Abstract
Determining protein-specific glycosylation in protein mixtures remains a difficult task. A common approach is to use gel electrophoresis to isolate the protein followed by glycan release from the identified band. However, gel bands are often composed of several proteins. Hence, release of glycans from specific bands often yields products not from a single protein but a composite. As an alternative, we present an approach whereby glycans are released with peptide tags allowing verification of glycans bound to specific proteins. We term the process in-gel nonspecific proteolysis for elucidating glycoproteins (INPEG). INPEG combines rapid gel separation of a protein mixture with in-gel nonspecific proteolysis of protein bands followed by tandem MS analysis of the resulting N- and O-glycopeptides. Here, in-gel digestion is shown for the first time with nonspecific and broad specific proteases such as pronase, proteinase K, pepsin, papain and subtilisin. Tandem MS analysis of the resulting glycopeptides separated on a porous graphitized carbon (PGC) chip was achieved via nanoflow liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (nano-LC/Q-TOF MS). In this study, rapid and automated glycopeptide assignment was achieved via an in -house software (Glycopeptide Finder) based on a combination of accurate mass measurement, tandem MS data and pre-determined protein I.D. (obtained via routine shotgun analysis). INPEG is here initially validated for O-glycosylation (kappa casein) and N-glycosylation (ribonuclease B). Applications of INPEG were further demonstrated for the rapid deter mination of detailed site-specific glycosylation of lactoferrin and transferrin following gel separation and INPEG analysis on crude bovine milk and human serum, respectively.
Introduction
Among the myriad of post-translational modifications (PTMs) of proteins, glycosylation is one of the most common types with about 70% of human proteins predicted to be glycosylated.1–2 There are multiple types of protein glycosylation; however, the two main forms of eukaryotic protein glycosylation are N-linked and O-linked.3 Although a prerequisite for the occurrence of N-linked glycosylation involves the Asn-Xaa-Ser/Thr consensus sequence (where Xaa can be any amino acid except proline), O-linked glycosylation lacks such a consensus sequence and can occur on any Ser or Thr on a protein’s backbone. Irrespective of the glycosylation type, the attached glycans are vital as they influence structural and functional features of the proteins they modify.4–14 It is therefore important to continuously develop techniques aimed at extensively characterizing protein glycosylation to further our understanding of the interplay between glycosylation and protein function.
Protein glycosylation analysis remains an arduous analytical challenge due to the diversity of glycan structures and the nature by which they associate with the protein backbones.15 The analysis is particularly complicated for biologically relevant protein mixtures, such as serum and milk, due to the broad dynamic range in protein concentrations and the diversity of the respective glycan structures. Consequently, separation techniques such as affinity chromatography, liquid chromatography and electrophoresis are typically incorporated in glycosylation analysis involving complex protein mixtures.16–17 Gel electrophoresis is readily available, relatively rapid, and generally robust for separating small amounts of protein mixtures based on their molecular weights and/or isoelectric points (PI). Following gel separation, the desired protein band is typically excised, cut into pieces and de-stained. The trapped protein(s) in the gel pieces are then deglycosylated either enzymatically or chemically before MS analysis of the recovered glycans.18 However as gel bands are often composed of several proteins, the released glycans even from single bands often yield products from multiple proteins. This method further fails to address site-specific glycosylation and site-heterogeneity.
Developing MS-based glycoproteomic strategies by analyzing glycopeptides is key to achieving detailed protein- and site-specific analysis. These strategies are best employed while incorporating existing methods such as gel electrophoresis. The typical method for extracting peptides from gel is with in-gel trypsin digestion of proteins.19 The yield for glycoproteins is a mixture of glycopeptides and peptides, which provide protein identification when analyzed by tandem MS and the subsequent database search. However; proteolysis with tryps in yields limited glycosylation information for several reasons. Many proteins are resistant to tryptic digestion.20–23 Trypsin digestion typically yields larger glycopeptides that are difficult to extract from gels. Additionally, the vast majority of the tryptic products are non-glycosylated peptides, with the glycopeptides further partitioned into many glycoforms effectively distributing their respective signals.24
To address these issues, we present a gel-based approach for determining site-specific glycoprotein analysis. “In-gel Nonspecific Proteolysis for Elucidating Glycoproteins” (INPEG) combines the simple and rapid 1-D gel separation of protein mixtures with a novel site-specific analysis of N- and O-glycans via in-gel nonspecific proteolysis. Glycopeptides are characterized using nanoflow liquid chromatography with micro fluidic porous graphitized carbon (PGC) chip and quadrupole time-of-flight mass spectrometry (nano-LC/Q-TOF MS). Rapid and automated glycopeptide assignment was achieved via in-house software (Glycopeptide Finder). The assignment is based on a combination of accurate mass measurement, tandem MS data and pre-determined protein I.D. (obtained via standard shotgun analysis). The output from Glycopeptide Finder yields glycan composition, glycosite and peptide I.D. for each glycopeptide. INPEG is here initially vetted using a cocktail of standard glycoproteins – ribonuclease B and kappa casein with the identified protein bands excised and digested with pronase. Both proteins were chosen as they represent N- and O-glycosylation, respectively, and serve as a proof of concept to illustrate the application of INPEG to both forms of protein glycosylation. Also, applications of INPEG are shown towards targeted protein-specific glycosylation analysis of bovine lactoferrin and human transferrin following gel-separation of bovine milk and human serum, respectively.
Materials and Methods
Materials and Reagents
Pronase, ribonuclease B (RNAse B), kappa casein (k-CN) and human serum were all purchased from Sigma Aldrich. The 10% polyacrylamide Tris gel was purchased from Bio -Rad. All reagents used were of analytical grade or better.
1-D Gel Analysis
35 μL of a 1 μg/μL ribonuclease B (RNAse B) and kappa casein (k-CN) cocktail, 35 μL of crude bovine milk and 35 μL of diluted human serum (x10 with water) were each prepared in separate vials. To each of these were added 35 μL of Laemmli buffer and 7 μL of 1 M dithiothreitol (DTT). The resulting mixtures were then heated for 7 min at 70 °C to reduce the proteins. Alkylation of the reduced proteins were achieved by adding 7.7 μL of 100 mM iodoacetamide (IAA) to each vial and leaving in the dark for 30 min. One -dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (1-D SDS-PAGE) of the alkylated protein mixtures were performed on 10% Tris-HCl polyacrylamide gels. For this analysis, 10 μg equivalent each of alkylated RNAse B and k-CN, 10 μL of bovine milk and 10 μL of human serum solutions were loaded onto separate gel lanes. A 10 μL solution of a molecular weight marker was loaded onto a separate gel lane to serve as molecular weight guide for the gel separations. The gel was allowed to run in a 10% SDS buffer for 60 mins at a constant voltage of 140 V. The gel was then stained with 50 mL of coomassie brilliant blue for 1 h and thoroughly rinsed in water.
In-gel Nonspecific Proteolysis
The targeted protein bands from each gel were excised, cut into 1mm3 pieces and placed in separate vials. Next, the gel pieces were rinsed and de-stained via alternating washes with 200 μL of 100 mM ammonium bicarbonate NH4HCO3 (pH 8.0) and 200 μL of acetonitrile (ACN) with gentle agitation. The wash step was repeated for a total of three washes. After the third wash, the gel pieces were further treated with 200 μL of a 50:50 ACN:NH4HCO3 for 30 min with gentle agitation and finally with 200 μL of ACN. After removal of the ACN, the gel pieces in each vial were completely dried in the speed vac followed by treatment with 40 μL of a 0.025 μg/μL pronase solution made in 100 mM NH4HCO3 (pH 8.0). Gel pieces were allowed to expand in the pronase solution for 30 min after which 200 μL of 100 mM NH4HCO3 (pH 8.0) digestion buffer was added to each vial to allow diffusion across gel pieces. Digestion was allowed to proceed overnight at 37°C in a water bath. Shotgun proteomic analysis to identify the protein(s) contained in each gel band was performed in parallel to the INPEG analysis. The pr oteomic experiment was achieved in similar way as the INPEG analysis except that in -gel tryptic digestion in each case was performed using 40 μL of a 0.0025 μg/μL trypsin solution made in 100 mM NH4HCO3 (pH 8.0).
Digest Recovery
The supernatants following in-gel proteolyses in the different vials were carefully drawn out into separate vials. Further recovery of glycopeptides still trapped in the gel pieces was achieved via treatment of the gel pieces in each vial with 200 μL extraction solution containing 5% formic acid (FA) in 60% ACN followed by sonication for 30 mins. The supernatants were again carefully drawn out from each vial and combined to the earlier recovered digest. This step was repeated one more time to ensure complete digest recovery. All the recovered digests were then completely dried in a speed vac prior to the MS analysis.
Instrumentation
After drying in the speed vac, glycopeptides were reconstituted in 10 μL of 0.1% FA in water prior to injection for mass spectrometric analyses. The glycopeptides were analyzed using an Agilent 1200 series LC system coupled to an Agilent 6520 Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA). The nano-LC/MS system is equipped with an auto sampler, a capillary loading pump (for sample enrichment), an analytical pump (for sample separation), a HPLC-Chip cube and the Agilent 6520 Q-TOF MS detector. Tandem MS analysis of glycopeptides was acquired in a data-dependent manner following LC separation on a microfluidic chip consisting of a 9 × 0.075 mm i.d. enrichment column and a 150 × 0.075 mm i.d. analytical column, both packed with 5 μm porous graphitized carbon (PGC) as the stationary phase. However, tryptic-generated peptide analyses for protein identification were carried out with a C18 chip. The two pumps in these analyses consist of a binary solvent: A, 3.0% ACN/water (v/v) with 0.1% formic acid (FA); B, 90% ACN/water (v/v) with 0.1% FA. A flow rate of 4 μL/min of solvent A was used for sample loading with 10 μL injection volume. A nano pump gradient was delivered at 0.4 μL/min using (A) 0.1% FA in 3.0% ACN/water (v/v) and (B) 0.1% FA in 90.0% ACN/water (v/v). Samples were eluted with 0% B (0.00–2.50 min); 0 to 16% B (2.50–20.00 min); 16 to 44% B (20.00–30.00 min); 44 to 100% B (30.00–35.00 min) and 100% B (35.00–45.00 min). The drying gas temperature was set at 325 °C with a flow rate of 4 L/min (2 L of filtered nitrogen gas and 2 L of filtered dry grade compressed air). MS and MS/MS spectra were acquired in the positive ionization mode with an acquisition time of 1587 milliseconds per spectrum and acquisition rate of 0.63 spectra per second. Also, while the MS data was acquired over a mass range of 500–3000 m/z, the tandem MS data was acquired over 50–3000 m/z mass range. Mass calibration was enabled using reference masses of m/z 922.010 and 1521.972 (ESI-TOF Tuning Mix G1969–85000, Agilent Technologies, Santa Clara, CA). Data analysis was performed on Agilent Mass Hunter (Agilent Technologies Inc.). For the tandem MS analysis, glycopeptides were subjected to collision induced fragmentation with nitrogen as the collision gas using a series of collision energies that were dependent on the m/z values of the different glycopeptides and peptides. The collision energies correspond to voltages (Vcollision) that were based on the equation: Vcollision = m/z (1/100 Da) Volts + 5 Volts; where the slope and offset of the voltages were set at (1.8/100 Da) and (− 2.4) respectively. The preferred charge states were set at 2, 3, >3 and unknown.
Data Processing
Glycopeptides were assigned based on a combination of accurate mass measurement, tandem MS data and protein I.D. (obtained via routine shotgun analysis). An in-house software (Glycopeptide Finder) was used to achieve rapid and automated assignment of the glycopeptides. The assignments were made within a specified tolerance level (≤ 20ppm). To identify glycopeptides in the tandem MS data, product ion spectra were sorted by the presence of carbohydrate-specific oxonium fragment ions such as m/z 163 [Hex+H]+1, m/z 204 [HexNAc+H]+1, m/z 366 [HexNAc+Hex+H]+1,m/z 292 [NeuAc+H]+1 and m/z 274 [NeuAc−H2O+H]+1. The glycan moieties of the glycopeptides were confirmed by the presence of B-type and Y-type ions derived from the sequence of glycan fragmentations in the product ion spectra. However, the peptide moieties were mainly confirmed via accurate measurement of their masses in the tandem MS data. Database search of the shotgun proteomic data was achieved using X! Tandem.
Results and Discussion
Validation of INPEG with Single Protein Standards
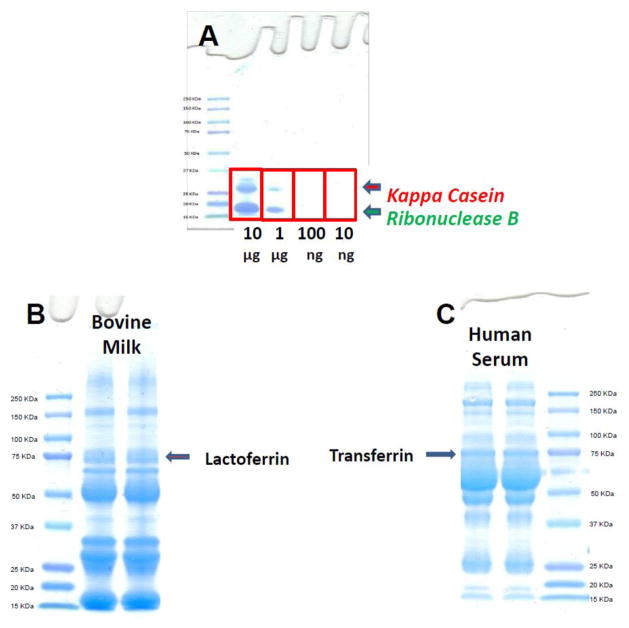
The following experiments establish that a mixture of proteases (pronase E) can be used to yield small glycopeptides and that glycopeptides of both N- and O-glycosylation types with and without sialylation can be reclaimed from the gel. To illustrate the capacity of the method to characterize N- and O-glycosylation, bovine lactoferrin (b-LF) and kappa casein (k-CN) were run on a gel (Figure 1A) and their associated glycopeptides were recovered from the gel bands after digestion with pronase E. O-glycosylation is particularly difficult due to the lack of consensus sequence for the glycosylation site. k-CN is a glycoprotein from bovine milk with six O-glycosites at Thr 142, Thr 152, Thr 154, Thr 157, Thr 163 and Thr 186 predominantly occupied with sialylated glycans.25 The site-specific heterogeneity has previously been well characterized 26, but the protein is important for illustrative purposes.
Figure 1.
1-D gel analysis of (A) various amounts of a mixture of kappa casein and ribonuclease B, (B) bovine milk proteins, and (C) human serum proteins.
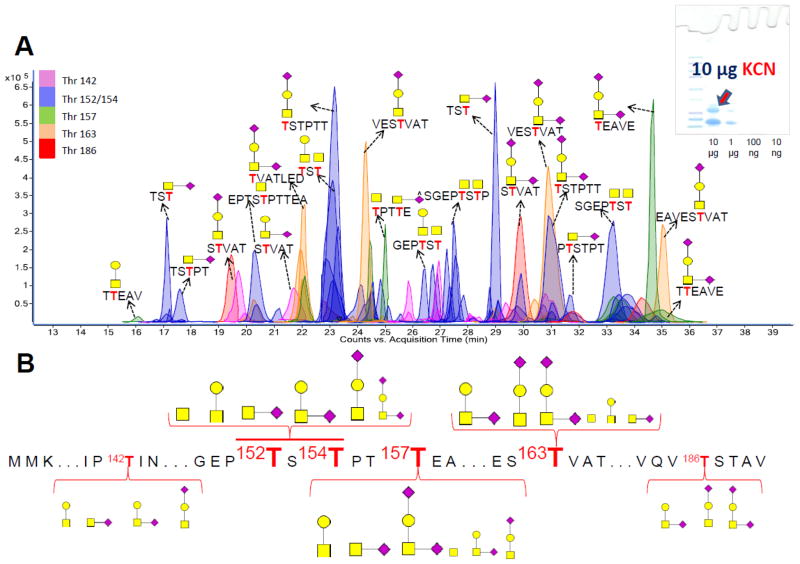
Figure 2A represents the extracted compound chromatogram (ECC) of glycopeptides resulting from INPEG analysis of the k-CN band. The glycopeptide assignments were achieved by accurate mass measurement using the Glycopeptide Finder software and further verified with tandem MS. For example, the deconvoluted tandem mass spectra for two k-CN glycopeptide isomers with m/z 567.69 and corresponding in mass to [S163TVAT+GalNAc+Gal+NeuAc+2H]+2 are shown in Supplementary Figures S1A and S1B. As expected, most of the observed glycopeptides in Figure 2A were sialylated with up to two sialic acid residues associated with a single O-glycosite. The glycosylation information obtained from this analysis is comprehensive as glycans associated with all six sites were observed.
Figure 2.
(A) Extracted compound chromatogram (ECC) of kappa casein glycopeptides generated following pronase digestion of 10 μg of the protein. (B) Detailed site-heterogeneity and detected glycoform abundance of kappa casein following INPEG analysis.
A summary of the detailed site-heterogeneity of kappa casein as observed with INPEG is shown in Figure 2B. The signal intensities of glycopeptides obtained from their MS analyses as previously described 27 yielded the abundances of glycoforms present on each observed glycosite as shown in the inset structure. The size of the amino acid letter and glycan structure designation is associated with the degree of site occupancy. High site occupancy is represented with large letters when the combined abundance of glycopeptides associated with a site is at least 10% of the total glycopeptide abundance for the entire protein. Small letters are used to signify occupancies below this threshold. Likewise, while large glycan structures are used when the combined abundance of glycopeptides associated with a specific glycan is at least 10% of the total glycopeptide abundance associated with a particular glycosite, small glycan structures are for glycoforms below this threshold. It is imperative to state that while the proposed scheme utilizes glycopeptide ion abundances to determine the extent of site occupancy; issues such as differences in ionization potential and dispersion of signal across peptide forms by nonspecific proteases could potentially influence the observed results and require further evaluat ion. However, using the proposed scheme, the Thr 152, Thr 154, Thr 157 and Thr 163 glycosites were classified as being highly glycosylated while only minor glycosylation was observed in the Thr 142 and Thr 186 glycosites.
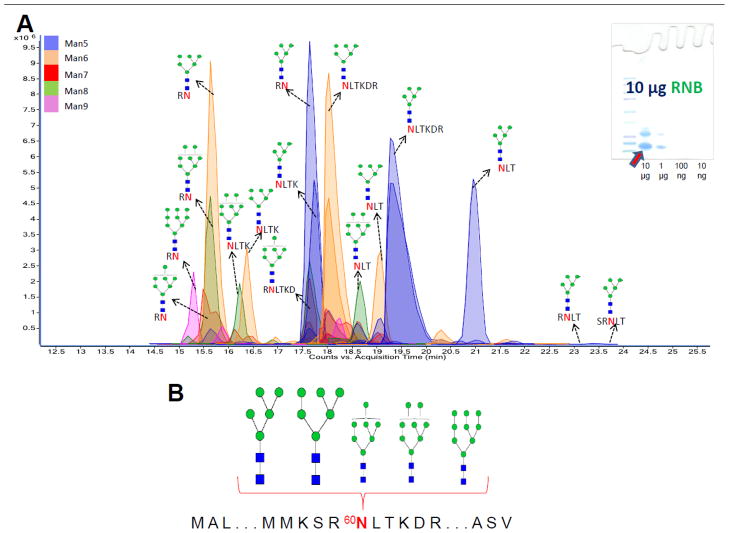
Ribonuclease B (RNAse B) was used to illustrate N-glycosylation as shown in Figure 1A. RNAse B is a bovine glycoprotein with a single known N-glycosite at Asn 60 exclusively occupied with high mannose glycans.28 Figure 3A represents the extracted compound chromatogram (ECC) of glycopeptides resulting from INPEG analysis of the RNAse B band. As expected, all the observed glycopeptides contained high mannose glycosylation. The glycosylation information obtained from the INPEG analysis is comprehensive as all five glycoforms (GlcNAc2:Man5, GlcNAc2:Man6, GlcNAc2:Man7, GlcNAc2:Man8 and GlcNAc2:Man9) of RNAse B were observed at the protein’s single N-glycosite. Figure 3B represents the detailed site-heterogeneity and observed site abundance of RNAse B glycosylation observed via INPEG. Again, the abundance of specific glycoforms present at each site was achieved using the MS signal intensities of glycopeptides. The GlcNAc2:Man5 and GlcNAc2:Man6 glycoforms were observed as being the major glycans of RNAse B with glycopeptides corresponding to both glycoforms accounting for at least 10% of the total RNAse B glycopeptide abundance. The GlcNAc2:Man7, GlcNAc2:Man8 and GlcNAc2:Man9 glycoforms were observed as relatively minor components as the combined abundance of their respective glycopeptides accounted for less than 10% of the total RNAse B glycopeptide abundance. This result is consistent with those of previous studies involving the quantification of RNAse B N-glycans using nuclear magnetic resonance (NMR) 29 and capillary gel electrophoresis 30 where GlcNAc2:Man5 and GlcNAc2:Man6 glycoforms were observed as being the major glycans of RNAse B. This consistency also points to the efficiency inherent with our glycopeptide extraction procedu re from gels. To confirm the RNAse B glycopeptide assignments made, two deconvoluted tandem mass spectra of RNAse B glycopeptides with m/z 846.359 (corresponding in mass to the glycopeptide [60NLTK+GlcNAc2+Man5+2H]+2) and m/z 1170.467 (corresponding in mass to the glycopeptide [60NLTK+GlcNAc2+Man9+2H]+2) are represented in Supplementary Figures S2A and S2B, respectively.
Figure 3.
(A) Extracted compound chromatogram (ECC) of ribonuclease B glycopeptides generated following pronase digestion of 10 μg of the protein. (B) Detailed site-heterogeneity and detected glycoform abundance of ribonuclease B following INPEG analysis.
Sensitivity of INPEG Analysis
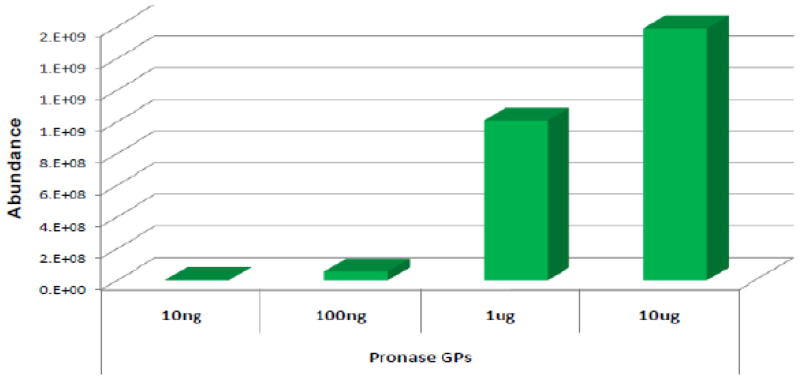
The results obtained with the k-CN and RNAse B show that INPEG yields extensive site-specific glycosylation with 10 μg of protein loaded on the gel. To determine the lowest amount of protein required to obtain extensive glycosylation information, a series of samples with varying amounts of RNAse B (1 μg, 100 ng and 10 ng) were loaded onto a gel, separated, excised and digested. The resulting glycopeptide digests in each case were extracted from the gel pieces and characterized by MS and tandem MS. Supplementary Figures S3A and S3B represent the ECCs of glycopeptides observed from the INPEG analyses of 1 μg and 100 ng RNAse B, respectively. Figure 4 shows the total ion abundances of RNAse B glycopeptides obtained from 10 μg, 1 μg, 100 ng and 10 ng of gel-loaded protein. Since RNAse B has a single glycosite, the combined glycopeptide ion abundance for each digestion was used in constructing the histogram. Although the total glycopeptide abundance decreased appropriately from 10 μg to 10 ng, similar glycoform information was obtained in the various digestions. The accompanying chromatograms (Figure 3A, S3A and S3B) supporting the histogram are annotated to show the number and types of the major glycopeptides observed in each digestion. Due to the nonspecificity of pronase, it is expected to obtain a mixed population of glycopeptide compounds during different digestions. However, there is consistency in the site-specific glycosylation information extracted from the data irrespective of the digestion. As expected the GlcNAc2:Man5 and GlcNAc2:Man6 glycoforms were predominant in the three chromatograms.
Figure 4.
Comparison of glycopeptide abundances following INPEG analysis of 10 μg, 1 μg, 100 ng and 10 ng of gel-loaded ribonuclease B.
Weak glycopeptide MS signals were obtained for the 10 ng digestion that could not be confirmed by tandem MS. However, tandem MS verification of glycopeptides was obtained with larger amounts of protein. Hence, these results show that INPEG is sensitive up to about 10 ng of gel-loaded protein for the MS detection while about 100 ng of gel-loaded protein is required to yield tandem MS data.
Compatibility of Proteases with Broad-specificity for INPEG Analysis
Separate from pronase E, other nonspecific proteases and proteases with broad specificity were tested with the INPEG strategy. Proteinase K, subtilisin, papain and pepsin are examples of such enzymes employed to determine the broad application of INPEG to an array of nonspecific and broadly specific proteases. The experiments with these enzymes were carried out in similar way as that with the pronase analysis except that the digestion buffer for the pepsin analysis was adjusted to pH 3.
Supplementary Figure S4 represents the ECC of RNAse B glycopeptides observed following INPEG analysis with subtilisin. With all the other proteases, glycopeptides corresponding to the five glycoforms of RNAse B were observed. However, the number and size (peptide moiety length) of glycopeptide compounds varied amongst the different enzymes. The difference is mainly due to the degree of nonspecificity of each enzyme. For example, fewer glycopeptides were observed in the subtilisin digestion of RNAse B compared to that obtained with the pronase analysis. Glycopeptides mainly containing RN, NLT, NLTK and NLTKD were observed following pronase digestion of RNAse B while the subtilisin digestion of RNAse B mainly yielded glycopeptides corresponding to NLTK and SRNLTKDR. With both enzymes, there is a unique advantage to the quality of data observed. As previously reported by our group 26, pronase digestion presents a unique advantage towards verifying glycosite and glycopeptide assignments as it yields a population of glycopeptides where various peptide moieties surround a particular glycosite. On the other hand, the fewer RNAse B glycopeptide population observed from the subtilisin digestion presents a less complicated data analysis scenario in that there is less glycopeptide signal splitting.
Application of INPEG towards Determining Targeted Protein-specific Glycosylation in Complex Mixtures
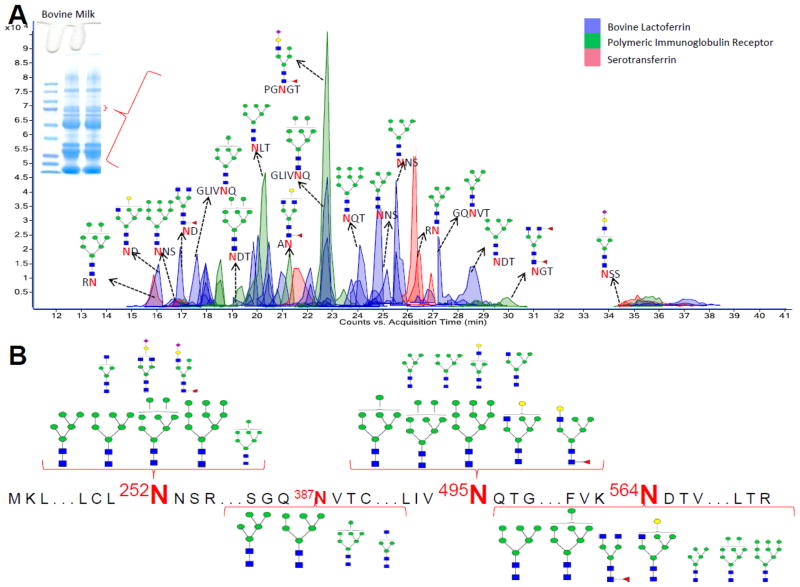
To demonstrate the utility of this method, a number of proteins were examined as part of a biological mixture. Bovine lactoferrin (b-LF) is a component of bovine milk with a molecular weight of 78 KDa and up to four known N-glycosites. Bovine milk is typical of most bodily fluid in that it contains hundreds of proteins with a broad dynamic range of protein concent rations.31 Lactoferrin being a targeted protein was partly isolated from bovine milk by using a 1-D gel and excising the gel band around the 78 KDa region (Figure 1B). To confirm the identity of the protein(s) in the band, a section of the excised band was treated with trypsin for protein identification while the rest of the band was treated with pronase E for site -specific glycosylation information of the identified protein(s). Protein identification was achieved via data base (X! Tandem) search of the resulting peptides’ tandem MS data. The analysis revealed that the band contained bovine lactoferrin, polymeric immunoglobulin receptor, albumin and serotransferrin with UniProtKB entries P24627, P81265, P02769, Q29443, respectively. Except for albumin, the other three proteins are glycosylated and were considered in the glycopeptide matching analysis with the Glycopeptide Finder software.
Figure 5A shows a representative ECC of the glycopeptides after pronase E digestion. Glycopeptides corresponding to lactoferrin (peaks shaded in blue), immunoglobulin receptor (peaks shaded in green) and serotransferrin (peaks shaded in red) were observed in the ECC. The majority of the observed glycopeptides were those containing high mannose type glycans, consistent with b-LF. Figure 5B is the site-heterogeneity information of b-LF determined by the analysis. Like with the k-CN and RNAse B experiments, Figure 5B also details the abundance of specific b-LF glycoforms present at each site observed. While the Asn 252, Asn 495 and Asn 564 glycosites were observed as being highly glycosylated, the Asn 387 glycosite was observed as only being slightly glycosylated. Supplementary Figures S5A and S5B represent the deconvoluted tandem MS data for two bovine lactoferrin glycopeptides with m/z 1026.295 and m/z 1107.321 corresponding in mass to the glycopeptides [564NDT+GlcNAc2+Man8+2H]+2 and [564NDT+GlcNAc2+Man9+2H]+2, respectively.
Figure 5.
(A) Extracted compound chromatogram (ECC) of glycopeptides generated from INPEG analysis of a bovine milk gel band containing bovine lactoferrin, polymeric immunoglobulin receptor and serotransferrin. (B) Detailed site-heterogeneity and detected glycoform abundance of bovine lactoferrin following INPEG analysis.
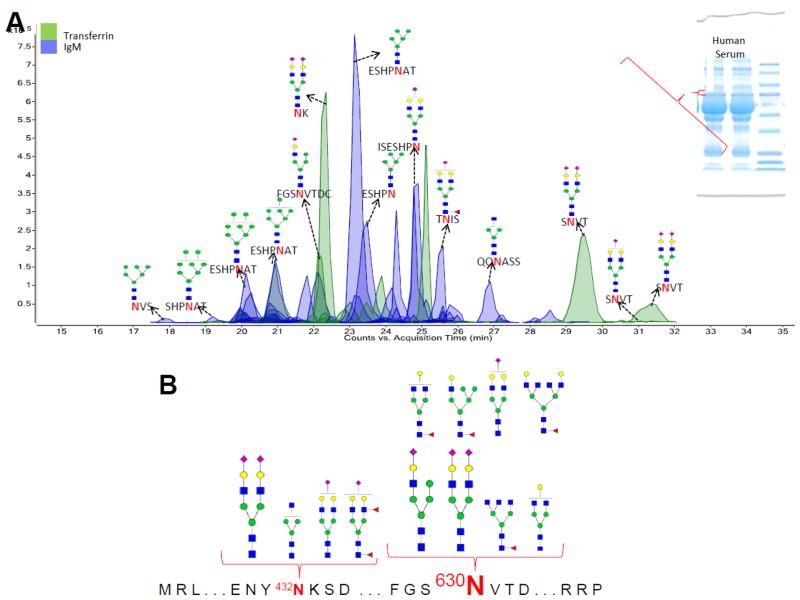
Following the success in determining the site-specific glycosylation of lactoferrin in bovine milk, INPEG is here further extended towards determining targeted protein-specific glycosylation in human serum. Human transferrin (h-TF) is one of the glycosylated components of human serum with a molecular weight of 78 KDa. Like bovine milk, human serum contains hundreds of proteins with a large dynamic range of protein concentrations. 32 Using 1-D gel separation, transferrin was partly isolated and excised around the 78 KDa region (Figure 1C). Tryptic digestion of a part of the excised gel band revealed transferrin, immunoglobulin M and albumin with UniProtKB entries P02787, P01871 and P02768, respectively. The remaining portion of the band was digested with pronase E to determine site-specific glycosylation of the identified protein(s). Besides albumin, the other two identified proteins are glycosylated and were included in the Glycopeptide Finder analysis towards matching the observed glycopeptides.
Figure 6A represents the ECC of glycopeptides observed following INPEG analysis of the band being investigated. Glycopeptides corresponding to transferrin (peaks shaded in green) and immunoglobulin M (peaks shaded in blue) were observed in the ECC. While the majority of the transferrin glycopeptides were observed with sialylated N-glycans, the immunoglobulin M glycopeptides were mainly observed with high mannose glycans.
Figure 6.
(A) Extracted compound chromatogram (ECC) of glycopeptides gener ated from INPEG analysis of a human serum gel band containing human transferring and immunoglobulin M. (B) Detailed site-heterogeneity and detected glycoform abundance of human transferrin.
Figure 6B represents the site-heterogeneity information of transferrin observed from the analysis. The site-specific abundance of transferrin glycoforms is detailed in the figure with the Asn 630 glycosite observed as being highly glycosylated while the Asn 432 glycosite was observed as only being sparingly glycosylated. Supplementary Figure S6 represents the site-heterogeneity and glycoform abundance information observed for IgM. While the IgM Asn 209 and Asn 279 glycosites were observed as being highly glycosylated, the Asn 46, Asn 272 and Asn 439 glycosites were considerably less glycosylated.
Discussion
Determination of protein-specific and site-specific glycosylation in complex mixtures is currently an extremely difficult task requiring large amounts of glycoproteins and considerable resources in time and funds. 1-D gel electrophoresis combined with glycomics strategies is typically insufficient for the analysis as individual gel bands typically contain a number of proteins that make site- and even protein-specific glycosylation analysis impossible. Protein isolation may often require the development of specific antibodies for protein capture and release. However, such methods are laborious and too expensive for routine analysis.
INPEG provides both protein-specific and site-specific glycosylation analysis with minimal separation of proteins. It employs an existing and readily available method for protein separation avoiding other more labor intensive separation techniques. INPEG can also be employed for biological fluids such as milk and serum. The analysis is generally sensitive with limits of detection slightly below that of coomassie blue stains. It can therefore be included in a standard workflow employing gel-electrophoresis. This means that less abundant proteins can be analyzed with the appropriate methods for depletion of more abundant proteins. The overall method is rapid allowing the characterization of several proteins in a mixture simultaneously. With this method, it will soon be possible to obtain a glycan map of a target protein from any biological sample at little cost and in a short amount of time (few hours).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health under Award Numbers (HD061923 and HD059127, ROIGM049077). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Refer to web version on PubMed Central for supplementary material
References
- 1.Apweiler R, Hermjakob H, Sharon N. Biochim Biophys Acta. 1999;1473(1):4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 2.An HJ, Froehlich JW, Lebrilla CB. Curr Opin Chem Biol. 2009;13(4):421–426. doi: 10.1016/j.cbpa.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiro RG. Glycobiology. 2002;12(14):43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 4.Newburg DS. Curr Med Chem. 1999;6(2):117–128. [PubMed] [Google Scholar]
- 5.Hamosh M. Biol Neonate. 1998;74(2):163–176. doi: 10.1159/000014021. [DOI] [PubMed] [Google Scholar]
- 6.Schroten H, Hanisch F, Plogmann R, Hacker J, Uhlenbruck G, Nobis-Bosch R, Wahn V. Infect Immun. 1992;60(7):2893–2899. doi: 10.1128/iai.60.7.2893-2899.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yolken R, Peterson J, Vonderfecht S, Fouts E, Midthun K, Newburg D. J Clin Invest. 1992;(90–95):1984–1991. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi K, Wakabayashi H, Shin K, Takase M. Biochem Cell Biol. 2006;84(3):291–296. doi: 10.1139/o06-054. [DOI] [PubMed] [Google Scholar]
- 9.Harmsen MC, Swart PJ, de Bethune MP, Pauwels R, De Clercq E, The TH, Meijer DK. J Infect Dis. 1995;172(2):380–388. doi: 10.1093/infdis/172.2.380. [DOI] [PubMed] [Google Scholar]
- 10.Hanson LA, Ahlstedt S, Andersson B, Carlsson B, Fallstrom SP, Mellander L, Porras O, Soderstrom T, Eden CS. Pediatrics. 1985;75(1):172–176. [PubMed] [Google Scholar]
- 11.Hamosh M, Peterson JA, Henderson TR, Scallan CD, Kiwan R, Ceriani RL, Armand M, Mehta NR, Hamosh P. Semin Perinatol. 1999;23(3):242–249. doi: 10.1016/s0146-0005(99)80069-x. [DOI] [PubMed] [Google Scholar]
- 12.Paterson JA, Patton S, Hamosh M. Biol Neonate. 1998;74(2):143–162. doi: 10.1159/000014020. [DOI] [PubMed] [Google Scholar]
- 13.Ofek I, Sharon N. Curr Top Microbiol Immunuol. 1990;151:91–113. doi: 10.1007/978-3-642-74703-8_5. [DOI] [PubMed] [Google Scholar]
- 14.Ashorn P, Vilja P, Shorn R, Rohn K. Mol Immunol. 1986;23(3):221–230. doi: 10.1016/0161-5890(86)90046-5. [DOI] [PubMed] [Google Scholar]
- 15.Stanley P, Schachter H, Taniguchi N. N-Glycans. In: Varki A, editor. Essentials of Glycobiology. 2. Vol. 1 Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. [Google Scholar]
- 16.Gravel P, Golaz O, Walzer C, Hochstrasser DF, Turler H, Balant LP. Anal Biochem. 1994;221:66–71. doi: 10.1006/abio.1994.1380. [DOI] [PubMed] [Google Scholar]
- 17.Mrochek JE, Dinsmore SR, Waalkes TP. Clin Chem. 1975;21:1314–1322. [PubMed] [Google Scholar]
- 18.Hansen R, Dickson AJ, Goodacre R, Stephens GM, Sellick CA. Biotechnology and Bioengineering. 2010;107(5):902–908. doi: 10.1002/bit.22879. [DOI] [PubMed] [Google Scholar]
- 19.Larsen MR, Hojrup P, Roepstorff P. Mol Cell Prot. 2005;4:107–119. doi: 10.1074/mcp.M400068-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Loo TW, Clarke DM. J Biol Chem. 1998;273(24):14671–14674. doi: 10.1074/jbc.273.24.14671. [DOI] [PubMed] [Google Scholar]
- 21.Loo TW, Clarke DM. The FASEB Journal. 1999;13(13):1724–1732. doi: 10.1096/fasebj.13.13.1724. [DOI] [PubMed] [Google Scholar]
- 22.Giancotti FG, Tarone G, Knudsen K, Damsky C, Comoglio PM. Exp Cell Res. 1985;156(1):182–190. doi: 10.1016/0014-4827(85)90272-1. [DOI] [PubMed] [Google Scholar]
- 23.Carter WG, Hakamori S. J Biol Chem. 1978;253(8):2867–2874. [PubMed] [Google Scholar]
- 24.Demelbauer UM, Plematl A, Josic D, Allmaier G, Rizzi A. J Chrom A. 2005;1080(1):15–21. doi: 10.1016/j.chroma.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 25.Holland JW, Deeth HC, Alewood PF. Proteomics. 2005;5:990–1002. doi: 10.1002/pmic.200401098. [DOI] [PubMed] [Google Scholar]
- 26.Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German JB, Lebrilla CB. J Proteome Res. 2011;10(5):2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua S, Nwosu CC, Strum JS, Seipert RR, An HJ, Zivkovic AM, German JB, Lebrilla CB. Anal and Bioanal Chem. 2012;403(5):1291–1302. doi: 10.1007/s00216-011-5109-x. [DOI] [PubMed] [Google Scholar]
- 28.An HJ, Peavy TR, Hedrick JL, Lebrilla CB. Anal Chem. 2003;75:5628–5637. doi: 10.1021/ac034414x. [DOI] [PubMed] [Google Scholar]
- 29.Fu D, Chen L, O’Neil RA. Carbohydr Res. 1994;261(2):173–186. doi: 10.1016/0008-6215(94)84015-6. [DOI] [PubMed] [Google Scholar]
- 30.Guttman A, Chen FA, Evangelista RA, Cooke N. Anal Biochem. 1996;233(2):234–242. doi: 10.1006/abio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 31.Le A, Barton LD, Sanders JT, Zhang Q. J Proteome Res. 2011;10(2):692–704. doi: 10.1021/pr100884z. [DOI] [PubMed] [Google Scholar]
- 32.Crichton RR, Charloteaux-Wauters M. Eur J Biochem. 1987;164(3):485–506. doi: 10.1111/j.1432-1033.1987.tb11155.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.