Abstract
The insulin/insulin growth factor-1(IGF1)/FOXO (IIF) signal transduction pathway plays a core role in the endocrine system. Although the components of this pathway have been well characterized, the evolutionary pattern remains poorly understood. Here, we perform a comprehensive analysis to study whether the differences of signaling transduction elements exist as well as to determine whether the genes are subject to equivalent evolutionary forces and how natural selection shapes the evolution pattern of proteins in an interacting system. Our results demonstrate that most IIF pathway components are present throughout all animal phyla investigated here, and they are under strong selective constraint. Remarkably, we detect that the components in the middle of the pathway undergo stronger purifying selection, which is different from previous similar reports. We also find that the dN/dS may be influenced by quite complicated factors including codon bias, protein length among others.
Keywords: gradient, codon bias, selective constraint, dN/dS
Introduction
The ability of organisms to respond appropriately to changes in their environment is one of the most important events in the evolution of life. Due to environmental changes, for higher eukaryotes it is essential to possess specialized mechanisms to maintain nutrition homeostasis between their food supply and energy demanding processes, such as growth, metabolism and reproduction. Indeed, the genes do not exert their functions separately, but form complex network, such as transcriptional network and biological signaling pathways. Some pathways play an important role in redirecting organism’s resources toward activity, maintenance, and survival after dietary intake by individual organisms.1–3 These pathways, therefore, represent key molecular candidates for the environmental constraints of adaption through large changes in their morphology and their developmental programs. This may also be reflected at the physiological and molecular level.4 In part, the organism’s hormones were conceived in accurate execution of such developmental programs.5,6 One of the most critical pathways in hormones signal transduction is IIF and its components have been well annotated. The connection topology and the molecular functions of the components have been well characterized in different organisms, including Drosophila melanogaster,7 and are highly conserved across metazoans.8,9 Mutation of genes in this signaling pathway can affect diverse traits such as glycogen and lipid storage.10,11 An important discovery stemming from Kuningas’ experiment was that genes in this signaling pathway also affect lifespan in yeast, nematodes, fruit flies and mice.12
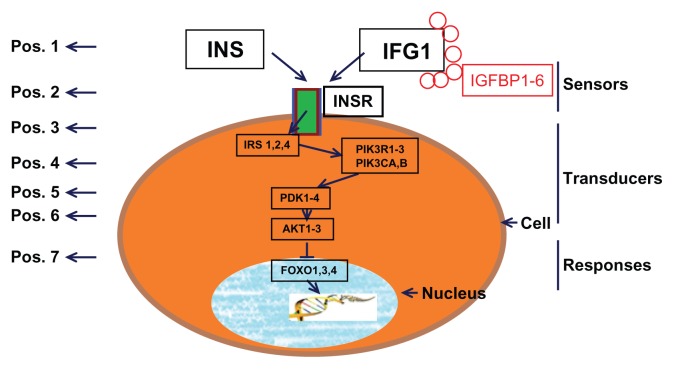
The components of the IIF pathway can be divided into three distinct phases according to interaction mechanisms: (1) Sensors (mainly InRs), the molecules responsible for insulin/IGF1 binding; (2) Transducers, the intracellular signaling components, which trigger a cascade of biochemical reactions to relay the signals (often through protein phosphorylation and dephosphorylation); and (3) Responses, the downstream effector elements. The transduction signal ultimately activates the effector elements in the pathway, which are involved in insulin- dependent cellular responses. Various signaling pathways are initiated by the binding of insulin/IGF1 to its receptor, and of these, the phosphoinositide 3-kinase–Akt/protein kinase B signaling pathway seems to be crucial in the ability of insulin to regulate metabolic homeostasis and protect from cell death.13 Activation of Akt/protein kinase B results in the phosphorylation of FOXO, causing FOXO to remain localized in the cytoplasm.14 As a result, FOXO cannot access its direct downstream binding targets. Here the FOXO proteins belong to “O” subfamily (FOXO1, FOXO3a, FOXO4 and FOXO6) of forkhead transcription factors. They regulate the transcription of target genes that are involved in metabolism. Apart from their roles in metabolism, FOXO members also mediate the survival- factor function of growth factors.15 A simple scheme of IIF pathway components is shown in Figure 1 (modified from16).
Figure 1.
An illustration of the IIF pathway genes.
Notes: Arrows indicate the direction of signal transduction. The numbers on the left side represent the position of the IIF pathway genes.
In previous studies, plenty of research on the insulin signaling pathway genes or proteins variation in their evolutionary rates have been illustrated.17 These studies, however, have only focused on individual genes or gene families; consequently, the properties and mechanisms underlying pathway/ network evolution remain largely unknown. Therefore, establishing the patterns of selection within a certain signaling transduction pathway allows us determine whether these genes are subject to equivalent evolutionary forces and how natural selection shapes the evolution of proteins in an interacting system.
A central question in the evolution of biochemical pathways concerns the role of the structure of signaling pathways. As known, some signaling pathways are structured in a way that the initial substrates in the pathway are ultimately transformed into several end products, based on the channeling of substrate into various branches of the pathway downstream. Therefore, Rausher et al18 arrive at the conclusion that upstream genes should be more pleiotropic than those downstream and so should face greater selective constraint.18–20 The negative correlation between selective constraint and topological position has also been explained by the hierarchical organization of these pathways. Namely, the upstream genes play more important roles than the downstream ones, therefore stronger selection constraints will be receive to remove deleterious mutations in upstream genes. However, a correlation between pathway position and selective constraint levels with downstream genes evolving under stronger purifying selection has been already observed in a number of pathways, including the insulin/TOR pathway in Drosophila and vertebrates.21,22 It is still perplexing whether this correlation is a general trend for all IGF pathways or specific for insulin/TOR. Therefore, we will address this question on a well-characterized IIF pathway.
In recent years, the sequencing of a large number of diverse metazoans genomes has demonstrated that, in the midst of this overall conservation, taxon- specific differences among pathway components exist, for example, the TLR pathway.23 Current knowledge of insulin signaling in model animals, with the wealth of genomic resources, offers the unique opportunity to answer whether there are subtle differences in the components that make up the IIF pathway. Therefore, this study is giving us the opportunity to unravel clues about the origin and the evolution of the IIF pathway by analyzing the genomes of different key metazoans species. Our specific goals are (1) to determine when IIF signaling pathway came into existence; (2) what is the selective forces operating on the different components and if evolutionary rate of the genes depend on their function or their position within the pathway; and (3) whether the IIF pathway genes exhibit evidence of positive selection.
Material and Method
Sequence data collection
If we are to get all members of a particular pathway for a given species, it is essential to have access to the genome sequence with good annotation. With this in mind, we have focused our analysis on the nuclear genomes of D. melanogaster and Caenorhabditis elegans, as model species for the Ecdysozoa, and those of Ciona intestinalis and Strongylocentrotus purpuratus, as representative chordates that predate the vertebrate lineage. Moreover, C. intestinalis and S. purpuratus are the closest known relatives of the chordates.24 Useful model organisms for the study of modern molecular, evolutionary, and cellular biology24,25 are included in this paper. The pathway components are also comprised of other vertebrates, including mammals, poultry, amphibians and fish. This allowed us to identify orthologous gene pairs and to show taxon-specific differences in the genomes of all these species.
A dataset of proteins with known involvement in the human IIF pathway was created according to the KEGG reference database (http://www.genome.jp/kegg/). Additional genes involved in IIF signaling were derived from searching relevant published literature. Consequently, we obtained a dataset of 27 genes with which to search the 19 metazoan species genomes. In order to test whether the evolutionary trend in the phylogeny was affected by taxonomic sampling used, we sorted the taxonomic samplings into 2 data sets: one for phylogenetically distant species (also called broad level species in this paper), including Homo sapiens (human), Mus musculus (mouse), Canis familiaris (dog), Monodelphis domestica (opossum), Gallus gallus (chicken), X. tropicalis (frog), D. rerio (zebrafish), T. nigroviridis (pufferfish), C. intestinalis (sea squirt), D. melanogaster (fly), C. elegans (worm) and sea urchin (S. purpuratus), and the other with a richer samplings from mammalian species composition of H. sapiens (human), Pan troglodytesa (Chimpanzee), Pongo pygmaeus (Orangutan), M. musculus (mouse), Rattus norvegicus (Rat), Cavia porcellus (Guinea Pig), Monodelphis domestica (opossum), Oryctolagus cuniculus (Rabbit), Equus caballus (Horse), Bos taurus (Cow) and C. familiaris (dog). We obtained the amino acids and protein coding sequences (CDS) of the IIF pathway genes from ENSEMBL (http://www.ensembl.org), UCSC and the National Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov). TBLASTN program26 was used to search for orthologous sequences displaying a high degree of similarity to human, mouse and zebrafish IIF components. Blast hits with high enough E- values (E-value < 10−5) were further reblasted with Blastp26 for confirmation. When a gene was found to have multiple alternative splice variants, the longest protein version was kept for further analysis.
Phylogenetic reconstruction
For each paralogous group/homologous group, we generated a multiple sequence alignment (MSA) of the amino acid sequences for each of our 2 data sets using the software Muscle with default settings.27 This amino acid MSA was further used to guide the alignment of the CDS using PAL2NAL.28 The resulting CDS alignments were manually improved using the software BioEdit 7.0.5.29 Unreliably aligned regions were excluded from further analysis. Next, by analyzing the topology of the gene family trees separately, we confirmed the orthologous/ paralogous relationships of the pathway components across each of our 2 data sets. DNA phylogenies for each gene family (representing a mixture of orthologous and paralogous genes) for the broad level species and mammals were constructed using MrBayes 3.1.2,30 applying the nucleotide substitution model that best fit the data according to the Akaike information criterion (AIC). We used Modeltest version 3.731 to select the best-fitting substitution model. At least 200,000 generations were run in 4 independent chains by MrBayes 3.1.2.30 A consensus tree was generated when the likelihood estimates had reached a stationary state (P < 0.001). The resulting trees of 12 broad level metazoan species were compared with those of mammalian species and were visually inspected for an orthologous group: a clade with 0.50 or greater Bayesian posterior probabilities (pps) support and containing at least one representative of each of the query sequences. If no such clade could be found, the gene would not be included in any further analysis. If more than one sequence of a species in the ortholog clade were found, the one with the most complete sequence was chosen. Finally, we gathered separate orthologous groups only if they could be easily distinguished by their gene sequences and orthologous phylogenetic trees. Furthermore, we concatenated the final set of genes from 1:1 orthologs from 8 broad level species (ranging from teleost to human) into a single alignment, because the sequence alignments and orthologous relationships of these 8 species were found more reliable than that of all broad level species. We then inferred a species tree using a BMCMC approach as implemented in MrBayes 3.1.230 as well. The 1:1 orthologs from mammalian species were easily identified with the help of UCSC and ENSEMBL. Finally, we formed 27 orthologous gene groups, and the orthologous genes from each group were further used to measure the evolutionary rates separately.
Codon-based analysis
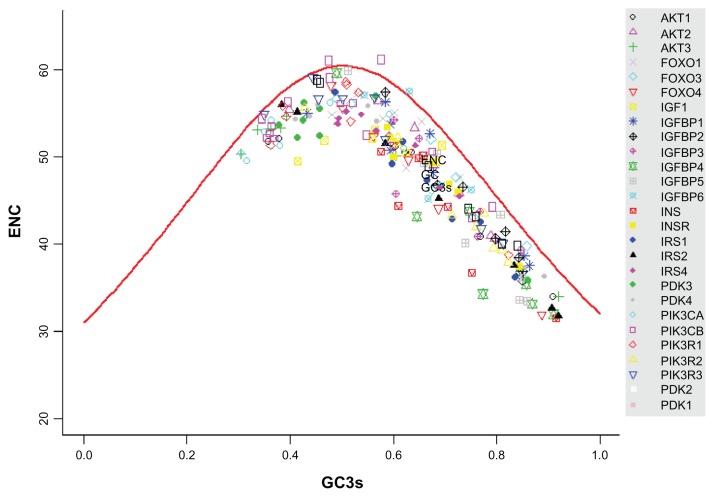
The degree of codon bias or codon preference is often used to reflect the level of selective constraint in a gene and the variation of synonymous substitution rates among genes, which may be related to codon usage.32 To measure the extent of codon usage bias in each orthologous group (27 groups totally), we estimated the effective number of codons (ENC) using DnaSP version 5.10 m,33 which varies between 20 and 61, where the lower the value, the more biased the codon usage.34 Because GC content particularly at their third position is strongly correlated with codon bias, a graphical comparison of ENC versus GC content at the 3rd codon position (GC3) was used to detect the possible bias. A scatter-plot of observed values of ENC versus GC3 was drawn, along with a curve describing the expected values using the Nc-plot technique.34
Tests for the impact of natural selection were carried out by estimating non-synonymous (dN) and synonymous (dS) divergence, and their ratio (ω = dN/dS) calculated by the codeml program from the PAML 4.4 package.35 We restricted this analysis to 2 data sets: one is composed of the 8 species (H. sapiens, M. musculus, C. familiaris, G. gallus, M. domestica, X. tropicalis, D. rerio and T. nigroviridis) and the other is the 11 mammalian species, because we could obtain reliable orthologous relationships and alignments. We used MSAs based on 1:1 ortholog sets, as described in the previous section.
The one ratio model (M0) which assumes that all branches and all sites underwent the same selective constraint, was first used to test the selective forces acting upon each orthologous gene group in this analysis. We also applied the free ratio model (Mf) which assumes individual ratios on each branch of the orthologous gene group. Using the free ratio model, saturation effects were avoided by discarding dS values that were less than 0.007 or more than 2.00, as values for which S*dS were less than 10. To determine whether some codon positions had evolved under positive selection in each orthologous group, we compared the M1a and M2a models36 and also the M7 and M8 models37 using the likelihood ratio test.38 Positive selection is invoked if the LRT is significant and the sites with ω > 1.39
Statistical analysis
We performed a multivariate analysis considering dN, ω, the pathway position, and some parameters influencing evolution selection levels (codon bias, protein length, analyzed codons and GC content21,40). All statistical analyses were performed by statistical package R (http://www.rproject.org). First, we evaluated whether these parameters were correlated using Kendall’s rank correlation coefficient (τ). Then, we analyzed the data using path analysis, an extension of multiple regression analysis that all variables are defined as random and new terminology is used. The first distinction made among variables in the model is between observed and unobserved random variables. Observed variables are called manifest variables and are directly observed. Before performing path analysis, the data were log-transformed and standardized to improve normality. All statistics related to path analysis was calculated by the sem package in R.
Results
Identification of IIF pathway genes in the investigated species
Aiming to identify all elements of the IIF signaling pathway in the investigated species, we initially gathered protein sequences for the selected species by using sequences of human, mouse and zebrafish as queries. Some sequences were readily available in GenBank, UCSC and ENSEMBL, and the others were from genomic alignments. We finally identified a total of 271 putative orthologs of the 27 IIF signaling pathway genes in genomes of 12 broad level species (Additional file 1); as well as 285 putative orthologs from 11 mammals (Additional file 2). Since current genomic projects include many unsequenced regions, this should be considered as the minimum number of actual genes.
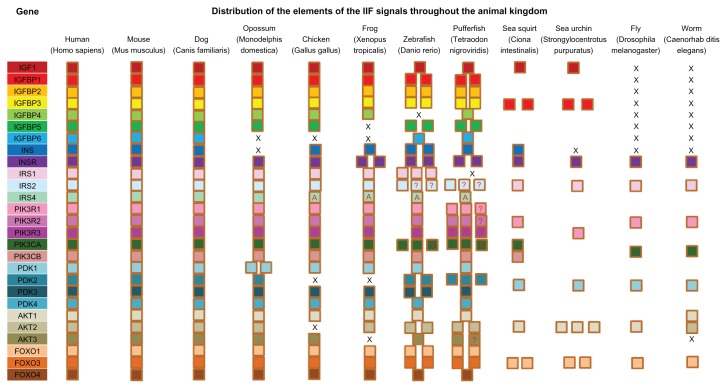
No insulin and IGF1 homologs proteins were found to be encoded for in the genomes of D. melanogaster and C. elegans. All other species, including C. intestinalis and S. purpuratus, had at least one putative protein similar to mammalian IGF1. Our data showed that most of the components in IIF pathway were present throughout all animal phyla investigated (Fig. 2), and further expanded by additional paralogs members in teleost fish. In addition, invertebrates encoded for a smaller subset of IIF pathway genes than vertebrates.
Figure 2.
Survey of the components of IIF pathway in selected metazoans.
Notes: Each square indicates a single pathway gene and squares with a question mark indicating uncertainty of the orthology, while with letter A indicating apparent orthology. Letter X’s represent absence of members.
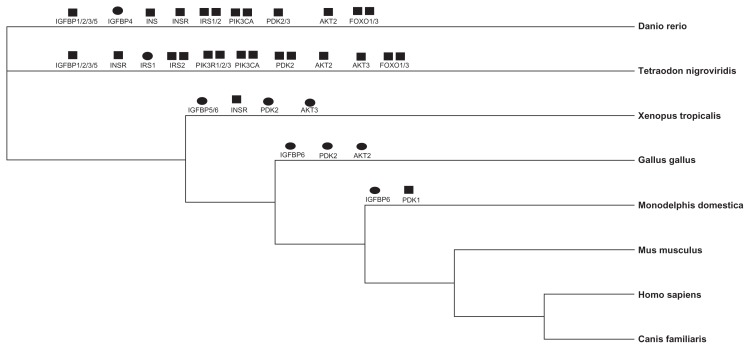
Phylogenies of the broad level species and mammals were presented in Additional file 3. Our 2 data sets provided slightly different topologies, especially when the major clades are considered. Taking advantage of the phylogenetic trees constructed using Bayesian methods, we readily obtained orthologous/ paralogous relationships of the different pathway genes for 12 broad level species as well as 11 mammals. According to the criterion described in the material and method, orthologous members were separated from each gene family and were used for species tree construction and evolutionary rates analysis. We found that the genomes of vertebrates, beginning with D. rerio, started to expand paralogous copies of the pathway components in this study. Six of IIF pathway genes had a 1:1 orthologous relationship in 8 genome (H. sapiens, M. musculus, C. familiaris, M. domestica, G. gallus, X. tropicalis, D. rerio and T. nigroviridis), while the remaining 21 genes underwent a number of duplication or loss events (44 duplications and 10 loss events; Fig. 3). However, only 7 loss events were detected in the 11 mammalian species genomes (Additional file 4).
Figure 3.
The phylogenetic tree showing the evolutionary relationships of the 8 higher species studied.
Notes: Gene duplication (black square) and loss (black ellipse) events detected in the IIF pathway across the 8 species were shown above each branch.
Codon usage bias and its correlation with GC3
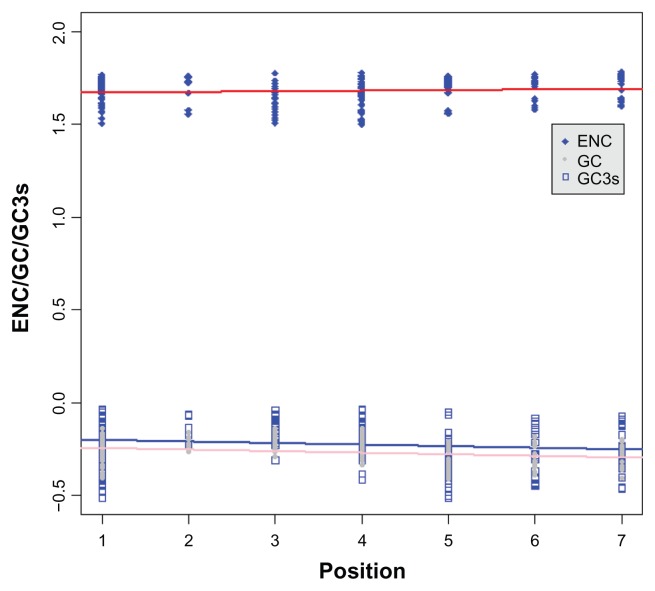
ENC is a measure of the degree of codon bias, ranging from 20, where codon bias is at a maximum and only 1 codon is used for each amino acid, to 61, where there is no codon bias and all codons are used. We first tested whether the ENC values and the GC contents for the total and 3rd codon position were influenced by gene positions in the broad level species and mammalian datasets. Codon bias showed a significant correlation with the positions of the elements (Kendall’s τ = 0.145, P = 0.004, measured as ENC) for 8 species (H. sapiens, M. musculus, C. familiaris, M. domestica, G. gallus, X. tropicalis, D. rerio and T. nigroviridis) from broad level dataset. As did the GC contents for the total and for the 3rd codon positions (τ = −0.220, P < 0.001 for the total GC content and τ = −0.161, P = 0.002 for GC3) (Fig. 4). Moreover, our results indicated that ENC was clearly associated with GC3 (τ = −0.661, P < 0.001). The results for mammalian species dataset (11 species) also had the same tendency (Additional file 5A–C). We subsequently graphed ENC as a function of GC3; along with a reference line (GCref) showing the expected position of genes whose codon usage is constrained solely by the nucleotide composition at the third codon position. From results shown in Figure 5 and Additional file 5D, it can be seen that most observed values of ENC track were below the reference line. We also performed t-test to assess whether the means of reference line and observe ENC values are statistically different from each other. We got significant difference between them (T = 6.99, P < 0.001). This indicates that the nucleotide composition at the third codon position is a major determinant of ENC.
Figure 4.
Correlations between different factors (codon bias, GC content for total and 3rd position) and the element position in the IIF pathway.
Notes: The positions of the IIF pathway genes were shown in Figure 1. Continuous lines represent regression lines. The ENC value has been log-transformed.
Figure 5.
Graph of the ENC versus the percent GC at third codon positions.
Note: The expectation of the ENC under the assumption of no selection on codon usage is given by the solid curve.
Relationship of dN/dS to pathway position
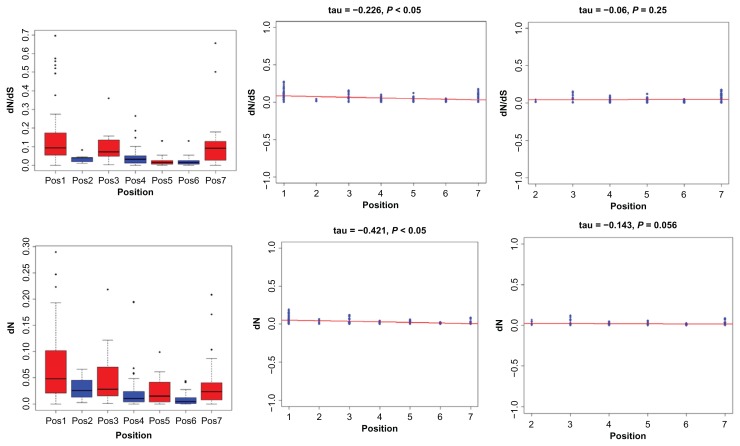
The strength of natural selection acting on genes is inferred by ω, the ratio of dN/dS (non-synonymous substitutions (dN)/synonymous substitutions (dS)). By using the M0 model, a single dN/dS value for all lineages was estimated for each orthologous gene group in the IIF pathway. The results showed that dN/ dS values were not uniformly distributed along the pathway, but varied greatly among genes, with ω values ranging from 0 to 0.21 for the broad level species (Table 1) and from 0.01 to 0.25 for mammalian species (Additional file 6), suggesting that purifying selection or selection constraint best explains the evolution of these genes. The free ratio model results demonstrated that the ω values for the IIF pathway genes varied substantially in each lineage, being estimated to be 0.0001~0.692 for the broad level species (Additional file 7) and 0.0001~0.773 for mammalian species (Additional file 8). In the further study the relationship between the ω values and the architecture of the IIF signaling pathway, we also performed other analysis. We first use boxplot graphical technique to display the distribution of selective constraints variables (dN, dS and dN/dS), which helps to see the difference of dN/dS and identify the outlying points (Fig. 6A). We found a significant negative correlation between ω estimates for IIF pathway genes and their positions in the pathway before removing the outlier (τ = −0.221; P < 0.001). After removing outlying points, we also detected negative correlation (τ = −0.226; P < 0.05; Fig. 6B). From boxplot, we found that dN/dS value of position 1 tended to be higher than most of the other positions. Next, we verified whether correlation was caused by genes in position 1. The results show that no correlation exists between them (τ = −0.06; P = 0.25) after removing position 1, as shown in Figure 6C. We also performed dN and dS analysis separately, and found that non-synonymous changes were similar to dN/dS pattern (Fig. 6D) and were the main contributors to the tendency (dN: τ = −0.421, P < 0.05, Fig. 6E). The negative linear relationship disappears again after removing position 1 (Fig. 6F). We got marginal negative correlation between dS and position before removing genes in position 1 (dS: τ = −0.138; P < 0.038; figure now shown) and correlation didn’t hold again if position 1 was exclude (data not shown). The results for mammals also have the similar tendency and corresponding results can be found in Additional file 9. Moreover, we compared the difference of dN/dS variable between broad level species and mammals, and we could not reject the null hypothesis that there is no difference between them (Wilcoxon rank test; P = 0.6108).
Table 1.
Summary statistics used in this study.
| Gene | Average protein length | Average GC content | Average GC3 content | Effective no. of codons | Percent of analyzed codons | dN | dS | ω (dN/dS) |
|---|---|---|---|---|---|---|---|---|
| IGF1 | 169.00 | 0.53 | 0.58 | 51.28 | 0.75 | 0.67 | 4.93 | 0.14 |
| IGFBP1 | 266.00 | 0.60 | 0.66 | 49.29 | 0.81 | 1.78 | 8.64 | 0.21 |
| IGFBP2 | 296.25 | 0.65 | 0.77 | 42.88 | 0.78 | 0.82 | 11.21 | 0.07 |
| IGFBP3 | 328.14 | 0.62 | 0.71 | 46.02 | 0.81 | 0.87 | 7.26 | 0.12 |
| IGFBP4 | 260.00 | 0.64 | 0.76 | 40.33 | 0.92 | 0.78 | 17.18 | 0.05 |
| IGFBP5 | 250.00 | 0.60 | 0.74 | 43.04 | 0.45 | 0.79 | 40.21 | 0.02 |
| IGFBP6 | 215.60 | 0.63 | 0.66 | 50.63 | 0.64 | 0.89 | 5.12 | 0.17 |
| INS | 122.14 | 0.60 | 0.70 | 44.00 | 0.82 | 1.14 | 8.57 | 0.13 |
| INSR | 1315.00 | 0.54 | 0.67 | 48.32 | 0.76 | 0.36 | 9.10 | 0.04 |
| IRS1 | 1074.57 | 0.61 | 0.67 | 46.61 | 0.48 | 0.80 | 15.39 | 0.05 |
| IRS2 | 1124.43 | 0.62 | 0.68 | 44.10 | 0.39 | 0.74 | 19.32 | 0.04 |
| IRS4 | 1048.38 | 0.57 | 0.58 | 52.22 | 0.50 | 1.72 | 15.98 | 0.11 |
| PIK3R1 | 725.00 | 0.47 | 0.50 | 53.64 | 0.99 | 0.40 | 7.99 | 0.05 |
| PIK3R2 | 713.13 | 0.59 | 0.72 | 43.97 | 0.82 | 0.77 | 18.96 | 0.04 |
| PIK3R3 | 493.50 | 0.49 | 0.56 | 52.48 | 0.75 | 0.47 | 17.84 | 0.03 |
| PIK3CA | 1068.50 | 0.43 | 0.43 | 52.61 | 1.00 | 0.12 | 6.38 | 0.02 |
| PIK3CB | 1049.57 | 0.45 | 0.45 | 53.37 | 0.87 | 0.35 | 8.17 | 0.04 |
| PDK1 | 423.88 | 0.50 | 0.55 | 53.40 | 0.86 | 0.43 | 10.51 | 0.04 |
| PDK2 | 412.29 | 0.54 | 0.68 | 47.12 | 0.98 | 0.37 | 14.54 | 0.03 |
| PDK3 | 411.25 | 0.47 | 0.50 | 52.24 | 0.98 | 0.26 | 7.04 | 0.04 |
| PDK4 | 405.71 | 0.50 | 0.59 | 49.89 | 0.85 | 0.46 | 8.16 | 0.06 |
| AKT1 | 471.25 | 0.52 | 0.64 | 45.93 | 0.91 | 0.15 | 9.21 | 0.02 |
| AKT2 | 482.43 | 0.53 | 0.66 | 48.85 | 0.99 | 0.21 | 8.60 | 0.02 |
| AKT3 | 471.43 | 0.45 | 0.47 | 49.86 | 0.90 | 0.06 | 23.59 | 0.00 |
| FOXO1 | 644.00 | 0.56 | 0.61 | 52.59 | 0.75 | 0.74 | 8.47 | 0.09 |
| FOXO3 | 617.88 | 0.59 | 0.69 | 47.26 | 0.63 | 0.61 | 15.38 | 0.04 |
| FOXO4 | 496.50 | 0.59 | 0.62 | 49.60 | 0.66 | 0.95 | 10.79 | 0.09 |
Figure 6.
The distribution of selective constraint variables and correlation between the position of the elements in the IIF pathway and selective constraint estimates.
Notes: A the distribution of dN/dS, B the linear relationship between dN/dS and position after removing outliers, C the linear relationship between dN/dS and position after removing genes from position 1, D the distribution of dN, E the linear relationship between dN and position after removing outliers and F the linear relationship between dN and position after removing genes from position 1. Continuous lines represent regression lines.
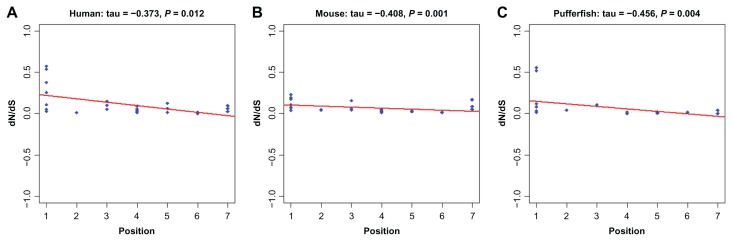
We wondered whether the correlation between ω and pathway position was a general trend in the phylogeny or whether it might only be attributable to some specific lineages. To test this, we adopted free-ratio model to estimate ω values along each linage for each orthologous gene group. ω correlated significantly with pathway position in H. sapiens, M. musculus and T. nigroviridis, but its pattern didn’t hold after removing position 1 genes. All the remaining lineages only obtain a negative τ statistics (Additional file 10), but do not arrive at a significant level even when considering the genes from position 1. This result demonstrated that the negative correlation between the ω values and the position of the elements in the pathway was a phylogeny-wide trend and was not caused by any lineage specific pattern. Scatterplots of data for H. sapiens, M. musculus and T. nigroviridis are given in Figure 7A–C, while plots of data from other lineages can be found in Additional file 10 (Figs. A–L).
Figure 7.
Correlation between the position of the elements in the IIF pathway and the dN/dS among different species. (A) for human, (B) for mouse and (C) for pufferfish and continuous lines represent regression lines.
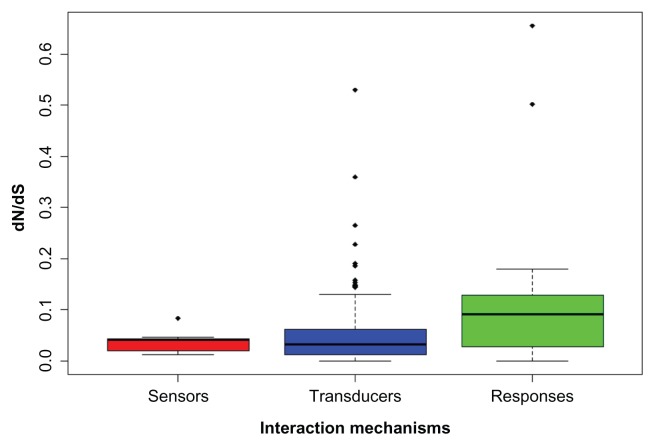
As introducing in the part of introduction, the components of the IIF pathway can be divided into three distinct phases according to interaction mechanisms: sensors (position 2), transducers (position 3 to 6) and responses (position 7). We also performed relationship analysis among the different functional elements in the pathway using broad level species. The results showed that the dN/dS values of the transducers genes were significantly smaller than downstream responses (Fig. 8; Wilcoxon rank test; P < 0.001). It is harder to get a significant difference when comparing the sensors with transducers, and comparing the sensors with responses, but the medium dN/dS in transducers is smaller than that in sensors.
Figure 8.
Box plot of the functional module of IIF pathway.
Notes: Sensors module comprised by genes from position 2, transducers by genes from position 3 to 6 and responses by genes from position 7. Dots represent the extreme dN/dS values (outliers), and black line represents the medium value.
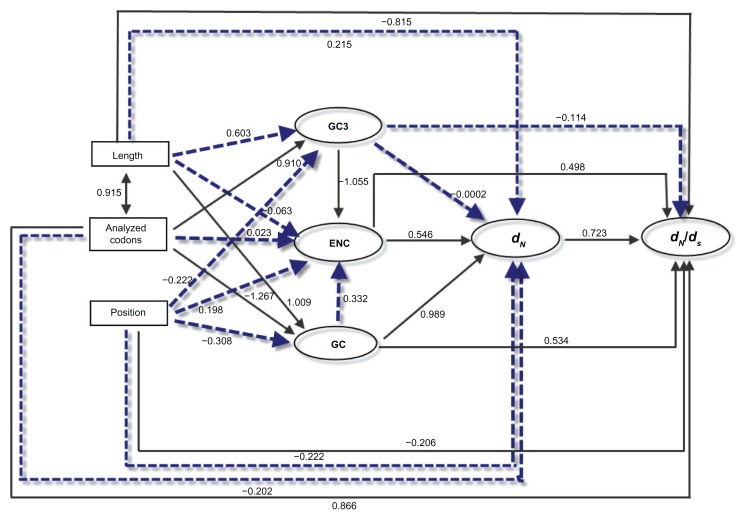
The selective constraint of a given gene is known to correlate with different factors, such as the protein length and the number of analyzed codons.21,40 We further examined the correlations of dN/dS with a series of potential factors. By Kendall’s rank correlation analyses, the ω (dN/dS) was shown to be marginally correlated with analyzed codons (τ = 0.284, P = 0.0498), but not significantly associated with protein length (τ = 0.254, P = 0.0804). Since codon bias, GC content, and protein length are intercorrelated, some of the observed correlations might actually result from indirect rather than direct effects. Path analysis can be used to better describe the directed dependencies among a set of variables: GC content, dN, ω, and the position in the pathway. As shown in Figure 9, dN/dS showed a significant correlation with the position of the elements in the pathway (standardized path coefficient, ß = −0.206; P = 0.03). Moreover, dN/dS were clearly affected by other factors, except GC3. Overall, the element position was not the unique factor to influence dN/dS. However, after removing genes at position 1, the correlation between dN/dS and position was no longer significant (ß = 0.204; P = 0.38), which is consistent with previous Kendall’s rank analysis. Nevertheless, it is difficult to summarize the key factor because the selective constraint often determined by the complex interplay among the large number of factors.
Figure 9.
Graphical representation of the path analysis used to analyze the relationships among pathway positions, non-synonymous substitution (dN), dN/dS ratio, codon bias (measured by ENC), protein length, analyzed codons and GC content.
Notes: Pathway position, protein length and analyzed codons were treated as exogenous variables, while the rest were treated as endogenous variables. The numbers on the arrows represent the standardized path coefficients (β). Solid and dotted lines represent significant and non-significant relationships, respectively.
Tests for positive selection
The results of the LRTs obtained from comparisons of M7 versus M8 and M1a versus M2a, which were conducted to examine positive selection in 27 gene orthologous groups in the broad level species, are shown in Table 2. LRTs for the comparison of M2a versus M1a were not statistically significant for any of the 27 genes. LRTs for the comparison of M8 versus M7, however, were significant for 2 of the 27 genes. IGFBP4 gene showed the footprint of positive selection, but LRTs were marginally significant (P = 0.06). In the mammalian dataset, only one positively selected gene was detected (Additional file 11). The similar phenomena of positive selected genes in conserved signal transduction also found in plant terpenoid biosynthesis pathway studying.41 Consequently, most genes in IIF pathway are under strong selective constraint, which does not exclude the possibility of past episodes of positive selection on these genes.
Table 2.
Genes showing evidence of positively selected codons.
| Gene | Model compared | ω value (M8) | |||
|---|---|---|---|---|---|
|
|
|||||
| M2 vs. M1 | M8 vs. M7 | ||||
|
|
|
||||
| 2ΔL = (L1−L0) | P value | 2ΔL = (L1−L0) | P value | ||
| IGF1 | 0 | 1 | 0 | 1 | |
| IGFBP1 | 0 | 1 | 0.99 | P > 0.05 | |
| IGFBP2 | 0 | 1 | 3.76 | P > 0.05 | |
| IGFBP3 | 0 | 1 | 0 | 1 | |
| IGFBP4 | 0 | 1 | 5.58 | 0.06 | 2.5 |
| IGFBP5 | 0 | 1 | 0.97 | P > 0.05 | |
| IGFBP6 | 0 | 1 | 0 | 1 | |
| INS | 0 | 1 | 10.73 | P < 0.05 | 218.92 |
| INSR | 0 | 1 | 1.57 | P > 0.05 | |
| IRS1 | 0 | 1 | 2.01 | P > 0.05 | |
| IRS2 | 0 | 1 | 0.44 | P > 0.05 | |
| IRS4 | 0 | 1 | 0.34 | P > 0.05 | |
| PIK3R1 | 0 | 1 | 0 | 1 | |
| PIK3R2 | 0 | 1 | 0.01 | P > 0.05 | |
| PIK3R3 | 0 | 1 | 0 | 1 | |
| PIK3CA | 0 | 1 | 0 | 1 | |
| PIK3CB | 0 | 1 | 0.34 | P > 0.05 | |
| PDK1 | 0 | 1 | 0 | 1 | |
| PDK2 | 0 | 1 | 2.79 | P > 0.05 | |
| PDK3 | 0 | 1 | 0 | 1 | |
| PDK4 | 0 | 1 | 0 | 1 | |
| AKT1 | 0 | 1 | 0 | 1 | |
| AKT2 | 0 | 1 | 0.96 | P > 0.05 | |
| AKT3 | 0 | 1 | 6.02 | 0.049 | 1 |
| FOXO1 | 0 | 1 | 0.62 | P > 0.05 | |
| FOXO3 | 0 | 1 | 0 | 1 | |
| FOXO4 | 0 | 1 | 1.16 | P > 0.05 | |
Note: Statistically significant results at 0.05 level are in boldface.
Discussion
Multicellular animals have their special mechanism to adapt to environmental changes. When environmental conditions are unfavorable to an organism’s processes, such as growth and reproduction, different nutrient-sensing pathways will be activted in response to nutrition shuffling. Most of these pathways exert their function by modulating the activity of insulin-like signaling. The IIF pathway is an important branch of insulin signaling and plays key roles in growth, metabolism, stress resistance, reproduction, and longevity in diverse organisms including mammals.7 Therefore, the evolution of IIF signaling transduction pathway was likely to be important to animals during evolutionary processes. The perception and transmission of signal of IIF pathway has been studied in detail in non-mammalian vertebrates and invertebrates since the 1970s.42–44 However, several key questions concerning the evolution of the IIF pathway remain unanswered. These include (1) which is the most distant animal species containing all the components necessary for IIF signaling?; and (2) whether the position of IIF component influences the evolutionary rate.
It has been reported that insulin-like peptides have been found in many invertebrate species including D. melanogaster and C. elegans.45–48 As for insulin and IGF1, the failure in detecting orthologs in D. melanogaster and C. elegans through BLAST searching may be explained by the known sequence divergence that existed among members of the insulin family. Previous phylogenetic analysis also confirmed that the insulin/IGF sequences formed one group and a more distantly related paralog confined within the insulin-like factor/relaxin (INSL/RLN) paralogon.47 In this study, both S. purpuratus and C. intestinalis were found to have genes encoding proteins similar to vertebrate insulin and IGF sequences, therefore, we deduced that the origins of IGF system, which may exert its roles in the IIF pathway, could date back to early stages in echinoderm evolution (520 MYA). Moreover, we observed several lineage specific gene duplications within the pathway components. For example, several additional IGFBPs homologs are present in the T. nigroviridis genome, compared to other vertebrates (see Additional file 1). A T. nigroviridis IGFBP1 gene can be found on both chromosomes 15 and 1 and duplicated sequences for IGFBP3 are positioned on the same two chromosomes. This result is consistent with the hypothesis that the teleost fishes are thought to have undergone additional genome duplication.49,50 A range of one to many orthologous genes in studied species may be explained by whole genome duplications occurring early in the vertebrate lineage,51 or that continuous gene duplications with the accumulation of new mutations for environmental adaption during vertebrate evolution occurred.52–55 In phylogenetic analysis, we identified 44 duplications and 10 loss events in the broad level dataset, while only 7 loss events were identified in mammalian species. The loss orthologs may be caused by the uncompleted genome or stringent searching criteria, and we have found partial sequences of the lost orthologs in the species’ contig sequence by BLAST searching.
Our study reveals a robust negative linear relationship between the value of dN/dS and the position of elements in the pathway at first. This result reaches the same conclusion for the analysis of insulin/ TOR pathway in Drosophila and vertebrates.21,22 However, we are not sure this correlation is a true trend along the insulin pathway, or rather caused by more outliers or high variation of selective constraint (Fig. 6A; two positive selected genes though one is marginally significant) in position 1. We first removed all the outlier (including the positive selective genes) along the pathway, and we still obtained the same conclusion (Fig. 6B). Therefore the outliers were not the main factor leading to the gradient in the strength of negative selection along the pathway. Next, we removed all genes in position 1 and the correlation vanished (Fig. 6C). We also applied the similar strategy to dN, dS and lineage specific dN/dS variables, and we arrived at the coincident conclusion that the variation in selective constraint does not depend on the pathway structure here. Therefore, the presented correlation comes from the relaxed selective constraint operated on genes of position 1, which give us an erroneous perception of truth.
The organisms will open or close sets of genes in response to environmental stresses. During this process, organisms and/or individual cells are equipped with special control systems to respond and acclimate to stress. We further studied the relationship between functional module (sensors, transducers, and responses) related to special control systems and selective constraints. From Figure 8, we found that the dN/dS values from transducers were lower than that from responses and sensor module. This result seems contrary to previous demonstrations that there is no linear relationship between selective constraint and component position after removing components from position 1. In fact, the inconsistence comes from unbalanced selective constraint values among groups and different statistical methods. In this paper, most of the lineages have strong selective constraints, and several higher dN/dS were assigned to outliers when we combined the components from positions 3 to 6. In this case, the smaller dN/dS dominated the rectangle box, which make it smaller than that of the sensor and response module. While in previous linear analysis, the dN/dS values distribute evenly among positions, we obtained no significant linear relationship after removing components in position 1. Moreover, functional module analysis considered Wilcoxon rank correlation, while Kendall’s rank analysis was used in positional analysis. This conclusion seems contrary to the pattern found in previous studies about other pathways. Rausher et al18 have shown that the selective constraint levels in the plant anthocyanin biosynthetic pathway correlate with the position of the elements in the pathway. Namely, the upstream genes evolved substantially more slowly than the downstream genes. They thus hypothesized that the upstream genes in a metabolic pathway evolved more slowly than the downstream genes due to stronger purifying selection for the upstream genes. Such a pattern has also been confirmed at the intragenic19 and population levels56 in the genus Ipomoea as well as by a molecular population study on regulatory and signal transduction genes20 but was not supported by other studies.21,57–59 Alvarez-Ponce et al21,22 concluded that downstream genes from the insulin/TOR pathway are selectively more constrained than upstream genes, no matter the species considered. Our study demonstrates that the medium of selective constraint in transducers is smaller than that in sensors and responses. This seems to be difficult to understand because the responses are an important transcription factor family, which belongs to “O” subfamily of forkhead transcription factors. They should undergo more severely selective constraint given that the downstream elements would be involved in a greater number of pathways and recruit more target members. One explanation could be that the elements of transducers are pivot genes of insulin pathway, and they are not only involve in IIF pathway, but also cross-talk with other important pathways such as c-Jun N-terminal kinase (JNK) and epidermal growth factor (EGF)/EGF receptor (EGFR). They should therefore have strong constraint. Just as Cork and Purugganan59 illustrated, the specific nature of selection on the component genes depended largely on the function of the pathway, but the dichotomy between upstream and downstream genes in a pathway was a crude differentiation of function.59
The selective constraints in molecular pathways were affected by diverse factors, such as connectivity,60,61 codon usage bias, and the length of the encoded proteins.21,40 Unfortunately, we could not obtain all higher animals’ interactomic data, thus we are not able to conclude that connectivity pattern was responsible for the correlation between selective constraints and the position of the elements in the pathway. Most importantly, we detected (1) a positive correlation between the position of the elements in the pathway and codon bias (Fig. 4), and (2) a positive correlation between analyzed codons length and the position of the elements in the pathway. Furthermore, codon bias and analyzed codons length are all correlated with dN/dS and dN in Figure 9. However, path analysis did not confirm that the relationship between selective constraint and the position is significant after factoring out the effects of position 1 (Fig. 9). This phenomenon demonstrates that the relaxed selective constraint at position 1 explains the purifying selection polarity along the IIF pathway.
Conclusions
Genomic sequence data has provided an opportunity to gain a better understanding about the evolution of pathway genes. In this study, we investigated the differences in signaling transduction elements among different animals, the variation of selection constraint along IIF signaling pathway genes, and the relationship between evolutionary forces and gene position in the pathway. Our study demonstrates that the IGF1 and insulin recruited by the IIF pathway can date back to early stages in echinoderm evolution. In addition, our results also indicated that the negative correlation between dN/dS estimates and position was caused by higher dN/dS values at position 1. Also, the evolutionary rate variation of the IIF pathway are correlated with codon bias, protein length and other factors, suggesting that the selective patterns may be influenced by complicated factors. Overall, further work will be needed to study the patterns of molecular evolution in pathways encompassing a wide range of topologies and to analyze the biological impact of the interconnection patterns to fully understand how pathway architecture constrains the evolution of its components.
Additional File
Gene_information_new1.xls
Acknowledgements
The authors wish to express their gratitude to the members of animal sciences laboratory of Shanghai Jiao Tong University.
Footnotes
Author Contributions
Conceived and designed the experiments: Minghui Wang and Qishan Wang. Analyzed the data: Minghui Wang. Wrote the first draft of the manuscript: Minghui Wang. Agree with manuscript results and conclusions: Zhen Wang and Qiong ping Wang. Made critical revisions and approved final version: Xiangzhe Zhang and Yuchun Pan. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
This work is supported by the National Natural Science Foundation of China (grant no. 31072003, 31000992, and 31272414), the National Key Technology R&D Program (grant no. 2008BADB2B11), National High Technology Research and Development Program of China (863) (grant no. 2008AA101002, 2008AA101009) and the National Key Basic Research Program (973) (grant no. 2006CB102102).
References
- 1.Goberdhan DC, Wilson C. The functions of insulin signaling: size isn’t everything, even in Drosophila. Differentiation. 2003 Sep;71(7):375–97. doi: 10.1046/j.1432-0436.2003.7107001.x. [DOI] [PubMed] [Google Scholar]
- 2.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 3.Nuzhdin SV, Brisson JA, Pickering A, Wayne ML, Harshman LG, McIntyre LM. Natural genetic variation in transcriptome reflects network structure inferred with major effect mutations: insulin/TOR and associated phenotypes in Drosophila melanogaster. BMC Genomics. 2009;10:124. doi: 10.1186/1471-2164-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters ER. Molecular adaptation and the origin of land plants. Mol Phylogenet Evol. 2003 Dec;29(3):456–63. doi: 10.1016/j.ympev.2003.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Chapman AB, Knight DM, Ringold GM. Glucocorticoid regulation of adipocyte differentiation: hormonal triggering of the developmental program and induction of a differentiation-dependent gene. J Cell Biol. 1985 Oct;101(4):1227–35. doi: 10.1083/jcb.101.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Power DM, Llewellyn L, Faustino M, et al. Thyroid hormones in growth and development of fish. Comp Biochem Physiol C Toxicol Pharmacol. 2001 Dec;130(4):447–59. doi: 10.1016/s1532-0456(01)00271-x. [DOI] [PubMed] [Google Scholar]
- 7.Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6(2):e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbieri M, Bonafe M, Franceschi C, Paolisso G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab. 2003 Nov;285(5):E1064–E71. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- 9.Pawlikowska L, Hu D, Huntsman S, et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009 Aug;8(4):460–72. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001 Dec 13;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo RS. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol Metab. 2002 May-Jun;13(4):156–62. doi: 10.1016/s1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- 12.Kuningas M, Mooijaart SP, van Heemst D, Zwaan BJ, Slagboom PE, Westendorp RG. Genes encoding longevity: from model organisms to humans. Aging Cell. 2008 Mar;7(2):270–80. doi: 10.1111/j.1474-9726.2008.00366.x. [DOI] [PubMed] [Google Scholar]
- 13.Burgering BM, Kops GJ. Cell cycle and death control: long live Forkheads. Trends Biochem Sci. 2002 Jul;27(7):352–60. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- 14.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117(4):421–6. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002 Oct 1;250(1):1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nature Reviews Molecular Cell Biology. 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Zhang X, Zhao H, Wang Q, Pan Y. FoxO gene family evolution in vertebrates. BMC Evol Biol. 2009;9:222. doi: 10.1186/1471-2148-9-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol Biol Evol. 1999;16(2):266–74. doi: 10.1093/oxfordjournals.molbev.a026108. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Rausher MD. Evolutionary rate variation in anthocyanin pathway genes. Mol Biol Evol. 2003 Nov;20(11):1844–53. doi: 10.1093/molbev/msg197. [DOI] [PubMed] [Google Scholar]
- 20.Riley RM, Jin W, Gibson G. Contrasting selection pressures on components of the Ras-mediated signal transduction pathway in Drosophila. Mol Ecol. 2003 May;12(5):1315–23. doi: 10.1046/j.1365-294x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 21.Alvarez-Ponce D, Aguade M, Rozas J. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Res. 2009 Feb;19(2):234–42. doi: 10.1101/gr.084038.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez-Ponce D, Aguade M, Rozas J. Comparative genomics of the vertebrate insulin/TOR signal transduction pathway: a network-level analysis of selective pressures. Genome Biol Evol. 2010 Jan;3:87–101. doi: 10.1093/gbe/evq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cormican P, Lloyd AT, Downing T, Connell SJ, Bradley D, O’Farrelly C. The avian Toll-Like receptor pathway—Subtle differences amidst general conformity. Developmental and Comparative Immunology. 2009;33(9):967–73. doi: 10.1016/j.dci.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Sodergren E, Weinstock GM, Davidson EH, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006 Nov 10;314(5801):941–52. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherwood NM, Tello JA, Roch GJ. Neuroendocrinology of protochordates: insights from Ciona genomics. Comp Biochem Physiol A Mol Integr Physiol. 2006 Jul;144(3):254–71. doi: 10.1016/j.cbpa.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 Sep 1;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609–W12. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. 1999. [Google Scholar]
- 30.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003 Aug 12;19(12):1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 31.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 32.Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–43. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999 Feb;15(2):174–5. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 34.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87(1):23–9. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007 Aug;24(8):1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 36.Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004 Oct;168(2):1041–51. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155(1):431–49. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whelan S, Goldman N. Distributions of statistics used for the comparison of models of sequence evolution in phylogenetics. Mol Biol Evol. 1999;16(9):1292–9. [Google Scholar]
- 39.Bielawski JP, Yang Z. Maximum likelihood methods for detecting adaptive evolution after gene duplication. J Struct Funct Genomics. 2003;3(1–4):201–12. [PubMed] [Google Scholar]
- 40.Ingvarsson PK. Gene expression and protein length influence codon usage and rates of sequence evolution in Populus tremula. Mol Biol Evol. 2007 Mar;24(3):836–44. doi: 10.1093/molbev/msl212. [DOI] [PubMed] [Google Scholar]
- 41.Ramsay H, Rieseberg LH, Ritland K. The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Mol Biol Evol. 2009 May;26(5):1045–53. doi: 10.1093/molbev/msp021. [DOI] [PubMed] [Google Scholar]
- 42.Kelley KM. Experimental diabetes mellitus in a teleost fish. I. Effect of complete isletectomy and subsequent hormonal treatment on metabolism in the goby, Gillichthys mirabilis. Endocrinology. 1993 Jun;132(6):2689–95. doi: 10.1210/endo.132.6.8504768. [DOI] [PubMed] [Google Scholar]
- 43.Nagasawa H, Kataoka H, Isogai A, et al. Amino-terminal amino acid sequence of the silkworm prothoracicotropic hormone: homology with insulin. Science. 1984;226:1344–5. doi: 10.1126/science.226.4680.1344. [DOI] [PubMed] [Google Scholar]
- 44.Wolkow CA, Munoz MJ, Riddle DL, Ruvkun G. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem. 2002 Dec 20;277(51):49591–7. doi: 10.1074/jbc.M207866200. [DOI] [PubMed] [Google Scholar]
- 45.Duret L, Guex N, Peitsch MC, Bairoch A. New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res. 1998 Apr;8(4):348–53. doi: 10.1101/gr.8.4.348. [DOI] [PubMed] [Google Scholar]
- 46.Krieger MJB, Jahan N, Riehle MA, Cao C, Brown MR. Molecular characterization of insulin-like peptide genes and their expression in the African malaria mosquito, Anopheles gambiae. Insect Mol Biol. 2004;13(3):305–15. doi: 10.1111/j.0962-1075.2004.00489.x. [DOI] [PubMed] [Google Scholar]
- 47.Olinski RP, Lundin LG, Hallbook F. Conserved synteny between the Ciona genome and human paralogons identifies large duplication events in the molecular evolution of the insulin-relaxin gene family. Mol Biol Evol. 2006;23(1):10–22. doi: 10.1093/molbev/msj002. [DOI] [PubMed] [Google Scholar]
- 48.Li C. The ever-expanding neuropeptide gene families in the nematode Caenorhabditis elegans. Parasitology. 2005;131(Suppl):S109–S27. doi: 10.1017/S0031182005009376. [DOI] [PubMed] [Google Scholar]
- 49.Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003 Mar;13(3):382–90. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001 Oct 29;356(1414):1661–79. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–33. [PubMed] [Google Scholar]
- 52.Friedman R, Hughes AL. Pattern and timing of gene duplication in animal genomes. Genome Res. 2001 Nov;11(11):1842–7. doi: 10.1101/gr.200601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes AL. Phylogenies of developmentally important proteins do not support the hypothesis of two rounds of genome duplication early in vertebrate history. J Mol Evol. 1999;48(5):565–76. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- 54.Hughes AL. Phylogenetic tests of the hypothesis of block duplication of homologous genes on human chromosomes 6, 9, and 1. Mol Biol Evol. 1998 Jul;15(7):854–70. doi: 10.1093/oxfordjournals.molbev.a025990. [DOI] [PubMed] [Google Scholar]
- 55.Martin A. Is tetralogy true? lack of support for the “One-to-Four Rule”. Mol Biol Evol. 2001;18(1):89–93. doi: 10.1093/oxfordjournals.molbev.a003723. [DOI] [PubMed] [Google Scholar]
- 56.Rausher MD, Lu Y, Meyer K. Variation in constraint versus positive selection as an explanation for evolutionary rate variation among anthocyanin genes. J Mol Evol. 2008;67(2):137–44. doi: 10.1007/s00239-008-9105-5. [DOI] [PubMed] [Google Scholar]
- 57.Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006;7(5):R39. doi: 10.1186/gb-2006-7-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsen KM, Womack A, Garrett AR, Suddith JI, Purugganan MD. Contrasting evolutionary forces in the Arabidopsis thaliana floral developmental pathway. Genetics. 2002 Apr;160(4):1641–50. doi: 10.1093/genetics/160.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cork JM, Purugganan MD. The evolution of molecular genetic pathways and networks. Bioessays. 2004 May;26(5):479–84. doi: 10.1002/bies.20026. [DOI] [PubMed] [Google Scholar]
- 60.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002 Apr 26;296(5568):750–2. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 61.Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol Biol Evol. 2005 Apr;22(4):803–6. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene_information_new1.xls