Abstract
MicroRNAs regulate gene expression through deadenylation, repression and mRNA decay. However, the contribution of each mechanism in non-steady-state situations remains unclear. We monitored the impact of miR-430 on ribosome occupancy of endogenous mRNAs in wild type and dicer mutants lacking mature miR-430. We find that miR-430 reduces the number of ribosomes on target mRNAs before causing mRNA decay. Translational repression occurs before complete deadenylation, and disrupting deadenylation using an internal poly(A) tail did not block target repression. Finally, we observe that ribosome density along the length of the message remains constant, suggesting that translational repression occurs by reducing the rate of initiation rather than affecting elongation or causing ribosomal drop-off. In summary, our results show that miR-430 regulates translation initiation before inducing mRNA decay.
MicroRNAs (miRNAs) control multiple processes including development, physiology and disease. These ~22nt RNAs regulate gene expression through translational repression, mRNA deadenylation and decay. The contribution and timing of these effects remain unclear (1, 2). While some studies show translational repression without mRNA decay (3–7), others point to decay as a primary effect (8–11). Ribosome profiling experiments, which quantify the number of ribosomes bound to a message (12), have suggested that the main effect of miRNAs is to accelerate decay, with only minor contribution from translational repression (8,13). This disparity may stem from the steady-state conditions used to assess the molecular effects of miRNAs, resulting in insufficient temporal resolution to identify the first step in miRNA-mediated repression (2).
To dissect the temporal effects of miRNA-mediated regulation, and distinguish between translational repression and mRNA decay, we have analyzed the ribosome profiles and RNA levels of endogenous messages in zebrafish embryos (fig. S1) (14). At the onset of zygotic transcription, zebrafish express a predominant miRNA (miR-430) that facilitates clearance of maternal mRNAs (15, 16) (Fig. 1A, B). By comparing the ribosome profile of wild type embryos with maternal and zygotic dicer mutants (MZdicer), before (2hpf) and after miR-430 expression (4 and 6hpf) (fig. S1), we can analyze the dynamics of translational repression and mRNA decay in vivo. We sequenced 54 million reads generated by ribosome profiling, which predominately map to ribosomes, tRNAs and coding sequences (CDS) compared to untranslated regions (UTRs), and 59 million reads generated by poly(A)+ selection (input RNA), which map equally to CDS and UTRs (Fig. 1C, Table S1, fig. S1, S2). We focused our analysis on 4476 genes, present and translated at 2hpf, before miR-430 is expressed (≥15 reads per kilobase, per million reads, RPKM) (fig. S2).
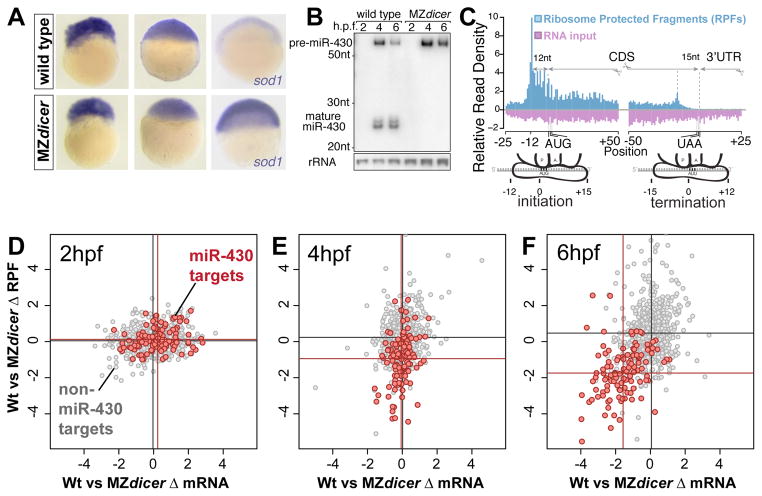
Figure 1. Temporal analysis of miR-430 mediated translational repression in zebrafish.
(A) In situ hybridization (purple) for miR-430 target gene sod1 in wild type and MZdicer embryos at 2, 4 and 6 hpf. Note that decay of the target is observed at 6hpf in a miRNA-dependent manner. (B) Northern blot showing miR-430 expression in wild type and MZdicer. (C) Ribosome protected fragments (RPF) and input reads mapped to a composite transcript. RPFs mainly map to the CDS. Input reads map to both the UTR and CDS. (D–F) Biplots show log2-fold RPKM differences of RPFs (y axis) and mRNA (x axis) between wild type and MZdicer at 2 (D), 4 (E) and 6 (F) hpf. Known miR-430 targets are in red (15), non-targets lacking miR-430 seeds in gray. Mean values per group are indicated as lines. Mean difference between targets and non targets:(E) RPF 2.26-fold, p=1.3e-24; RNA, 1.05-fold, p=0.12; (F) RPF 4.6-fold, p=1.5e-44; RNA 3.1-fold, p=8.1e-44, by two-sided Wilcoxon rank sum test.
We reasoned that if mRNA decay is the main effect of miR-430 on their targets, a reduction in ribosome protected fragments (RPFs) should parallel the loss of input reads. Alternatively, if translation repression precedes decay, RPF loss should precede loss of input (fig. S1). Before miR-430 is expressed (2hpf), we find no significant reduction of either RPFs or mRNA in wild type compared to MZdicer embryos for known miR-430 targets (15) or non-targets (Fig. 1D, 2A). At 4hpf, once miR-430 is expressed, we observed a significant decrease in RPF number for miR-430 targets in wild type compared to MZdicer (p=1.3e-24, Wilcoxon rank-sum test), without a corresponding decrease in mRNA (Fig. 1E, 2A). Thus, translation repression is occurring independent of RNA decay. However, at 6 hpf, we observe a significant reduction in the number of RPFs that coincides with a reduction in input reads (p<1e-43), suggesting that by 6hpf, miR-430 targets have undergone mRNA decay with a mild contribution from additional translational repression (Fig. 1F, 2A). These results show that for targets experimentally identified by their miR-430-dependent decay (15), translation repression occurs before the decay.
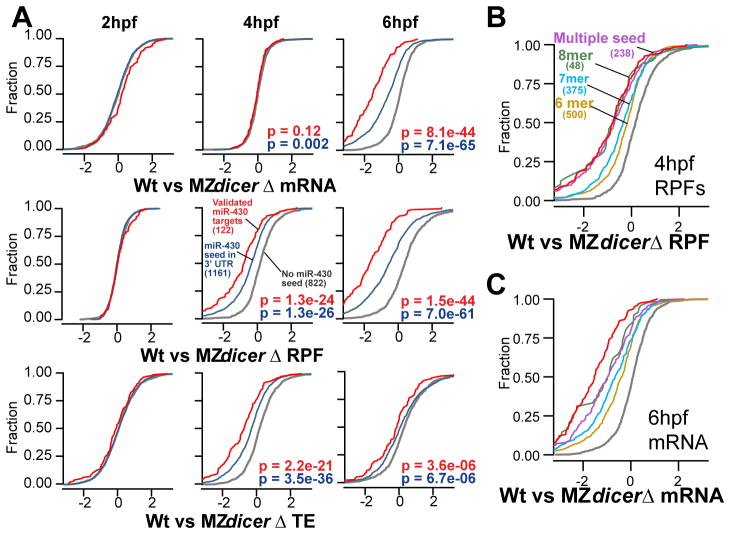
Figure 2. miR-430 induces translation repression prior to RNA decay.
(A)Cumulative distributions of mRNA, RPF, and translation efficiency differences (Δ) between wild type and MZdicer for known miR-430 targets (red), all genes with 3′UTR miR-430 seed sites (blue), and non targets (gray), with number of genes in parentheses. P values for rank-sum tests are shown for non-targets vs. known targets (red) and vs. all predicted targets (blue). (B, C) Cumulative distribution plots with predicted targets separated by seed type as indicated.
To determine the predominant effect of miRNA mediated regulation independent of these experimentally identified targets, we asked whether translational repression or mRNA decay is i) more associated with miRNA target sites and ii) better correlated with miRNA seed strength. First, we found that miR-430 target sites are enriched in both transcripts with lower RPF at 4hpf (p=1.6e-19 for 7mers, Fisher’s exact test) and lower input at 6hpf (p=8.9e-41) in wild type compared to MZdicer, but not lower input at 4hpf (p>0.33) (fig. S3, S4). Second, when we identified all putative miR-430 targets that contain 3′UTR seed matches, the level of regulation followed the order of seed strength (multiple sites>8mer>7mer>6mer) for both translational repression at 4hpf (p=1.2e-6, Kruskal-Wallis) and mRNA decay at 6hpf (p=2.0e-10) (Fig. 2B, C, and fig. S5, S6). Further analysis confirmed that targets that are first translationally repressed predominantly coincide with those that undergo mRNA decay later, suggesting that most targets undergo both regulatory effects (Fig. 3A). These mRNAs correspond to the most strongly regulated targets (p=5.0e-10, Kruskal-Wallis) (Fig. 3B), are the most significantly enriched for miR-430 target sites (p<2.2e-16, Chi-squared test, 4 d.o.f.) (Fig. 3C), and are correlated with various 3′ UTR sequence characteristics (fig. S8).
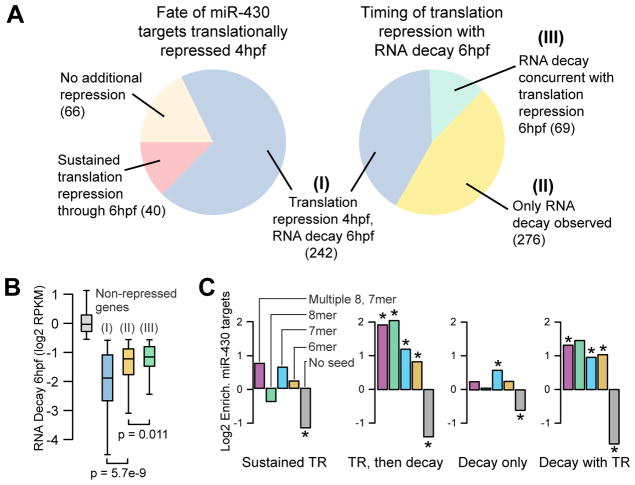
Figure 3. miR-430 induces translation repression followed by RNA decay.
(A) Pie charts of different repression categories (cutoffs defined in Fig. S7). 70% of the targets translationally repressed at 4hpf go on to be deadenylated or degraded at 6hpf (Group I). Among transcripts decayed 6hpf, 41% were translationally repressed 4hpf (I), 47% were not observed to be translationally repressed (II), and the remainder experience concurrent translation repression 6hpf not explained by the decay (III). (B) Box and whisker plot showing that the level of RNA decay at 6hpf is highest among genes that are translationally repressed early. (C) The different modes of repression induce significant enrichment in miR-430 target seeds (* indicates p<0.05, Fisher’s exact test). See Table S2 for counts.
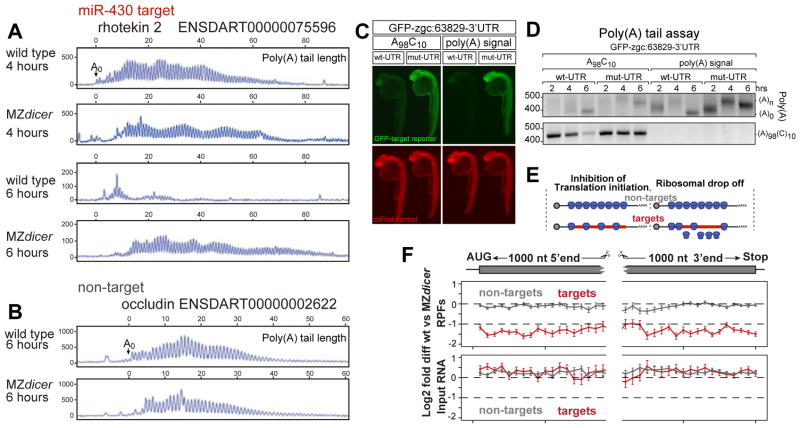
It has been postulated that loss of the poly(A) tail may be the underlying cause of miRNA-mediated repression (1, 2, 15, 17, 18). Since the input was poly(A)+ selected, lower mRNA levels for miR-430 targets at 6hpf could arise from mRNA deadenylation or decay (14)(fig S3). Conversely, the similar mRNA levels at 4hpf between wild type and MZdicer suggest that full deadenylation has not occurred by the onset of translational repression. To analyze the dynamics of deadenylation of individual endogenous mRNAs, we first combined poly(A) tail analysis with high resolution gel electrophoresis (Fig. 4A, fig. S9). We found that the endogenous miR-430 target rhotekin2 (repressed ~80 percent in wild type) is fully deadenylated by 6hpf but is still polyadenylated at 4hpf in wild type and MZdicer (albeit slightly shorter in wild type embryos). There was no difference in poly(A) tail length at 2hpf, before miR-430 is expressed, nor for a non-target between wild type and MZdicer (Fig. 4B, fig. S10). Second, when we analyzed adipor1a that undergoes translational repression at 4 and 6hpf without decay, we found no apparent deadenylation by 6hpf (fig. S10). While these results indicate that loss of RPFs occur before complete deadenylation, they cannot exclude that initial deadenylation might be responsible for the translational repression observed. Next, to determine whether repression requires deadenylation, we disrupted deadenylation of a GFP-zgc:63829-3′UTR reporter mRNA using an internal poly(A) tail followed by 10C (A98C10)(19,20) and compared repression of this reporter with one containing a polyadenylation signal(Fig. 4C, D). We observed repression of both reporters compared to versions where the miR-430 site was mutated GCACTT to GGTCTT, even when deadenylation was reduced (Fig. 4C). These results indicate that translational repression observed at 4hpf occurs before and independently of complete deadenylation.
Figure 4. Poly(A) length and ribosome distribution.
(A, B) Single nucleotide resolution electrophoresis for poly(A) length for a target (A) and non-target (B) in wild type and MZdicer (14). A0 represents the polyadenylation site confirmed by DNA sequencing (fig. S9). (C)GFP expression (green) from an injected miR-430 reporter mRNA containing the 3′UTR for zgc:63829 with wild type (wt-UTR) or mutated (mut-UTR) miR-430 sites. The 3′UTRs are followed by an internal poly(A) tail (A98C10) or a polyadenylation signal. Expression of a co-injected dsRed control mRNA is shown in red. Note the repression of the wild-type reporter compared to the mutant reporter when a internal polyA tail is used. (D) Gel electrophoresis of a PCR to determine the length of the poly(A) tail (ending in A, upper panel) and A98C10 (below). Note the polyadenylation by 2hpf and deadenylation by 6hpf, but deadenylation of the mRNA with the internal polyA tail (A98C10)is delayed. Estimated size of the deadenylated product is shown as (A0). (E) Two models for translational repression: reducing translation initiation (left) or causing ribosome drop off/slower elongation rate (right). (F) Plot showing relative RPF read density (top) and mRNA density (bottom) along the length of miR-430 targets (red) undergoing >1.5-fold translation repression at 4hpf; and non-targets (gray). Points show mean +/− SEM log2 fold differences between wild type and MZdicer expression in 50nt bins spanning the first and last 1000 nts of the genes (14). Bins represent 41≥N≥277 genes for targets, 107≥N≥906 genes for non targets. Bin values do not significantly differ (p=0.53, Friedman rank sum test).
miRNAs have been proposed to influence protein translation by either reducing the rate of translation initiation, reducing elongation, or accelerating ribosome drop-off (1, 2, 18). Ribosome profiling allows us to quantify ribosome position by examining the RPF distribution along the length of the message. If miR-430 functions primarily by reducing translation initiation, then we would expect lower ribosome occupancy, but uniform density along repressed messages. In contrast, if miR-430 causes ribosomal drop-off or reduced translation elongation with the same initiation rate, we would expect a graded distribution of ribosomes, with fewer RPFs in the 3′ end than the 5′ end (Fig. 4E). When we aggregate the reads of miR-430 targets translationally repressed at 4hpf, we find uniform loss of RPF density along the length of the target mRNAs, suggesting that miR-430 inhibits translation initiation (Fig. 4F).
In summary, we show that miR-430 first induces translational repression by reducing the rate of translation initiation, and then induces mRNA decay through deadenylation (fig. S11). Our results reconcile observations in vitro (6, 7, 21–23), which typically employ short time courses and observed repression before deadenylation, with observations in vivo (8–11, 24), which are carried out over longer timescales after perturbing miRNA function, where the strongest effect appears to be deadenylation and decay. Most of the miR-430 regulated genes undergo translational repression followed by decay. A small group of targets appear to be primarily regulated only at the level of translation; however, because of the limited number of time points analyzed, it is possible that those targets could undergo decay at later time points (15).
Previously, several laboratories including ours identified deadenylation as a main effect of miRNA-mediated regulation (15, 17, 24, 25), leading to our initial hypothesis that complete deadenylation (at 6hpf) disrupts the interaction between the poly(A)-binding protein and the cap through eIF4G, thus reducing translation of the message (15, 18). Our findings here show instead that initial repression occurs before complete deadenylation, and disrupting deadenylation does not block translational repression. These data align with the observation that miRNAs can induce repression in transcripts that lack a poly(A) but include instead a histone tail or a self cleaving ribozyme (17, 24). Recent studies have reported that the CCR4-NOT complex is recruited by GW182/TNRC6 to target mRNAs (26–28) and can repress translation independent of its deadenylase activity (26–29). Furthermore, it appears that GW182 has two distinct domains that are independently required to elicit repression and deadenylation (19,20). These results suggest that repression can occur independent of deadenylation in vivo and that miRNAs trigger these two mechanisms in parallel to ensure target mRNA repression and decay. Yet, the degree by which each mechanism regulates different genes may vary depending on the 3′UTR context, as shown in C. elegans (30), the miRNA or even the cell type.
Supplementary Material
Acknowledgments
We thank Sandra Wolin and her laboratory for vital intellectual and technical support and access to equipment and reagents. D Schoenberg, D Patil and Y Mishima for reagents and discussions. S Baserga, D Cazalla, D Cifuentes and S Moxon for discussions. Supported by the Pew Fellows Program in Biomedical Sciences (A.A.B.), The Pew Scholars Program in the Biomedical Sciences, the Yale Scholar Program and NIH grants R01GM081602-05 (A.J.G).
Footnotes
Contributions: A.A.B and A.J.G designed the project. AAB performed the experiments. A.A.B and M.T.L performed data analysis. A.A.B., M.T.L. and A.J.G interpreted the data and wrote the manuscript. Sequencing data are deposited in GEO with accession number GSE34743.
References and notes
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 4.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007 Sep 21;317:1764. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 7.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005 Feb 17;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005 Aug 26;122:553. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.See supporting material on Science Online.
- 15.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 16.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Rev Genet. 2011;12:99. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 19.Fukaya T, Tomari Y. PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J. 2011;30:4998. doi: 10.1038/emboj.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabian MR, Svitkin YV, Sonenberg N. An efficient system for let-7 microRNA and GW182 protein-mediated deadenylation in vitro. Methods Mol Biol. 2011;725:207. doi: 10.1007/978-1-61779-046-1_14. [DOI] [PubMed] [Google Scholar]
- 23.Zdanowicz A, et al. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell. 2009;35:881. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beilharz TH, et al. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS ONE. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nature Struct Biol. 2011;18:1218. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nature Struct Biol. 2011;18:1211. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 28.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Mol Cell. 2011;44:120. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu E, et al. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol Cell. 2010;40:558. doi: 10.1016/j.molcel.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.