinger-Ness A, Ozbun MA. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. J Virol. 2008;82:9505–12. doi: 10.1128/JVI.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, Fassati A. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 2011;7:e1002194. doi: 10.1371/journal.ppat.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Heino P, Zhou J, Lambert PF. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology. 2000;276:304–14. doi: 10.1006/viro.2000.0342. [DOI] [PubMed] [Google Scholar]
- 100. Pereira R, Hitzeroth II, Rybicki EP. Insights into the role and function of L2, the minor capsid protein of papillomaviruses. Arch Virol. 2009;154:187–97. doi: 10.1007/s00705-009-0310-3. [DOI] [PubMed] [Google Scholar]
- 101. Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78:751–7. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhao KN, Hengst K, Liu WJ, et al. BPV1 E2 protein enhances packaging of full-length plasmid DNA in BPV1 pseudovirions. Virology. 2000;272:382–93. doi: 10.1006/viro.2000.0348. [DOI] [PubMed] [Google Scholar]
- 103. Hadaschik D, Hinterkeuser K, Oldak M, Pfister HJ, Smola-Hess S. The Papillomavirus E2 protein binds to and synergizes with C/EBP factors involved in keratinocyte differentiation. J Virol. 2003;77:5253–65. doi: 10.1128/JVI.77.9.5253-5265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ramirez-Salazar E, Centeno F, Nieto K, Valencia-Hernandez A, Salcedo M, Garrido E. HPV16 E2 could act as down-regulator in cellular genes implicated in apoptosis, proliferation and cell differentiation. Virol J. 2011;8:247. doi: 10.1186/1743-422X-8-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Burns JE, Walker HF, Schmitz C, Maitland NJ. Phenotypic effects of HPV-16 E2 protein expression in human keratinocytes. Virology. 2010;401:314–21. doi: 10.1016/j.virol.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 106. Basile JR, Zacny V, Munger K. The cytokines tumor necrosis factor-alpha (TNF-alpha ) and TNF-related apoptosis-inducing ligand differentially modulate proliferation and apoptotic pathways in human keratinocytes expressing the human papillomavirus-16 E7 oncoprotein. J Biol Chem. 2001;276:22522–8. doi: 10.1074/jbc.M010505200. [DOI] [PubMed] [Google Scholar]
- 107. Tiala I, Suomela S, Huuhtanen J, et al. The CCHCR1 (HCR) gene is relevant for skin steroidogenesis and downregulated in cultured psoriatic keratinocytes. J Mol Med (Berl) 2007;85:589–601. doi: 10.1007/s00109-006-0155-0. [DOI] [PubMed] [Google Scholar]
- 108. Suomela S, Elomaa O, Skoog T, et al. CCHCR1 is up-regulated in skin cancer and associated with EGFR expression. PLoS One. 2009;4:e6030. doi: 10.1371/journal.pone.0006030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cao XX, Xu JD, Xu JW, et al. RACK1 promotes breast carcinoma migration/metastasis via activation of the RhoA/Rho kinase pathway. Breast Cancer Res Treat. 2011;126:555–63. doi: 10.1007/s10549-010-0955-3. [DOI] [PubMed] [Google Scholar]
- 110. Ogita H, Rikitake Y, Miyoshi J, Takai Y. Cell adhesion molecules nectins and associating proteins: Implications for physiology and pathology. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:621–9. doi: 10.2183/pjab.86.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Park SY, Avraham H, Avraham S. Characterization of the tyrosine kinases RAFTK/Pyk2 and FAK in nerve growth factor-induced neuronal differentiation. J Biol Chem. 2000;275:19768–77. doi: 10.1074/jbc.M909932199. [DOI] [PubMed] [Google Scholar]
- 112. McDonald JA. Extracellular matrix assembly. Annu Rev Cell Biol. 1988;4:183–207. doi: 10.1146/annurev.cb.04.110188.001151. [DOI] [PubMed] [Google Scholar]
- 113. Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem Sci. 2011;36:159–69. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 114. Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–98. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cassonnet P, Rolloy C, Neveu G, et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat Methods. 2011;8:990–2. doi: 10.1038/nmeth.1773. [DOI] [PubMed] [Google Scholar]
- 116. Venkatesan K, Rual JF, Vazquez A, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Calderwood MA, Venkatesan K, Xing L, et al. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci USA. 2007;104:7606–11. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. de Chassey B, Navratil V, Tafforeau L, et al. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:30. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Uetz P, Dong YA, Zeretzke C, et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–42. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 120. D'Abramo CM, Archambault J. Small molecule inhibitors of human papillomavirus protein - protein interactions. Open Virol J. 2011;5:80–95. doi: 10.2174/1874357901105010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Natur. 200;450:1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 122. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Boner W, Morgan IM. Novel cellular interacting partners of the human papillomavirus 16 transcription/replication factor E2. Virus Res. 2002;90:113–8. doi: 10.1016/s0168-1702(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 124. Kim J, Lee D, Gwan Hwang S, Hwang ES, Choe J. BRCA1 associates with human papillomavirus type 18 E2 and stimulates E2-dependent transcription. Biochem Biophys Res Commun. 2003;305:1008–16. doi: 10.1016/s0006-291x(03)00880-5. [DOI] [PubMed] [Google Scholar]
- 125. McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol. 2006;80:9530–43. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wu SY, Lee AY, Hou SY, et al. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–96. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Breiding DE, Sverdrup F, Grossel MJ, Moscufo N, Boonchai W, Androphy EJ. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol Cell Biol. 1997;17:7208–19. doi: 10.1128/mcb.17.12.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Benson JD, Lawande R, Howley PM. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J Virol. 1997;71:8041–7. doi: 10.1128/jvi.71.10.8041-8047.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gammoh N, Gardiol D, Massimi P, Banks L. The Mdm2 ubiquitin ligase enhances transcriptional activity of human papillomavirus E2. J Virol. 2009;83:1538–43. doi: 10.1128/JVI.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lee D, Kim JW, Kim K, et al. Functional interaction between human papillomavirus type 18 E2 and poly(ADP-ribose) polymerase 1. Oncogene. 2002;21:5877–85. doi: 10.1038/sj.onc.1205723. [DOI] [PubMed] [Google Scholar]
- 131. Wang WS, Lee MS, Tseng CE, et al. Interaction between human papillomavirus type 5 E2 and polo-like kinase 1. J Med Virol. 2009;81:536–44. doi: 10.1002/jmv.21404. [DOI] [PubMed] [Google Scholar]
- 132. Li R, Knight JD, Jackson SP, Tjian R, Botchan MR. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991;65:493–5. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- 133. Centeno F, Ramirez-Salazar E, Garcia-Villa E, Gariglio P, Garrido E. TAF1 interacts with and modulates human papillomavirus 16 E2-dependent transcriptional regulation. Intervirology. 2008;51:137–43. doi: 10.1159/000141706. [DOI] [PubMed] [Google Scholar]
- 134. Carrillo E, Garrido E, Gariglio P. Specific in vitro interaction between papillomavirus E2 proteins and TBP-associated factors. Intervirology. 2004;47:342–9. doi: 10.1159/000080878. [DOI] [PubMed] [Google Scholar]
- 135. Enzenauer C, Mengus G, Lavigne A, Davidson I, Pfister H, May M. Interaction of human papillomavirus 8 regulatory proteins E2, E6 and E7 with components of the TFIID complex. Intervirology. 1998;41:80–90. doi: 10.1159/000024918. [DOI] [PubMed] [Google Scholar]
- 136. Boner W, Taylor ER, Tsirimonaki E, Yamane K, Campo MS, Morgan IM. A Functional interaction between the human papillomavirus 16 transcription/replication factor E2 and the DNA damage response protein TopBP1. J Biol Chem. 2002;277:22297–303. doi: 10.1074/jbc.M202163200. [DOI] [PubMed] [Google Scholar]
- 137. Massimi P, Pim D, Bertoli C, Bouvard V, Banks L. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene. 1999;18:7748–54. doi: 10.1038/sj.onc.1203208. [DOI] [PubMed] [Google Scholar]
- 138. Neveu G, Cassonnet P, Vidalain PO, et al. Comparative analysis of virus-host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase. Methods. 2012 doi: 10.1016/j.ymeth.2012.07.029. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]