Introduction
Herein we present a brief historical review of our approach to identify T cell stimulatory allergen epitopes relevant to allergic disease. The intent was to develop an approach to thoroughly map allergenic epitopes recognized by T cells, and phenotype the different cell types potentially involved in stimulating and inhibiting allergic responses. However, the number of allergen epitopes is extremely large and the studies required for mapping would be too complex. However, in order for allergen specific T cells to be activated, HLA molecules are required to form bimolecular complexes with the peptides derived from allergen molecules. In vitro binding assays and bioinformatic predictions can identify these peptides. A series of pivotal studies, described herein, delineates the feasibility of the approach to map the highly diverse repertoire of allergen epitopes recognizable to responsive T cells.
Rationale for large-scale allergen epitope identification
Allergic disease involves both adaptive and innate immunity, as recently highlighted in several excellent reviews 1, 2. At the level of adaptive responses, a key component are IgE antibodies produced by allergen-specific B cells 3. However, T cells also play a key role both as regulators of antibody responses as well as directly contributing to pathogenesis by secretion of various cytokines 4, 5. Furthermore, several studies implicate T cell responses as a determinant 6, 7 of the clinical efficacy of SIT (Specific Immuno-Therapy).
Despite their importance, the exact molecular structures (epitopes) recognized by T cells specific for common allergens have not been thoroughly mapped. Mapping these epitopes is of importance, to allow the measurement and characterization of allergen specific T cell responses, to directly assess the phenotype of responding cells, monitor efficacy of therapeutic treatments and probe mechanisms orchestrating pathogenic events.
The need to broadly define specific epitopes recognized in the context of allergic disease was emphasized by a meta-analysis of allergy-related data from the Immune Epitope Database (IEDB). The IEDB is a NIAID-sponsored resource that catalogs all data related to immune epitopes associated with infectious diseases, autoimmunity, transplantation and allergy 8 and makes it freely accessible to the scientific community. The majority of allergy-related epitopes found in the database are recognized by B cells / IgE antibodies, and the number of T-cell epitopes recognized by CD4+ T cells in the context of HLA class II is relatively low. Overall, coverage of known allergens was sparse, with data available for only ~17% of all allergens listed by the International Union Immunological Society (IUIS) database. Thus, further research was required to provide a more balanced representation across different allergen categories.
The complex nature of T helper subsets in allergic disease
It is well appreciated that human T helper (Th) cell responses are mediated by several different cell subsets, each associated with distinct functions and phenotypes. A large body of research has been devoted to the characterization of these subsets, their function in different types of immune responses and the complex interplay that occurs between each subset. The classic Th1, Th2, Th17 and Treg (Tr1) subsets are defined on the basis of the patterns of cytokine production and expression of specific chemokine receptors. In addition to these cell subsets, more recent investigations have also described Th22 and Th9 subsets 9.
Th2 cells have a clear established role in allergies, characterized by the production of IL-4, IL-5 and IL-13, and promote production of IgE. The role of the other Th subsets such as Th1, Th17 and regulatory T cells is less clearly defined. Th1 cells in allergies and asthma 10, 11 are associated with the production of IFNg, which is antagonistic to Th2 responses, and therefore it has been speculated that Th1 responses may protect against allergy and asthma. Indeed, Th1 cells have been shown to inhibit the proliferation and development of Th2 cells 12 and even IgE production in some instances 13.
Th17 cells were first described as key mediators of inflammation and autoimmune disease 14, 15. Though a role for IL-17 in allergy and asthma has been established, several conflicting reports exist regarding its involvement in the development and pathogenesis of allergic disease 16. A number of studies have described IL-17 as playing a proinflammatory role in allergy and asthma, whereas other studies have assigned IL-17 a more protective role 17, 18.
IL-10 has been well characterized as a mediator capable of suppressing T cell activation 19, 20. It is produced in large part by a subset of regulatory T cells known as Tr1s 21. Tr1s are characterized by the expression of CD4, CD25 and Foxp3, and the presence of these cells in allergic and asthmatic patients is associated with suppression of symptoms 19, 22. In fact, a number of studies suggest the presence or absence of Tr1s and/or IL-10 may contribute to seasonal exacerbations and efficacy of SIT 23-25.
Several observations highlight the complex interactions and plasticity of Th subsets 26. A subset of Th2 cells in PBMC from allergic individuals stimulated with anti-CD3 27 produces both Th17 and Th2 cytokines and is characterized by expression of CCR4 and CCR6. The production of IL-17 is induced from Th2 cells and Th2 cytokines synergize with IL-17 in inducing the production of various chemokines. Several studies suggest that Th17 cells have the capacity to transform into other lineages 26 or become dual IL-4/IL-17 producing cells 28. Indeed the interplay between Th cells producing these two cytokines has been implicated in the heterogeneity of the pathology of severe asthma 29. Similarly, several studies 30 suggest that Th1 polarizing stimuli can induce Th2 cells to produce IFNγ. Finally, besides the well-appreciated regulatory activity of IL-10 in terms of production from Tr1s and resulting in inhibition of other Th subsets, it has been suggested that Th1 and IFNγ producing cells regulate themselves by inducing IL-10 production 31, 32.
These reports highlight the plasticity and interplay of Th cell subsets producing different cytokines. However, the lack of information regarding the actual epitopes recognized by Th cells derived from common allergens severely limits our capacity to probe these interactions in more depth.
A broad approach to the study and characterization of HLA class II restricted epitopes
Clearly defined T cell epitopes are the basis for the study and characterization of the allergen-specific T cell population. To address this issue, we set out to investigate a large panel of common allergens and to identify their T cell epitopes 33.
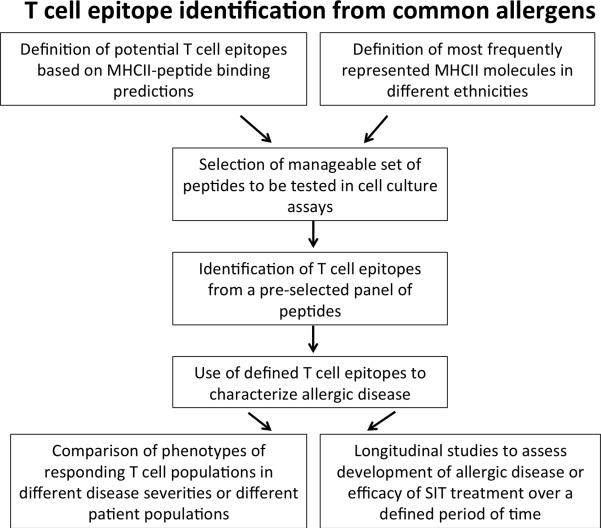
CD4 T cells recognize a bimolecular complex of the specific epitopes bound to specific HLA class II molecules (HLA restriction). HLA class II molecules are encoded by three different loci, designated as HLA DR, DP or DQ. Each of these loci is extremely polymorphic. This complexity must be taken into account in the design of a general strategy for identification and characterization of T cell epitopes in allergens. As outlined in Figure 1, we first selected a comprehensive, yet manageable, set of HLA DR, DP, and DQ molecules, representative of the most frequently expressed variants in different ethnicities worldwide. A panel of 25 different HLA class II molecules was selected for the experiments described below.
Figure 1.
A schematic overview of processes involved in large-scale T cell epitope identification from common allergens.
The number of peptides that could be tested to define epitopes from common allergens is very large, which poses significant challenges in terms of amounts of patient derived blood samples required. One method to reduce the complexity is to rely on the fact that epitopes have to bind specific HLA molecules, and the binding specificity of these molecules can be predicted. Accordingly, for each of the selected HLA molecules we developed high throughput binding assays and derived detailed peptide binding motifs. While assays and motifs were generally already available for DR molecules 34-37, the efforts directed towards DP and DQ specificities were largely novel.38,39.
Next, large panels of synthetic peptides were tested for binding to the various HLA class II molecules. This generated a dataset of over 40,000 HLA-peptide binding affinities for the various HLA DR, DP and DQ allelic variants. Utilizing this dataset, we generated prediction tools utilizing several machine learning algorithms and made them freely available to the scientific community through the IEDB website 40. The large datasets of measured binding affinities were also utilized to develop a functional classification of HLA class II on the basis of shared binding repertoires. Seven different main “supertypes” (main DR, DR4, DRB3, main DQ, DQ7, main DP, and DP2) were defined, corresponding to groups of alleles associated with similar peptide binding specificities. These results indeed highlighted that in general a high degree of peptide binding repertoire overlap exists amongst the various different HLA class II molecules.
Identification of T cell epitopes in the Timothy grass (TG) system
In approaching the problem of defining T cell epitopes from common allergens we expected to encounter a significant degree of heterogeneity due to HLA polymorphism. However, because of the overlap in repertoire of peptides bound by different HLA molecules, we also anticipated that some allergen-derived peptides are capable of binding several different HLA molecules (promiscuous binding). Our general hypothesis was that promiscuous epitopes account for a large fraction of reactivities, and that such promiscuous epitopes could be identified based on predicted HLA class II binding affinities.
This hypothesis was tested in the context of responses from patients allergic to Timothy grass. We considered 10 different proteins (Phl p 1, 2, 3, 4, 5, 6, 7, 11, 12, 13), corresponding to well-described allergens previously shown to elicit IgE reactivity. A total of 687 overlapping peptides spanning the sequences of these allergens were synthesized and tested for reactivity with T cell lines obtained by in vitro restimulation of PBMC with TG pollen extract. These studies utilized a cohort of 43 donors that included 10 non-allergic, 25 allergic, and 8 TG SIT individuals. A total of 70 unique peptides, corresponding to 43 unique Phl p antigenic regions were recognized 41. Hence, as expected, the repertoire of epitopes recognized by TG-specific T cell responses is highly diverse. This data is also consistent with several previous studies characterizing TG-derived T cell epitopes, which also highlighted a highly diverse repertoire 42, 43.
Validation of an approach for epitope identification based on bioinformatic prediction of promiscuous HLA binding
Next, the restricting HLA locus was identified using anti-DR, DP or DQ antibodies. Interestingly, we found that while most of the responses are DR restricted, DP and DQ also account for a significant fraction of total spot forming cells (SFCs). The actual allelic variant restricting the response was determined by the use of fibroblasts expressing a single HLA class II molecule, and/or matched and mismatched EBV transformed B cell lines.
A key issue for these validation studies was whether we could account for the majority of the responses with relatively few promiscuous regions. Analysis of the data relating to the response magnitude revealed that this was indeed the case. More specifically, the top 20 antigenic regions accounted for about 80% of the responses against the TG allergens considered. We further found that all of these most dominant antigenic regions are promiscuous in that multiple HLA class II molecules can bind and present them. This observation provides at least a partial explanation for their dominance. The next key issue for our validation studies was whether we would be able to predict the allergen epitopes in silico. We found that in silico bioinformatic prediction of promiscuous binding identified about half of the responses in individual donors 41.
Additional data supporting the validity of bioinformatic predictions to identifiy allergen-derived epitopes was recently provided in the Bla g (Blattella germanica) cockroach allergy system 44. In those studies, analysis of Bla g 1, 2, 4, 5, 6, and 7 allergen sequences led to the identification of 25 unique T cell epitopes, with 5 epitopes accounting for over half of the total response. These studies determined that Bla g 5 was the most dominant allergen for T cell responses. SIT resulted in down-modulation of IL-5 production, without induction of IFN-γ Bla g derived peptides.
Overall, we concluded that our initial hypotheses that promiscuous epitopes account for a large fraction of reactivities, and that such promiscuous epitopes could be identified based on predicted HLA class II binding affinities were valid. Consequently, we concluded that the approach for identification of allergen epitopes based on HLA predictions could be implemented to address a larger scale screen of allergenic proteins.
Further large-scale identification of allergen-derived epitopes
Based on the results obtained in the TG system, we embarked on a large scale screen of airborne allergens derived from a variety of different allergen sources. For each allergen system, all available sequences were collected. It was apparent that in the case of certain allergen sources many (in excess of 10) sequences were available, while in other cases only few protein sequences were reported.
Next, the predictive strategy defined in the TG model system was utilized to identify promiscuous HLA class II binding peptides from the various allergen sources. As a result, a total of 1736 peptides were synthesized33 and tested for recognition in allergic donor PBMCs (Figure 1). Allergen extracts effectively stimulated T cell responses in most donors, with an average of 95.3% positivity over the different allergen extracts considered. However, when the percent of extract response captured by pools of predicted epitopes was examined, we observed a significant degree of variability33, which correlated with the number of allergen sequences available for each allergen extract. On average, in the case of allergen extracts for which only one defined protein sequence was available, the peptides accounted for only 4% of the total response observed with the extract. By contrast, in the case of allergen extracts for which 6 or more sequences were available, the peptides accounted for approximately 40% of the total extract response.
A total of 87 different antigenic regions were recognized in 2 or more donors. Further experiments characterized the phenotype and restriction of these antigenic regions. In many cases the epitopes identified were the first defined T cell epitopes for that particular allergen source. In other cases, where T cell epitopes were previously known, our analysis greatly enhanced the breadth of T cell epitopes available to the scientific community 33.
Discussion
The ultimate goal of the identification of allergen epitopes is to utilize these molecularly defined reagents to illuminate and probe the mechanisms underlying T cell responses in allergic disease. We have initiated studies aimed at characterizing different disease states and correlated them with different epitope specific responses. In particular, several reports from the literature suggest that distinct patterns of cytokine production are associated with allergic asthma. A body of data has implicated, beyond Th2 cytokines, roles for IFNγ, IL-17 and IL-10 in allergic reactions 10, 11, 17-19, 22-25, 46, 47. Notably most reports have utilized mitogens or model antigens for stimulations. Since the actual T cell populations specifically responding to the allergen epitopes were not analyzed, this raises doubts regarding the physiological relevance of these stimulations. We anticipate that definition of the actual epitopes recognized from allergen specific T cells will allow probing the interplay of different Th cell subsets in allergic disease and improve the understanding of their effects on the disease profile and progression. Furthermore, we are in the process of characterizing T cell responses from patients undergoing the course of specific Immunotherapy to TG allergy by analyzing in detail the cytokines and functional phenotype of the allergen-specific T cells generated, and monitor for significant changes over the course of SIT.
Conclusion
Recent years witnessed a significant expansion of knowledge related to epitopes recognized in allergic disease, and to the complex interplay between different Th cell subsets. We anticipate that the combination of the advances in these two fields will yield new insights regarding the molecular mechanisms of disease, and thereby translate into potential novel intervention strategies, as well as novel ways to determine the magnitude and phenotype of allergen-specific responses for diagnostic purposes.
Acknowledgments
Financial support: This work was supported by National Institutes of Allergy and Infectious diseases Contract HSN272200700048C (to A.S.) and Grant U19AI100275 (to A.S.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author's contribution: All authors contributed to the preparation and critical reviewing and editing of the manuscript.
References
- 1.Locksley RM. Asthma and allergic inflammation. Cell. 2010;140:777–83. doi: 10.1016/j.cell.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holgate ST. Innate and adaptive immune responses in asthma. Nature medicine. 2012;18:673–83. doi: 10.1038/nm.2731. [DOI] [PubMed] [Google Scholar]
- 3.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nature reviews. Immunology. 2008;8:205–17. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 4.Romagnani S. The role of lymphocytes in allergic disease. The Journal of allergy and clinical immunology. 2000;105:399–408. doi: 10.1067/mai.2000.104575. [DOI] [PubMed] [Google Scholar]
- 5.Del Prete G. Human Th1 and Th2 lymphocytes: their role in the pathophysiology of atopy. Allergy. 1992;47:450–5. doi: 10.1111/j.1398-9995.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 6.Ebner C, Siemann U, Bohle B, et al. Immunological changes during specific immunotherapy of grass pollen allergy: reduced lymphoproliferative responses to allergen and shift from TH2 to TH1 in T-cell clones specific for Phl p 1, a major grass pollen allergen. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 1997;27:1007–15. doi: 10.1111/j.1365-2222.1997.tb01252.x. [DOI] [PubMed] [Google Scholar]
- 7.Bohle B, Kinaciyan T, Gerstmayr M, et al. Sublingual immunotherapy induces IL-10-producing T regulatory cells, allergen-specific T-cell tolerance, and immune deviation. The Journal of allergy and clinical immunology. 2007;120:707–13. doi: 10.1016/j.jaci.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Wang P, Kim Y, et al. Immune epitope database analysis resource (IEDB-AR). Nucleic acids research. 2008;36:W513–8. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zielinski CE, Corti D, Mele F, et al. Dissecting the human immunologic memory for pathogens. Immunological reviews. 2011;240:40–51. doi: 10.1111/j.1600-065X.2010.01000.x. [DOI] [PubMed] [Google Scholar]
- 10.Mazzarella G, Bianco A, Catena E, De Palma R, Abbate GF. Th1/Th2 lymphocyte polarization in asthma. Allergy. 2000;55(Suppl 61):6–9. doi: 10.1034/j.1398-9995.2000.00511.x. [DOI] [PubMed] [Google Scholar]
- 11.Santarlasci V, Maggi L, Capone M, et al. TGF-beta indirectly favors the development of human Th17 cells by inhibiting Th1 cells. European journal of immunology. 2009;39:207–15. doi: 10.1002/eji.200838748. [DOI] [PubMed] [Google Scholar]
- 12.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 13.Coffman RL, Seymour BW, Lebman DA, et al. The role of helper T cell products in mouse B cell differentiation and isotype regulation. Immunological reviews. 1988;102:5–28. doi: 10.1111/j.1600-065x.1988.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 14.Annunziato F, Cosmi L, Liotta F, Maggi E, Romagnani S. The phenotype of human Th17 cells and their precursors, the cytokines that mediate their differentiation and the role of Th17 cells in inflammation. International immunology. 2008;20:1361–8. doi: 10.1093/intimm/dxn106. [DOI] [PubMed] [Google Scholar]
- 15.Romagnani S. Human Th17 cells. Arthritis research & therapy. 2008;10:206. doi: 10.1186/ar2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcorn JF, Crowe CR, Kolls JK. TH17 cells in asthma and COPD. Annual review of physiology. 2010;72:495–516. doi: 10.1146/annurev-physiol-021909-135926. [DOI] [PubMed] [Google Scholar]
- 17.Beriou G, Costantino CM, Ashley CW, et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113:4240–9. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi M, Takahashi D, Hizawa N, et al. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. The Journal of allergy and clinical immunology. 2006;117:795–801. doi: 10.1016/j.jaci.2005.12.1346. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd CM, Hawrylowicz CM. Regulatory T cells in asthma. Immunity. 2009;31:438–49. doi: 10.1016/j.immuni.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryanna K, Stratigou V, Safinia N, Hawrylowicz C. Regulatory T cells in bronchial asthma. Allergy. 2009;64:335–47. doi: 10.1111/j.1398-9995.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Yang A, Huang H, et al. Induction of type 2 T helper cell allergen tolerance by IL-10-differentiated regulatory dendritic cells. American journal of respiratory cell and molecular biology. 2010;42:190–9. doi: 10.1165/rcmb.2009-0023OC. [DOI] [PubMed] [Google Scholar]
- 22.Han D, Wang C, Lou W, et al. Allergen-specific IL-10-secreting type I T regulatory cells, but not CD4(+)CD25(+)Foxp3(+) T cells, are decreased in peripheral blood of patients with persistent allergic rhinitis. Clinical immunology. 2010;136:292–301. doi: 10.1016/j.clim.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Akdis CA, Akdis M. Mechanisms of allergen-specific immunotherapy. The Journal of allergy and clinical immunology. 2011;127:18–27. doi: 10.1016/j.jaci.2010.11.030. quiz 8-9. [DOI] [PubMed] [Google Scholar]
- 24.Anderson AE, Mackerness KJ, Aizen M, et al. Seasonal changes in suppressive capacity of CD4+ CD25+ T cells from patients with hayfever are allergen-specific and may result in part from expansion of effector T cells among the CD25+ population. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2009;39:1693–9. doi: 10.1111/j.1365-2222.2009.03320.x. [DOI] [PubMed] [Google Scholar]
- 25.Domdey A, Liu A, Millner A, et al. The T cell response to major grass allergens is regulated and includes IL-10 production in atopic but not in non-atopic subjects. International archives of allergy and immunology. 2010;152:243–54. doi: 10.1159/000283033. [DOI] [PubMed] [Google Scholar]
- 26.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell research. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YH, Voo KS, Liu B, et al. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. The Journal of experimental medicine. 2010;207:2479–91. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosmi L, Maggi L, Santarlasci V, et al. Identification of a novel subset of human circulating memory CD4(+) T cells that produce both IL-17A and IL-4. The Journal of allergy and clinical immunology. 2010;125:222–30. e1–4. doi: 10.1016/j.jaci.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt-Weber CB, Akdis M, Akdis CA. TH17 cells in the big picture of immunology. The Journal of allergy and clinical immunology. 2007;120:247–54. doi: 10.1016/j.jaci.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 30.Hegazy AN, Peine M, Helmstetter C, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–28. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nature immunology. 2011;12:327–34. doi: 10.1038/ni.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stumhofer JS, Silver JS, Laurence A, et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nature immunology. 2007;8:1363–71. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 33.Oseroff C, Sidney J, Vita R, et al. T Cell Responses to Known Allergen Proteins Are Differently Polarized and Account for a Variable Fraction of Total Response to Allergen Extracts. Journal of immunology. 2012 doi: 10.4049/jimmunol.1200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenbaum J, Sidney J, Chung J, et al. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–35. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapin N, Hoof I, Lund O, Nielsen M. The MHC motif viewer: a visualization tool for MHC binding motifs. In: Coligan John E., editor. Current protocols in immunology. 2010. Chapter 18:Unit 18 7. [DOI] [PubMed] [Google Scholar]
- 36.Sidney J, Southwood S, Oseroff C, et al. Measurement of MHC/peptide interactions by gel filtration. In: Coligan John E., editor. Current protocols in immunology. 2001. Chapter 18:Unit 18 3. [DOI] [PubMed] [Google Scholar]
- 37.Southwood S, Sidney J, Kondo A, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. Journal of immunology. 1998;160:3363–73. [PubMed] [Google Scholar]
- 38.Sidney J, Steen A, Moore C, et al. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. Journal of immunology. 2010;184:2492–503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidney J, Steen A, Moore C, et al. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. Journal of immunology. 2010;185:4189–98. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P, Sidney J, Kim Y, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oseroff C, Sidney J, Kotturi MF, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. Journal of immunology. 2010;185:943–55. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller WD, Karamfilov T, Kahlert H, et al. Mapping of T-cell epitopes of Phl p 5: evidence for crossreacting and non-crossreacting T-cell epitopes within Phl p 5 isoallergens. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 1998;28:1538–48. doi: 10.1046/j.1365-2222.1998.00432.x. [DOI] [PubMed] [Google Scholar]
- 43.Schenk S, Breiteneder H, Susani M, et al. T-cell epitopes of Phl p 1, major pollen allergen of timothy grass (Phleum pratense): evidence for crossreacting and non-crossreacting T-cell epitopes within grass group I allergens. The Journal of allergy and clinical immunology. 1995;96:986–96. doi: 10.1016/s0091-6749(95)70237-7. [DOI] [PubMed] [Google Scholar]
- 44.Oseroff C, Sidney J, Tripple V, et al. Analysis of T Cell Responses to the Major Allergens from German Cockroach: Epitope Specificity and Relationship to IgE Production. Journal of immunology. 2012;189:679–88. doi: 10.4049/jimmunol.1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King TP, Hoffman D, Lowenstein H, et al. Allergen nomenclature. WHO/IUIS Allergen Nomenclature Subcommittee. International archives of allergy and immunology. 1994;105:224–33. doi: 10.1159/000236761. [DOI] [PubMed] [Google Scholar]
- 46.Ling EM, Smith T, Nguyen XD, et al. Relation of CD4+CD25+ regulatory T-cell suppression of allergen-driven T-cell activation to atopic status and expression of allergic disease. Lancet. 2004;363:608–15. doi: 10.1016/S0140-6736(04)15592-X. [DOI] [PubMed] [Google Scholar]
- 47.Voo KS, Wang YH, Santori FR, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4793–8. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]