Summary
Histone methyltransferases (HMTases), as chromatin modifiers, regulate the transcriptomic landscape in normal development as well in diseases such as cancer. Here, we molecularly order two HMTases, EZH2 and MMSET that have established genetic links to oncogenesis. EZH2, which mediates histone H3K27 trimethylation and is associated with gene silencing, was shown to be coordinately expressed and function upstream of MMSET, which mediates H3K36 dimethylation and is associated with active transcription. We found that the EZH2-MMSET HMTase axis is coordinated by a microRNA network and that the oncogenic functions of EZH2 require MMSET activity. Together, these results suggest that the EZH2-MMSET HMTase axis coordinately functions as a master regulator of transcriptional repression, activation, and oncogenesis and may represent an attractive therapeutic target in cancer.
Introduction
Accumulating evidence supports a central role for epigenetic processes in the development of cancer (Jones and Baylin, 2007). Altered chromatin states are hallmarks of tumor progression (Baylin and Jones, 2011; Seligson et al., 2005) and next-generation sequencing based approaches have revealed characteristic genetic alterations in several chromatin modifiers including histone methyltransferases (HMTases) (Jiao et al., 2011; Morin et al., 2010; Varela et al., 2011). For example, recurrent rearrangements or activating point mutations of HMTases have been described in malignancies involving MLL (Krivtsov and Armstrong, 2007), DOT1L (Nguyen and Zhang, 2011), NSD1 (Jaju et al., 2001), MMSET/NSD2 (Chesi et al., 1998; Santra et al., 2003) and EZH2 (Chase and Cross, 2011; Morin et al., 2010). HMTases have gained a special interest in oncology due to their enzymatic activity and thus potential therapeutic tractability (Albert and Helin, 2010; Spannhoff et al., 2009). Little is known about how these HMTases might be linked together. The HMTase EZH2, or enhancer of zeste 2, is the SET-domain containing catalytic subunit of the Polycomb Repressive Complex 2 (PRC2), which is comprised of SUZ12, EED, RbAp48 and JARID2 among other interacting proteins that function to represses transcription through histone H3K27 trimethylation (Cao et al., 2002; Kondo et al., 2008; Margueron and Reinberg, 2011). EZH2 is overexpressed in diverse solid tumors and is associated with poor prognosis and metastatic progression (Bachmann et al., 2006; Berezovska et al., 2006; Gong et al., 2011; Kleer et al., 2003; Margueron and Reinberg, 2011; Varambally et al., 2002; Yamada et al., 2011). EZH2 upregulation in cancer occurs in part through the loss of microRNA, miR-101, that post-transcriptionally represses EZH2 via binding to its 3′UTR (Friedman et al., 2009; Varambally et al., 2008). Knock-down of EZH2 has been shown to block cell proliferation, cell invasion, tumor growth and metastasis (Richter et al., 2009; Takeshita et al., 2005; Varambally et al., 2008), while overexpression of EZH2 produces an oncogenic phenotype (Herrera-Merchan et al., 2012; Min et al., 2010). Efforts are underway by a number of drug companies to develop small molecule inhibitors of EZH2 enzymatic activity (Crea et al., 2011; Creasy et al., 2012; Tan et al., 2007). Direct targeting of EZH2 may present challenges due to its role in normal stem and hematologic cells. Furthermore, recurrent inactivating mutations of EZH2 have been found in diseases such as myelodysplasia and T-cell precursor acute lymphoblastic leukaemia (Ernst et al., 2010; Nikoloski et al., 2010; Zhang et al., 2012), suggesting that in certain contexts, EZH2 can have tumor suppressive activities. Thus, identifying alternative enzymatic targets downstream of EZH2 may present opportunities for selective therapeutic modulation of EZH2 function.
MMSET (multiple myeloma SET domain), also known as WHSC1 (Wolf-Hirschhorn syndrome candidate 1) or NSD2 (nuclear SET domain-containing 2) is a SET-domain containing HMTase that is involved in the recurrent chromosomal translocation t(4;14) resulting in IgH enhancer- driven overexpression of MMSET in up to 20% of multiple myeloma (Chesi et al., 1998; Stec et al., 1998). MMSET binds to transcriptionally active regions of the genome (Ram et al., 2011) and specifically catalyzes H3K36 dimethylation, a mark associated with regions of open chromatin (Kuo et al., 2011; Li et al., 2009; Wagner and Carpenter, 2012). The oncogenic functions of MMSET in multiple myeloma are associated with its HMTase catalytic activity (Brito et al., 2009; Kuo et al., 2011; Lauring et al., 2008), making it amenable to therapeutic intervention. Recent reports suggest that like EZH2, MMSET is overexpressed in diverse solid tumors however the consequence and mechanism of this overexpression is not well characterized (Hudlebusch et al., 2011; Kassambara et al., 2009).
Here, we show that EZH2 and MMSET expression are tightly correlated in cancers. This correlation can partly be explained by the existence of an ordered HMTase axis, linked through microRNAs and histone marks. EZH2 functions upstream of MMSET and is directly responsible for H3K27me3 and associated transcriptional repression. Through the attenuation of selected microRNAs, however, EZH2 mediates overexpression of MMSET protein and indirect histone H3K36me2 which is associated with transcriptional activation.
Results
Coordinated expression of EZH2 and MMSET in cancer progression
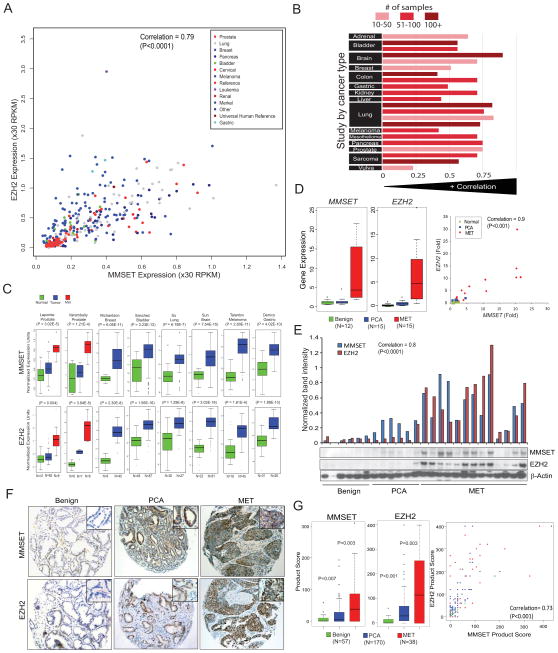
Since its initial association with prostate cancer progression (Varambally et al., 2002), the HMTase EZH2 has been the most well studied HMTase in cancer and has been shown to be consistently over-expressed across different cancer types and often associated with aggressiveness (Gong et al., 2011; Kleer et al., 2003; Lu et al., 2010; Min et al., 2010; Yamada et al., 2011). Thus, to look for HMTases that may be functionally linked to EZH2, we analyzed several published and unpublished transcriptomic datasets. EZH2 transcript expression levels were compared to all HMTases (n=51) in our compendium of RNA-seq data comprising 474 malignant and benign samples, representing 14 different tissue types. MMSET displayed the strongest correlation with EZH2 expression (Spearman correlation coefficient ρ= 0.79, p=7.27E-99) (Figure 1a; Table S1). The only other HMTases with a correlation coefficient of >0.5 were SUV39H1, PRMT3, SUV39H2, PRMT5 and NSD1 (ρ=0.68, 0.68, 0.64, 0.62 and 0.59 respectively) (Table S1). MMSET and EZH2 expression were the most correlated in colon/gastric cancer (n=46; ρ=0.89), pancreatic cancer (n=40; ρ=0.87), lymphoma (n=11; ρ=0.86) prostate cancer (n=100; ρ=0.83) and least correlated in cervical (n=10; ρ= −0.04) and oral cancer (n=7; ρ= −0.39) by RNA-seq analysis. Similarly, a meta-analysis of 1755 samples from 22 published microarray gene expression studies in Oncomine revealed co-expression of MMSET and EZH2 in 15 different cancers (Figure 1b; Table S2). In a majority of studies (82%) (Table S2), MMSET displayed the most correlated transcript expression with EZH2. Figure 1C depicts the levels of EZH2 and MMSET transcripts across a number of published microarray datasets.
Figure 1. MMSET and EZH2 are coordinately overexpressed and associated with tumor progression across cancer types.
(A) MMSET and EZH2 transcript expression levels are positively correlated across an RNA-seq (transcriptome) compendium of human tumors and cancer cell lines (n=474). The scatterplot displays the expression values in RPKM (Reads Per Kilobase per Million). (B) MMSET and EZH2 transcript expression across 22 microarray-based gene expression studies (n = 1,755) representing 15 tumor types. The histogram displays the correlation score within an individual study while bar shading indicates the number of samples used to calculate each correlation score. (C) MMSET and EZH2 transcript expression in multiple solid tumors based on Oncomine analysis. First author and statistical significance for respective studies are indicated. (D) qRT-PCR and (E) immunoblot analysis of MMSET and EZH2 transcripts and protein in a test cohort of benign prostate, clinically localized prostate cancer (PCA, and metastatic prostate cancer tissue (MET). Band intensity by densitometry is represented as a bar graph above the immunoblot. (F) Representative tissue microarray images depicting immunostaining pattern for MMSET and EZH2. Inset represents 40X magnifications. (G) Tissue microarray analysis of MMSET and EZH2 protein expression (left panel). Overall staining for each marker was measured by multiplying staining percentage (0–100%) by staining intensity on a numerical scale (none=1, weak=2, moderate=3, strong=4), resulting in an overall product score. Also see Table S1, S2, Figure S1.
To corroborate this observation experimentally, we tested 42 prostate samples including benign, clinically localized prostate cancer (PCA) and metastatic prostate cancer tissues (MET) for the expression of EZH2 and MMSET by qRT-PCR. While metastatic prostate cancer samples showed the highest expression of EZH2 as previously reported, a similar pattern of expression was observed for MMSET (Figure 1D) with a correlation of ρ=0.9 (p<0.001) for EZH2 and MMSET levels. EZH2 and MMSET transcript levels were 10-fold and 7-fold higher in metastatic tissues as compared to clinically localized prostate cancer, respectively (p= 0.007 and 0.002). To explore the EZH2-MMSET association at the level of protein expression, we carried out immunoblot analyses of cell lines and tissues. In a panel of prostate and breast cell lines, MMSET and EZH2 exhibited a correlation of 0.738 (p<0.0001) at the protein level (Figure S1A). Interestingly normal prostate primary cells (PrEC) and two independent primary normal melanocytes displayed low to undetectable levels of EZH2 and MMSET proteins (Figure S1A and data not shown). A similar correlation at the protein level was observed in a series of benign adjacent prostate, clinically localized prostate cancer and metastatic prostate cancer tissues (Figure 1E), with a correlation of 0.8 (p<0.0001).
To study the expression of these proteins in situ, we analyzed prostate cancer tissue microarrays (TMA) consisting of benign adjacent (n=57), PCA (n=170) and MET (n=38) samples and found that the product score of MMSET and EZH2 staining showed progressive increase from benign, localized and metastatic samples (p-value <0.01, <0.001 and ≤0.003 respectively) (Figures 1F and 1G), with a significant correlation between MMSET and EZH2 product scores (correlation coefficient = 0.73, p-value <0.001) (Figure 1G, right panel). Additionally, in cases of elevated expression, both MMSET and EZH2 were localized to the nucleus (Figure 1F). To assess whether MMSET expression, like EZH2 as reported in the literature (Bachmann et al., 2006; Varambally et al., 2002), is associated with poor prognosis, we used Oncomine Powertools to carry out Kaplan-Meier analysis of published studies (multiple myeloma, lymphoma, lung cancer and breast cancer) and observed poor outcome for patients with increased expression of MMSET transcript (Figure S1B). Taken together, these data establish MMSET as a highly expressed HMTase in multiple solid cancers that is correlated with the expression of EZH2 and is independently associated with poor patient survival in multiple cancer types.
MMSET functions downstream of EZH2 in an HMTase regulatory axis coordinated by microRNAs
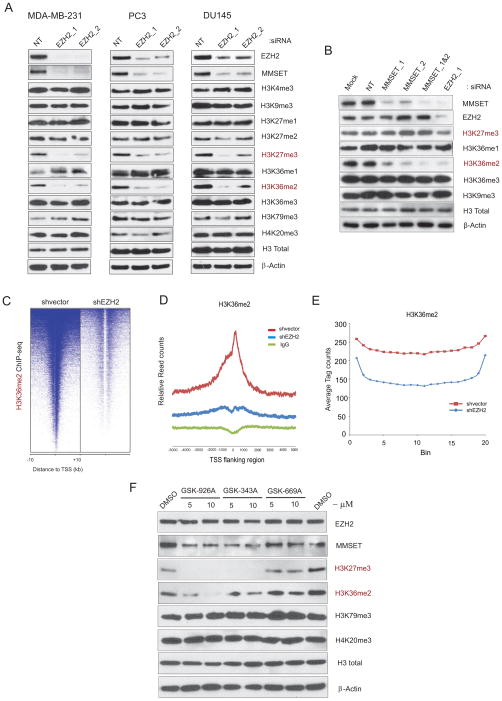
To investigate the functional relationship between EZH2 and MMSET, we knocked down EZH2 expression using two independent well characterized EZH2 specific siRNA and examined the effect on MMSET expression in prostate and breast cancer cell lines that harbor high levels of endogenous EZH2 and MMSET. Interestingly, MMSET expression was dramatically reduced upon EZH2 depletion in all cell lines tested (Figure 2A). We also interrogated various histone methylation marks for alterations resulting from EZH2 depletion and as expected, observed a reduction in the EZH2 mark H3K27me3, and remarkably, of all the other histone methylation marks tested, only the MMSET-associated histone mark, H3K36me2 (Kuo et al., 2011; Li et al., 2009) was also decreased (Figure 2A). Conversely, knockdown of MMSET did not affect the levels of EZH2 or its target H3K27me3, but the MMSET mark H3K36me2 was reduced as expected; H3K36me1 and H3K36me3 levels were unchanged (Figure 2B). Together, these observations suggest that MMSET functions downstream of EZH2 and knockdown of EHZ2, indirectly leads to the reduction of H3K36me2 -a histone mark generally associated with transcriptional activation.
Figure 2. EZH2 knockdown affects MMSET and its associated transcription activation mark H3K36me2.
(A) Immunoblot analysis of histone marks affected by EZH2 knockdown in cancer cell lines. NT, non-targeting siRNA. Total histone H3 and β-Actin served as loading controls. (B) As in (A) except MMSET knockdown. (C) EZH2 depletion leads to diminished global transcription activation mark H3K36me2 as determined by genome-wide ChIP-seq analysis. Heatmap representation of ChIP-seq binding peaks for H3K36me2 in stable shvector control compared with shEZH2 MDA-MB-231 breast cancer cells. Genomic target regions are rank ordered based on the level of H3K36me2 enrichment at each gene displayed from −10kb to +10kb surrounding annotated transcription start sites. (D) Average TSS-aligned profiles of H3K36me2 occupancy is shown for all annotated genes in control and EZH2 knockdown cells. (E) Average H3K36me2 occupancy in the gene bodies is shown. Genes containing H3K36me2 in control and EZH2 knockdown were each divided in to 20 bins and the H3K36me2 ChIP-seq tags within each bin was counted and averaged. (F) DU145 cells treated with EZH2 specific lead inhibitor compounds (GSK-926A and GSK-343A) and inactive analogue (GSK-669A) for 4 days and the levels of EZH2, MMSET, H3K27me3, H3K36me2, and H3K79me3 along with H4K20me3 as control were examined by immunoblotting using total cell extracts. Total H3 and β-Actin were used as a loading control. Also see Figure S2.
To study global cellular effects of EZH2 depletion on H3K36me2 mark at the chromatin level, we created stable EZH2 knockdown cells (Figure S2A) and performed chromatin immunoprecipitation with anti-H3K36me2 antibody followed by next-generation sequencing (ChIP-seq) (Figure S2B). We observed a global reduction in H3K36me2 enrichment at transcription start sites (TSS) and throughout gene bodies (Figures 2C–2E); 2,934 genes were enriched for H3K36me2 in EZH2 knockdown cells compared to 8,635 genes in the control cells, suggesting a loss of the H3K36me2 mark in more than 6,000 genes upon EZH2 knockdown (Table S3). Focusing on genes in which the H3K36me2 mark was lost throughout the gene body, we validated several differentially enriched genes by ChIP-qPCR including GLS2, a gene previously shown to harbor H3K36me2 marks (Martinez-Garcia et al., 2011), as well as genes such as Bola1, Plekhb1 etc. (Figures S2C, S2D and S2E). Knockdown of the transcriptional repressor EZH2 effectively reduces a histone mark associated with open chromatin indicating a broader role for EZH2 in chromatin homeostasis. In order to test the effect of EZH2-specific inhibitors on MMSET expression and its H3K36me2 mark, we employed EZH2-specific SAM-competitive lead inhibitor compounds identified through highthroughput biochemical screen by GSK. As expected, treatment of cells with these lead compounds (GSK-926A and GSK-343A) showed complete loss of EZH2 associated H3K27me3 mark without any change in EZH2 protein levels (Figure 2F). Interestingly, MMSET and its associated H3K36me2 mark also displayed reduction upon treatment with GSK-926A and GSK-343A. However, GSK-669A which is an inactive analogue had no effect on any of the histone marks. Next, we tested the effect of 3-Deazaneplanocin A (DZNep) (a compound known to deplete cellular levels of PRC2 components and its associated histone mark) on MMSET and its histone mark (Tan et al., 2007). As expected, DZNep treatment reduced EZH2 levels as well as its associated H3K27me3 mark in DU145 cells. Interestingly, the level of MMSET and its associated H3K36me2 mark were markedly attenuated (Figure S2F). However, DZNep-treated cells that were briefly exposed to proteasome inhibitor MG132 led to restoration of EZH2 but not MMSET levels (Figure S2G). Thus, DZNep treatment leads to EZH2 depletion through proteasome mediated degradation that subsequently results in reduction of MMSET levels. Further supporting our hypothesis, long term exposure to MG132 slightly relieved the repression of both EZH2 and MMSET (Figure S2H) demonstrating that MMSET expression is dependent on EZH2 activity.
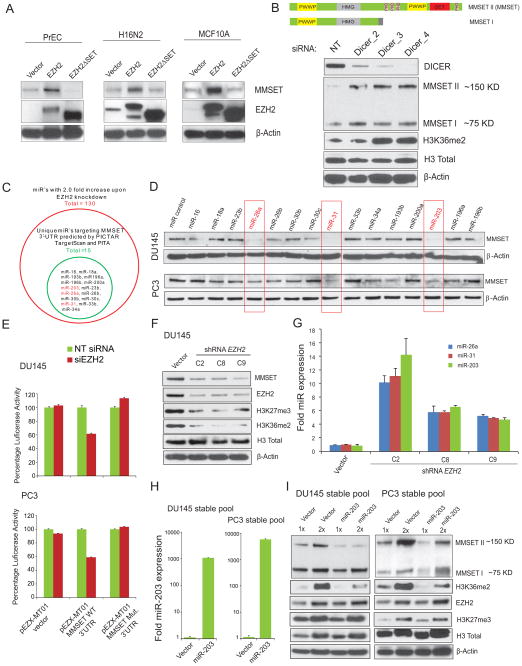
To explore the mechanism by which EZH2 regulates MMSET expression, we overexpressed EZH2 in multiple normal prostate and benign breast epithelial cells by adenoviral infection. A significant induction of MMSET protein was seen in cells overexpressing EZH2 compared to cells expressing EZH23SET mutant lacking HMTase activity (Figure 3A). By contrast, MMSET transcript levels were unchanged by EZH2 overexpression (data not shown). Taken together, this led us to hypothesize that MMSET regulation may occur at post-transcriptional level through microRNAs (miRNAs) in these normal epithelial cells. To test this hypothesis, we first knocked down Dicer, a key protein required for miRNA biogenesis, and observed a marked increase in MMSET protein (Figure 3B) but no significant increase of transcript levels in Dicer-depleted PC3 cells (Figure S3A). We recently reported that EZH2 represses the transcription of several miRNAs that act as tumor suppressors in cancer through epigenetic silencing mediated by H3K27me3 (Cao et al., 2011). Here, we investigated whether a subset of miRNAs identified in that study might target the MMSET 3′UTR. Using miRNA target analysis (PicTar, TargetScan and PITA), multiple binding sites were found in the MMSET 3′UTR (longer isoform) for 15 miRNAs out of a total of 130 previously defined EZH2-repressed miRNAs (Cao et al., 2011) (Figure 3C). To identify the specific miRNAs that target MMSET, we overexpressed each of these 15 miRNAs in DU145 and PC3 cells and evaluated MMSET protein expression. Overexpression of miR-26a, miR-31 and miR-203, that bind to multiple sites in the 3′UTR of MMSET (Figure S3B), led to a decrease in MMSET protein level in both cell lines (Figure 3D). Moreover, these miRNAs repressed a luciferase reporter containing the full length 3′UTR of MMSET in HeLa and HEK293 cells, corroborating the interaction of these miRNAs with MMSET 3′UTR (Figure S3C). To evaluate the EZH2-microRNA-MMSET axis further, we generated MMSET 3′UTR mutated for all of the 20 potential miR-203, miR-26a and miR-31 binding sites (Figure S3B) and performed luciferase reporter assay upon EZH2 knockdown. Depletion of EZH2 led to substantial repression of MMSET wild type 3′UTR reporter activity, whereas mutant 3′UTR reporter activity was resistant to repression by EZH2 knockdown in two different prostate cancer cell lines DU145 and PC3 (Figure 3E), clearly demonstrating the regulation of MMSET expression by EZH2 repressed miRNAs. Additionally, three different stable clones of EZH2 knockdown DU145 cells exhibiting reduced MMSET and H3K36me2 (Figure 3F), displayed increased expression of miR-26a, miR-31 and miR-203 (Figure 3G). Finally, DU145 and PC3 cells stably overexpressing miR-203 displayed reduced MMSET protein and H3K36me2 mark without any significant change in EZH2 and H3K27me3 levels (Figure 3H and 3I). Taken together, these data strongly suggest the existence of an HMTase axis linking EZH2 and MMSET, where EZH2 induces MMSET expression by repressing a set of miRNAs, resulting in coordinated transcriptional repression and activation in cancer cells.
Figure 3. EZH2 regulates MMSET expression coordinated by microRNAs.
(A) EZH2 overexpression induces MMSET protein expression but not transcript expression in benign epithelial cells. Immunoblot analysis of MMSET and EZH2 in the primary prostate cells (PrEC) and benign breast cell lines H16N2 and MCF10A following transduction with vector control, EZH2, or EZH23SET mutant adenovirus. (B) Top, Schematic of two major isoforms of MMSET with different functional domains. MMSET I is a shorter isoform and MMSET II is a full length SET domain containing enzymatically active isoform. Bottom, Dicer knockdown enhances MMSET protein expression. Immunoblot analysis of Dicer and MMSET following knockdown of Dicer in PC3 cells (C) Venn diagram displaying miRs which are increased upon EZH2 knockdown (red circle) and miRs that are predicted to target the 3′UTR of MMSET by multiple algorithms (green circle). (D) Immunoblot analysis of MMSET in DU145 and PC3 prostate cancer cells transfected with 15 predicted MMSET 3′UTR targeting miRs. (E) microRNAs binding mutant 3′UTR of MMSET is resistant to EZH2 mediated regulation. Luciferase Reporter assays with MMSET wild-type or mutant 3′UTR co-transfected with either NT siRNA or siEZH2 in DU145 and PC3 cells. pEZX-MT01 without 3′UTR served as control. Luciferase activity was measured 48hr post transfection, normalized using Renilla luciferase activity and the results are plotted as mean ±SEM (n=6) percentage luciferase activity. (F) Stable knockdown of EZH2 reduces MMSET protein level with a concomitant increase in miR-203, miR-26a and miR-31 expression. Immunoblot analysis of MMSET, EZH2, and their respective histone methylation marks in 3 stable DU145-shEZH2 clones. Total H3 and β-Actin were used as loading controls. (G) TaqMan qRT-PCR analysis showing upregulation of miR-203, 26a and 31; expression was normalized using U6 small RNA in stable vector and EZH2 knockdown clones. (H) TaqMan qRT-PCR of miR-203 in DU145 and PC3 pools stably expressing miR-203. (I) Stable overexpression of miR-203 reduces MMSET but not EZH2 levels. Immunoblot analysis of MMSET, EZH2 and their respective histone methylation marks. Note the lack of change to shorter MMSET isoform upon miR-203 overexpression. Also see Figures S3.
EZH2-mediated cell invasion and intravasation requires MMSET expression
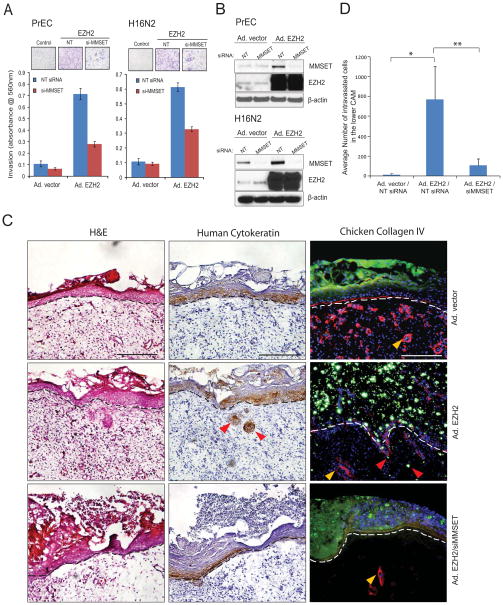
Earlier we reported that overexpression of EZH2 confers high invasive potential to normal prostate and breast epithelial cells (Cao et al., 2008; Kleer et al., 2003; Yu et al., 2007). To test the role of MMSET in mediating the EZH2 oncogenic program, we performed Matrigel invasion assays using primary prostate epithelial cells (PrEC) and benign immortalized breast epithelial cells (H16N2) infected with control or EZH2 adenoviruses along with control or MMSET specific siRNA duplexes. As expected, cells overexpressing EZH2 displayed high invasive potential that was dramatically attenuated upon MMSET knockdown, suggesting that MMSET is critical for EZH2-mediated cell invasion (Figure 4A). Overexpression of EZH2 induced MMSET protein level which was then depleted upon MMSET siRNA treatment (Figure 4B). To examine the effect of MMSET knockdown on EZH2-mediated invasion and intravasation in vivo, we employed a chicken chorioallantoic membrane (CAM) model. Breast epithelial cells overexpressing EZH2 consistently invaded the CAM, whereas knockdown of MMSET abolished this effect (Figure 4C), and the relative number of EZH2 overexpressing cells that intravasate into the vasculature of the lower CAM was significantly reduced upon MMSET knockdown, (p= 0.02 and 0.03 for control vs. EZH2 overexpression vs. EZH2 overexpression/MMSET knockdown) (Figure 4D). In a reciprocal approach to test which pro-metastatic effect of EZH2 requires MMSET, we performed cell proliferation, migration and invasion assay in DU145 cells transfected with EZH2 specific siRNA duplexes along with control or MMSET adenovirus infection (Figure S4A). As expected knockdown of EZH2 led to reduced cell proliferation but this was not rescued by MMSET overexpression (Figure S4B). However, migration and matrigel invasion phenotypes were significantly rescued by reintroduction of MMSET (Figure S4C). Taken together, these data clearly demonstrates MMSET as a downstream mediator of EZH2 regulated cell migration and invasion phenotypes.
Figure 4. MMSET expression is required for EZH2 mediated invasion and intravasation.
(A) Knockdown of MMSET attenuates EZH2 mediated cell invasion. Cell invasion was carried out using a modified Boyden chamber assay. PrEC and H16N2 benign cells were transduced with EZH2 adenovirus with or without siRNA to MMSET. Representative images of invading cells (inset). (B) Immunoblot analysis using the protein lysates from A showing induction of MMSET upon EZH2 overexpression. (C) EZH2 induced cell invasion in vivo was attenuated with siRNA to MMSET. H16N2 cells transduced with adeno-vector control, adeno-EZH2 or adeno-EZH2/siMMSET and labeled with green fluorescent microspheres were cultured atop the embryonic chick CAM for 3 days. The upper CAM was harvested 3 days post-engraftment of the cells and frozen sections were stained for hematoxylin and eosin (left panels), human-specific cytokeratin (immunohistochemistry, middle panels) or chicken-specific collagen IV (red immunofluorescence, right panels). The basement membrane is represented by dotted line, yellow arrow-heads indicate blood vessels in mesenchymal CAM tissue and red arrow-heads indicate cells invading through the basement membrane. Representative images are shown with a 200 μm scale bar. (D) Genomic DNA isolated from the lower CAM from C was used to measure the intravasated human cells by qPCR using human-specific Alu primers. Mean cell number ±SEM from 8 eggs per group shown, with * and ** indicates p=0.02 or p=0.03, Student’s t-test. Also see Figure S4.
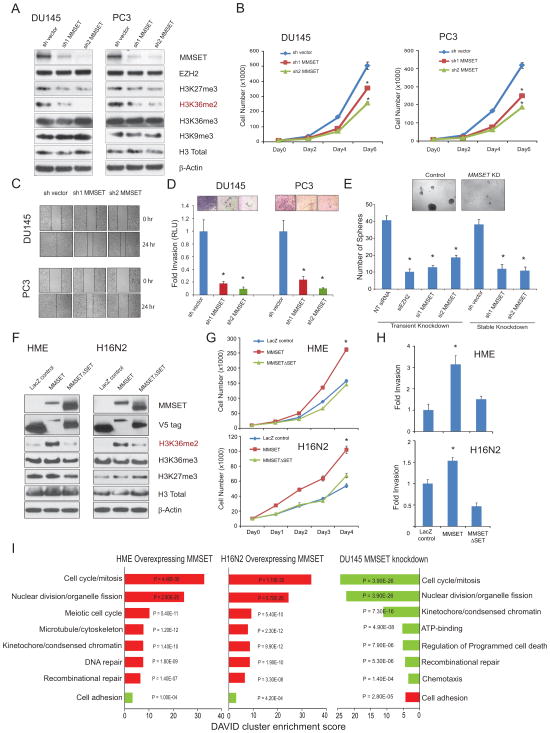
MMSET overexpression promotes diverse oncogenic phenotypes in vitro
To study the role of MMSET during cancer progression, we created stable knockdowns of MMSET in DU145 and PC3 prostate cancer cell lines using two different lentivirus-based shRNAs. Both cell lines express high levels of endogenous MMSET and upon knockdown, clearly show reduction in its H3K36me2 chromatin mark without affecting other H3 histone methylation marks or EZH2 levels (Figure 5A). Stable knockdown of MMSET reduced DU145 and PC3 cell proliferation in vitro as well as wound healing ability, as measured by cell count and scratch assay respectively (Figures 5B and 5C) and showed a dramatic reduction in their ability to invade through Matrigel as assessed by Boyden chamber assay (Figure 5D). Additionally, we evaluated the potential of DU145 cells to form prostatospheres in sphere-promoting cell media which serves as a surrogate measure of stem cell-like phenotypes; cells that are able to form spheres in general have enhanced stem cell characteristics (Lawson et al., 2007). Here, we found that both transient and stable knockdown of MMSET significantly inhibited the ability of DU145 cells to form spheres (Figure 5E). EZH2 knockdown of DU145 cells, which are known to inhibit prostatosphere formation, were used as a positive control (Cao et al., 2011). Conversely, overexpression of MMSET in HME and H16N2 benign immortalized breast epithelial cells specifically induced H3K36me2 compared to lacZ control and by contrast, overexpression of MMSET SET domain deletion mutant (MMSET3SET) had no effect (Figure 5F). MMSET overexpression induced significantly higher cell proliferation and invasion in vitro as measured by cell count and Boyden chamber assay compared to control cells (Figures 5G and 5H). MMSET overexpressing HME and H16N2 cells were profiled by gene expression microarrays and subsequent gene ontology analysis revealed enrichment of gene set concepts such as cell cycle, apoptosis, DNA repair, and cell adhesion (Figure 5I). Importantly, when MMSET was knocked down in DU145 prostate cancer cell lines many of these same concepts were repressed (Figure 5I, right panel). Furthermore, Gene Set Enrichment Analysis (GSEA) revealed enrichment for multiple cancer specific gene profiles in HME cells overexpressing MMSET and not in MMSET3SET (Figures S5).
Figure 5. MMSET overexpression promotes cell proliferation, self-renewal and cell invasive phenotype.
(A) Immunoblot analysis of MMSET, EZH2 and different histone methylation marks in stable MMSET knockdown cells. (B–D) MMSET knockdown in DU145 and PC3 cells exhibit reduced cell proliferation (B), reduced cell migration as assessed by wound healing assay (C), and reduced cell invasion as monitored by Matrigel invasion assay (D) with representative images of invaded cells shown in the inset. (E) Prostatosphere formation by DU145 cells is decreased by transient or stable knockdown of MMSET. Cells transfected with siEZH2 were used as a positive control. Representative images of prostatospheres formed by control or MMSET knockdown cells are shown. (F) Overexpression of MMSET but not its SET domain mutant specifically induces H3K36me2 in HME and H16N2 benign epithelial cells. LacZ overexpression was used a control. (G, H) MMSET overexpression induces increased cell proliferation and cell invasion respectively in HME or H16N2 cells. Mean ± SD shown with *p<0.01 (Student’s t-test) for all experiments. (I) Gene ontology analysis of MMSET overexpression and knockdown microarray data using the DAVID program. Red bars represent the top significant hits for upregulated genes. Green bars represent the top hits for downregulated genes. DAVID enrichment scores are plotted with P values. Also see Figure S5.
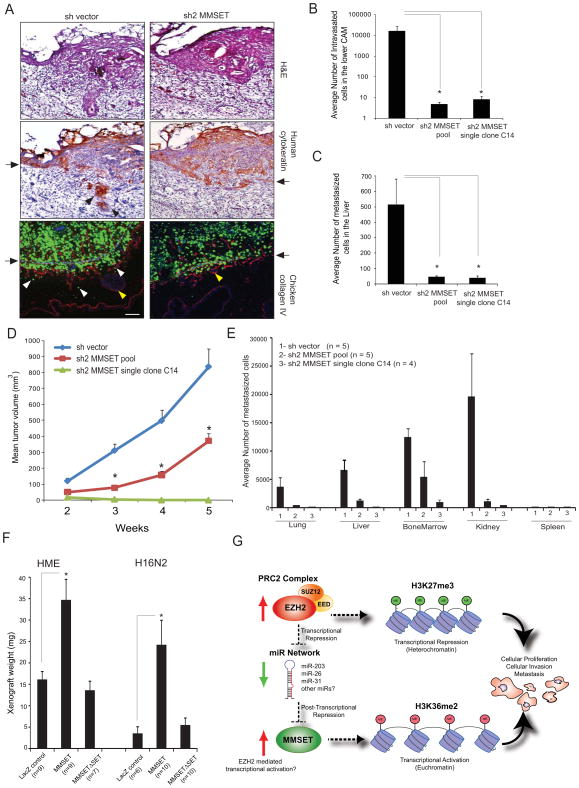
MMSET overexpression affects tumor formation and metastasis in vivo
In order to test the tumorigenic functions of MMSET in vivo, the chicken CAM model was employed in which three major steps leading to spontaneous metastasis can be assessed including local invasion, intravasation and metastasis to distant organs. MMSET knockdown of DU145 cells impaired their ability to invade the CAM basement membrane and resulted in a significantly decreased number of intravasated cells in the lower CAM compared to control cells (Figures 6A and 6B). To eliminate the possibility of potential biases inherent in experiments with pooled population of cells or single clones, we carried out in vivo experiments using both models in parallel. Spontaneous metastasis of MMSET knockdown cells to the chicken embryonic liver was also dramatically reduced (Figure 6C). We next tested the effect of MMSET knockdown in the murine DU145 xenograft model and observed a significantly reduced tumor growth (p<0.0001) (Figure 6D). Interestingly, the MMSET single clone knockdown cells did not form measurable tumors in mice. As expected, a significant decrease in Ki-67-positive immunostained nuclei was observed in the tumors collected from MMSET knockdown DU145 cells compared to control cells (Figure S6). Multiple internal organs such as lung, liver, kidney and bone marrow isolated from xenografted mice showed significantly reduced metastatic burden (as assessed by human Alu-qPCR for spontaneous micrometastases) in the MMSET knockdown group (Figure 6E). Notably, the spleen did not show any sign of spontaneous metastasis even in the control group, indicating that this organ is a poor site for forming secondary metastasis by prostate cancer cells. Finally, HME and H16N2 cells ectopically overexpressing MMSET showed a significant increase in xenograft weight compared to the lacZ control or MMSET3SET mutant as assessed by CAM assay (Figure 6F).
Figure 6. MMSET knockdown attenuates tumorigenesis in vivo.
(A) MMSET knockdown attenuates basement membrane invasion in a chicken CAM model. DU145 cells expressing GFP shvector control or shMMSET were engrafted atop the CAM of 11-day-old chick embryo and cultured for 3 days. Representative H&E, IHC with human specific cytokeratin, and immunofluorescence (IF) micrographs of CAM cross-sections showing DU145 cells (green) crossing the basement membrane (arrows). Red, chicken collagen IV; blue, DAPI staining for cell nuclei; arrows, basement membrane; red and white arrowheads, invaded cells; yellow arrowheads, blood vessels in CAM tissue. Scale bar (100 μm). (B) MMSET knockdown reduces cancer cell intravasation. Genomic DNA from lower CAM was isolated 3 days post-engraftment and number of intravasated cells was measured by human Alu-qPCR. (C) MMSET knockdown attenuates distant metastasis. Control or MMSET knockdown pool or single clone cells were inoculated atop CAM of 11-day-old chick embryos. Genomic DNA from lungs was extracted seven days post-inoculation and analyzed by human Alu-qPCR. Bar graph represents average cell number with ±SEM from N=7 per group. (D) MMSET knockdown attenuates DU145 tumor growth in mice. N=7 mice per group were injected subcutaneously. (E) MMSET knockdown reduces spontaneous metastasis in mouse xenograft model. Various organs and bone marrow cells were isolated from the DU145 xenografted mice after 5 weeks. Genomic DNA isolated from these organs was analyzed for metastasized cells by human Alu-qPCR. (F) Increased xenograft growth in vivo in CAM assay by HME and H16N2 cells overexpressing MMSET.*p<0.05, Student’s t-test. (G) Proposed model for the EZH2-MMSET histone methyltransferase axis in cancer. Red and green arrows indicate upregulation and downregulation respectively. Also see Figure S6.
Discussion
In this study, we characterize a remarkably tight association and functional link between the HMTases EZH2 and MMSET across different cancer types. Like EZH2, MMSET is elevated in aggressive forms of cancer and is often associated with poor prognosis. Somewhat akin to kinase phosphorylation cascades, in which an upstream kinase mediates the phosphorylation and activation of a downstream kinase, our data suggests the existence of a histone methylation axis, in which EZH2 mediated H3K27 trimethylation leads to H3K36 dimethylation via regulation of MMSET (Figure 6G). The link between EZH2 and MMSET is mediated by a network of microRNAs, including miR-203, miR-26a and miR-31. The high levels of MMSET transcripts in diverse cancers could be due to its increased stability owing to the loss of MMSET 3′-UTR targeting microRNAs such as miR-203, miR-26 and miR-31 which are known tumor suppressor microRNAs also involved in negative regulation of, for instance PRC1/PRC2 components and the NF-kappaB pathway (Cao et al., 2011; Sander et al., 2008; Yamagishi et al., 2012). EZH2, as the catalytic component of PRC2, has primarily been linked to transcriptional repression of target genes through its H3K27 trimethylation activity. However, these studies suggest that EZH2 can in parallel initiate transcriptional activation of key target genes through its induction of MMSET. The inter-connected repressive and activating histone methylation marks, generated by the interplay and coordinated regulation of the EZH2-MMSET axis, highlights yet another level of complexity in the dynamics of the epigenetic landscape of cells in disease and normal development. Interestingly, multiple myeloma cells naturally harboring the t(4;14) translocation of MMSET appear to bypass the EZH2-MMSET axis, whereas t(4;14) translocation negative multiple myeloma cells exhibit an intact EZH2-microRNA-MMSET axis (data not shown).
EZH2 has thus far been shown to manifest its oncogenic activity primarily through repression of target genes including p16INK4alpha (Bracken et al., 2007; Kotake et al., 2007) DAB2IP (Min et al., 2010), ADRB2 (Yu et al., 2007), WNT pathway antagonists (Cheng et al., 2011), VASH1 (Lu et al., 2010) and CDH1 (Cao et al., 2008), among others, a majority of these being tumor suppressors. Downstream effectors of EZH2 specifically linked with cancer cell metastasis have not been previously identified. Here, our data implicates MMSET as a major effector of EZH2, involved in key steps of the tumorigenic pathway: cell proliferation, invasion, migration, intravasation, and metastasis. Thus, MMSET represents a bona fide oncogene and a chromatin modifier that is associated with EZH2 activity and involved in tumor cell metastasis.
While there is tremendous interest in the pharmaceutical and biotech industry to develop inhibitors of EZH2 for cancer, the genomic data provide contrasting roles for EZH2 in cancer development. For example, in follicular and diffuse large B cell lymphoma, activating mutations of EZH2 have been identified suggesting oncogenic activities of EZH2, while in myelodysplasia and T-cell precursor acute lymphoblastic leukemia inactivating mutations of EZH2 have been identified, suggesting tumor suppressive activity of EZH2. Thus, total inactivation of the transcriptional repressive activity of EZH2 may not be desirable in cancer. Approaching downstream oncogenic targets of EZH2 such as MMSET, which is involved in transcriptional activation, may provide an alternate therapeutic strategy.
In summary, we provide compelling evidence for the existence of an HMTase axis involving the transcriptional repressor EZH2 and the transcriptional activator MMSET which is partly interconnected by microRNAs. An analogous HMTase axes likely exist and also play a role in epigenetic regulation in normal development and disease progression.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures can be found in supplemental information.
Cell cultures, antibodies and immunoblot analyses
Primary human prostate epithelial cells, PrEC, and immortalized human prostate epithelial cells, RWPE1, were obtained from Lonza and American Type Culture Collection (ATCC) respectively and were maintained according to the supplier’s instructions. Immortalized breast cell lines, HME and H16N2, were grown in MEBM medium with supplements (Lonza). Human prostate and breast cancer cell lines were all obtained from ATCC and maintained according to supplier’s instructions. Antibodies used in the study are listed in Table S4 and provided as an excel file. All antibodies were employed at dilutions suggested by the manufacturers. For immunoblot analysis, 200μg total protein extract was boiled in sample buffer and 10–15ug aliquots were separated by SDS-PAGE and transferred onto PVDF membrane (GE Healthcare). The membrane was incubated for half an hour in blocking buffer [Tris-buffered saline, 0.1% Tween (TBS-T), 5% nonfat dry milk] followed by incubation overnight at 4°C with the primary antibody. Following a wash with TBS-T, the blot was incubated with horseradish peroxidase-conjugated secondary antibody and signals were visualized by enhanced chemiluminescence system as per manufacturer’s protocol (GE Healthcare).
Supplementary Material
Highlights.
Coordinated regulation of EZH2 and MMSET in cancer progression
EZH2 regulates MMSET and its H3K36me2 mark through microRNAs
MMSET is a downstream mediator of EZH2 oncogenic function
MMSET overexpression promotes diverse oncogenic phenotypes in vitro and in vivo
Acknowledgments
We thank Catherine Grasso for RNA-seq portal, T. Barrette for database management, Vijaya Lakshmi Dommeti, Xiaojun Jing and Khalid Suleman for technical assistance, Dashyant Dhanak, Caretha Creasy and Peter Tummino from GlaxoSmithKline for their lead EZH2 inhibitor compounds, Jyoti Athanikar and Karen Giles for critically reading the manuscript and submission of documents. This work was supported in part by the Early Detection Research Network grant (UO1 CA111275), Prostate SPORE grant (P50CA69568), the Department of Defense (DOD) Era of Hope grant (BC075023) to A.M.C. A.M.C. is also supported by the Doris Duke Charitable Foundation Clinical Scientist Award and the Prostate Cancer Foundation. A.M.C. is an American Cancer Society Research Professor and A. Alfred Taubman Scholar. Q.C. is supported by DOD postdoctoral training award (PC09472). Q.Z. is supported by NIH (RO1HG005119). S.V. is supported by NIH (R01CA157845).
Footnotes
ACCESSION NUMBERS
Gene expression and ChIP-seq coordinates have been deposited in the GEO # (submitted and pending)
Supplemental information includes Supplemental Experimental Procedures, six figures, two tables; two excel files and Supplemental References and can be found with this article online at …
Author contributions
I.A.A. and A.M.C. conceived the study and experiments. I.A.A. performed majority of the experiments with assistance from Q.C., L.D., M.P. and S.V. B.A performed mouse xenograft studies. L.P.K., R.M., R.J.L., J.S. and N.P. performed TMA staining and analysis. Y.-M. W., and X.C. constructed ChIP-seq library and carried out sequencing. J.H.K., Z.M. and Z.S.Q performed ChIP-seq analysis. M.K.I and C.A.M. performed gene expression analysis and miRNA predictions. I.A.A, C.K.-S. and A.M.C wrote the manuscript that was reviewed by all the authors.
Conflict of interest
A.M.C. is a co-founder and SAB member of Compendia Biosciences which supports Oncomine and associated bioinformatics tools. A.M.C. serves on the SAB of Constellation Pharmaceuticals and serves as an advisor to Ventana/Roche and Glaxo Smith Kline. None of these companies were involved in these studies or approved of its content.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5:1886–1901. doi: 10.4161/cc.5.16.3222. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, Avramidou A, Irving JA, Gonzalez D, Davies FE, et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica. 2009;94:78–86. doi: 10.3324/haematol.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20:187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, Mehra R, Laxman B, Cao X, Kleer CG, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS, Feng H, Ching AK, Cheung KF, Wong HK, Tong JH, et al. EZH2-mediated concordant repression of Wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–4039. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: Time for clinical application? Crit Rev Oncol Hematol. 2011 doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Creasy CL, McCabe MT, Korenchuk S, Diaz E, Ott H, Thompson CS, Ganji G, Gorman SA, LaFrance LV, Brandt M, et al. A novel selective EZH2 inhibitor exhibits anti-tumor activity in lymphoma with EZH2 activating mutations. Paper presented at: American Association for Cancer Research Annual Conference; Chicago, AACR. 2012. [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- Gong Y, Huo L, Liu P, Sneige N, Sun X, Ueno NT, Lucci A, Buchholz TA, Valero V, Cristofanilli M. Polycomb group protein EZH2 is frequently expressed in inflammatory breast cancer and is predictive of worse clinical outcome. Cancer. 2011;117:5476–5484. doi: 10.1002/cncr.26179. [DOI] [PubMed] [Google Scholar]
- Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiaer E, Rossing HH, Johansen JV, Jorgensen M, Sauter G, Helin K. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–2933. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K, Cross NC, Cheng JF, Aplan PD, Kearney L, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98:1264–1267. doi: 10.1182/blood.v98.4.1264. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, Klein B, Moreaux J. MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun. 2009;379:840–845. doi: 10.1016/j.bbrc.2008.12.093. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. NSD2 Links Dimethylation of Histone H3 at Lysine 36 to Oncogenic Programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, Arceci RJ, Matsui W, Park BH. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci U S A. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, Armaiz-Pena GN, Hu W, Stone RL, Munkarah A, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt LM, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial Patterning of Chromatin Regulators Uncovered by Genome-wide Location Analysis in Human Cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, Hotfilder M, Lowel D, von Luettichau I, Mossbrugger I, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Spannhoff A, Sippl W, Jung M. Cancer treatment of the future: inhibitors of histone methyltransferases. Int J Biochem Cell Biol. 2009;41:4–11. doi: 10.1016/j.biocel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, Teratani T, Namatame N, Yamamoto Y, Hanai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci U S A. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Fujii S, Daiko H, Nishimura M, Chiba T, Ochiai A. Aberrant expression of EZH2 is associated with a poor outcome and P53 alteration in squamous cell carcinoma of the esophagus. Int J Oncol. 2011;38:345–353. doi: 10.3892/ijo.2010.868. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, Matsuda Y, Sato-Otsubo A, Muto S, Utsunomiya A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012;21:121–135. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Yu J, Cao Q, Mehra R, Laxman B, Tomlins SA, Creighton CJ, Dhanasekaran SM, Shen R, Chen G, Morris DS, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.