Abstract
Amide proton transfer (APT) imaging is a new MRI technique that detects endogenous mobile proteins and peptides in tissue via saturation of the amide protons in the peptide bonds. Initial studies have shown promise in detecting tumor and stroke, but this technique was hampered by magnetic field inhomogeneity and a low signal-to-noise ratio. Several important prerequisites for performing APT imaging experiments include designing an effective APT imaging pulse sequence based on the hardware capability, optimizing the experimental protocol for the best clinical imaging quality, and developing data processing approaches for effective image assessment. In this chapter, technical issues, such as pulse sequence design and optimization, magnetic field inhomogeneity correction, specific absorption rate minimization, and scan duration, are addressed.
Keywords: APT, magnetization transfer, protein, brain tumor, field inhomogeneity, MRI
1. Introduction
Proteins constitute 18% of the total mass of a typical mammalian cell. From an MRI point of view, these cellular proteins can be divided into two broad types: bound proteins, which possess solid-like properties and have protons with short T2(~ μs), and mobile proteins and peptides, which rotate rapidly and whose protons have relatively long T2(~ tens of ms). Solid-like macromolecules can be detected by conventional magnetization transfer (MT) (1, 2). It was recently demonstrated that it is possible to produce endogenous mobile protein- and peptide-based MRI contrast (3) using a chemical exchange saturation transfer (CEST) enhancement scheme (4). This approach, called amide proton transfer (APT) imaging (3), was shown to be sensitive to pH changes in stroke (3, 5, 6) due to the effect of pH on proton exchange, and to be able to provide brain tumor contrast (7-9), based on the increased cellular content of proteins and peptides in malignant tumors (10, 11).
APT imaging has the potential to expand the range of molecular MRI techniques to the endogenous protein and peptide level. It is a safe, non-invasive technology that can be easily implemented using existing hardware for clinical neuro-imaging of brain tumors, stroke, and other neurologic disorders. The APT approach is in early development, and the technique is far from optimal. In this chapter, the initial experience and the prerequisites for performing an effective APT imaging experiment on the human brain at 3T are provided, using imaging of a human glioma as an example.
2. Materials
2.1. Hardware and Software
A Philips 3T MRI scanner (Philips Medical Systems, Best, The Netherlands).
An eight-channel phased-array head coil.
Two earplugs, a foam head holder, and a headband.
A personal computer loaded with the Philips pulse-programming environment (PPE), and interactive data language (IDL, Research Systems, Inc., Boulder, CO, USA).
2.2. Phantoms, Human Subjects, and Other Materials
A bottle of liquid egg whites from the supermarket (see Note 1).
Several large bottles of water (3-5 L).
Healthy human subjects.
Patients with brain tumors.
Gadolinium contrast agents (ProHance, Bracco Diagnostic Inc., Princeton, NJ, USA).
3. Methods
The effect of APT is associated with a low-concentration proton pool of mobile proteins and peptides. Therefore, the first step in conducting a successful APT imaging experiment is to design an effective imaging pulse sequence for the maximal APT enhancement. Because APT occurs in a small offset range around the water resonance frequency, a weak RF field should be applied to avoid too much water signal intensity attenuation due to direct water saturation and conventional MT. After the pulse sequence is programmed, it must be tested and optimized for the best image and z-spectrum (see Note 2) quality.
APT imaging is confounded by local magnetic field (B0) inhomogeneity (see Note 3). The initial human studies used a two-offset approach, symmetric around the global water center frequency, causing significant field-inhomogeneity-based image artifacts in many regions, especially in the frontal lobe and near the skull (8, 9). To solve this problem, a practical, six-offset, multi-acquisition method, combined with a single-acquisition z-spectrum, may be used (see Fig. 1). As a compromise between optimum signal-to-noise ratio (SNR) and the ability for some correction, this method can acquire high-SNR APT images with B0 inhomogeneity correction within an acceptable scanning time (a few minutes).
Fig. 1.
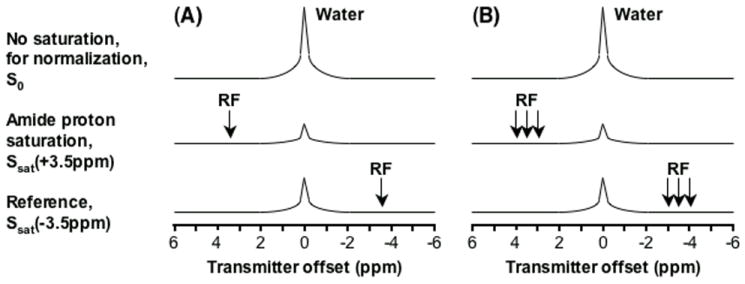
Schemes for APT-image acquisition. (A) Standard two-offset APT scan (+3.5 ppm for label, -3.5 ppm for reference). (B) Six-offset APT scan (±3, ±3.5, ±4 ppm). The effects of conventional MT and direct water saturation reduce the water signal intensities at all offsets (±3,±3.5, ±4 ppm), and the existence of APT causes an extra reduction around the offset of 3.5 ppm. (Reproduced from ref. 9 with permission from Wiley-Liss, Inc.)
Finally, it is important to develop an APT data processing approach for effective image assessment. This includes two steps: determination of the water-center-frequency shifts for each voxel, and B0 inhomogeneity correction for z-spectra or APT images. The flow chart for APT-image data processing is shown in Fig. 2.
Fig. 2.
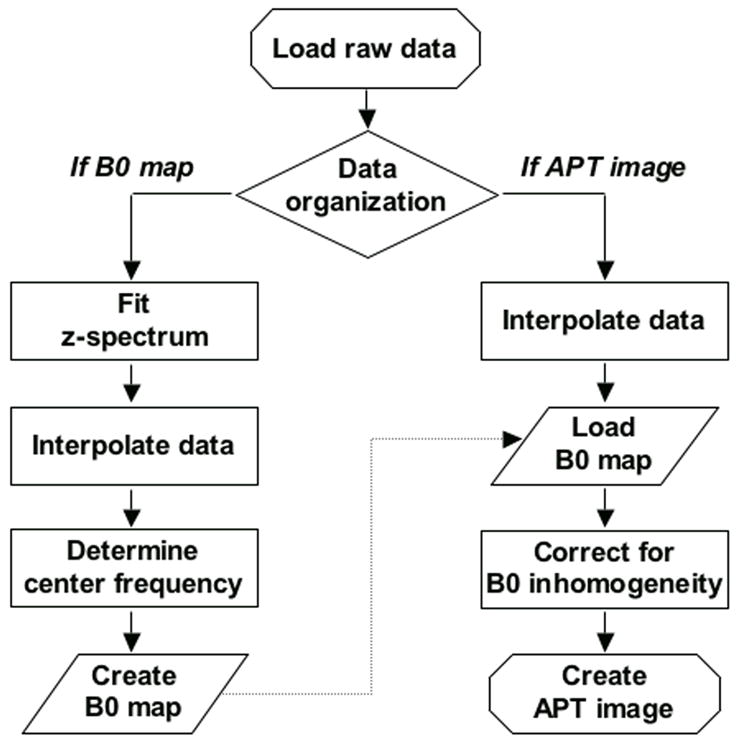
Flow chart of APT-image data processing. The procedure is divided into two steps: 1) the generation of a water-frequency shift map, and 2) the correction of APT-image data using the obtained shift map. Both steps are performed on a voxel-by-voxel basis.
3.1. Design of an APT Imaging Pulse Sequence
These instructions assume the use of a Philips 3T MRI scanner, together with a body coil for RF transmission and an eight-channel phased-array coil for reception. Adjustments should be made according to the capability and limitations of the scanner hardware, particularly the duty cycle of radiofrequency (RF) power amplifiers (see Note 4).
Add a low-power long block pulse for proton saturation (up to 4 μT and 500 ms). The weak continuous-wave (CW) RF saturation scheme has been widely used for various CEST imaging experiments. The RF saturation power and time are the most important pulse sequence parameters that must be optimized.
Use the turbo spin-echo (TSE) for imaging readout. A single slice is acquired (see Note 5). Single-shot acquisition should be used. Sensitivity encoding (SENSE) is used to reduce the TSE factor and specific absorption rate (SAR).
Add lipid suppression, such as selective partial inversion recovery (SPIR).
Adjust all pulse sequence parameters. This should also include the repetition time (TR), echo time (TE), field of view (FOV), image matrix, and slice thickness. The SAR should be kept below the U.S. Food and Drug Administration (FDA) limit for head (3.0 W/kg).
3.2. Test on Phantoms
Buy a bottle of egg whites from the supermarket. Shake the phantom for five minutes and put the phantom into the center of the magnet. To increase the loading, add several large bottles of water around the phantom.
Acquire localizer images and a SENSE reference scan.
Perform a z-spectrum experiment using an offset range of 8 to -8 ppm with an interval of 0.5 ppm (one signal average). One image without RF saturation is acquired for normalization. To protect the scanner, conservative MRI parameters should be used for the first test: saturation time 100 ms, saturation power 1 μT, and TR 10 s. Higher-order (up to second order) volume shimming should be used.
Improve image quality and remove any image artifacts by modifying the imaging parameters, including the SENSE factor.
Repeat the z-spectrum experiments using the other pulse sequence parameters: saturation time 500 ms, saturation power 2-4 μT, and TR 3 s.
Examine the z-spectrum characteristics from a small region of interest (ROI). An ideal z-spectrum should be very smooth across all frequency offsets, with the lowest signal intensity at the water frequency, but the z-spectrum is generally shifted by the B0 inhomogeneity.
Perform the B0 inhomogeneity correction on a voxel-by-voxel basis for each z-spectrum experiment (see Section 3.6).
Plot the z-spectrum and MTRasym-spectrum (see Note 2) for a small ROI, and compare the curves at the different power levels. The maximal APT effect at 3.5 ppm is about 5-8%.
3.3. Optimization on Healthy Normal Subjects
Complete the initial screening and the consent form.
Put the subject into the center of the magnet. The head of the subject should be restrained to avoid motion artifacts. This is accomplished using a very comfortable foam head holder and a headband.
Acquire localizer images and a SENSE reference scan.
Perform a z-spectrum experiment using an offset range of 8 to -8 ppm with an interval of 0.5 ppm (saturation time 500 ms, power 1 μT, FOV 212×212 mm2, matrix 128×64, slice thickness 5 mm, TR 3 s, TE 11 ms, one signal average). One image without RF saturation is acquired for normalization. Higher-order (up to second order) volume shimming should be used.
Improve image quality and remove any image artifacts by modifying the imaging parameters, including the SENSE factor.
Repeat the z-spectrum experiments with higher saturation power levels at 2-4 μT.
Move the subject out of the magnet.
Examine the z-spectrum characteristics from a small ROI. An ideal z-spectrum should be very smooth across all frequency offsets with the lowest signal intensity at the water frequency, but the z-spectrum may be shifted by the B0 inhomogeneity.
Perform the B0 inhomogeneity correction on a voxel-by-voxel basis for each z-spectrum experiment (see Section 3.6).
Plot the z-spectra and MTRasym-spectra (see Note 2) from gray matter, white matter, and cerebrospinal fluid (CSF). Compare the curves at the different power levels. Determine a characteristic RF saturation power at which MTRasym(3.5ppm) is approximately zero for the whole brain (see Note 6).
3.4. Two- and Six-Offset Acquisition Scheme for APT Images
These instructions assume that the APT pulse sequence has been optimized with the z-spectrum experiments (see Section 3.3). The characteristic RF saturation power is 3 μT. The other imaging parameters are: saturation time 500 ms, TR 3 s, FOV 212×212 mm2, matrix 128×64, slice thickness 5 mm, TR 3 s, and TE 11 ms.
Acquire the saturation images at +3.5 ppm twice (eight signal averages).
Determine the signal intensity from a small ROI in one saturation image. Subtract the two saturation images and determine the noise from the same ROI in the difference image. Calculate the SNR. The SNR should be 100:1 or better to see the APT effect that is a few percent of the water intensity.
Perform an APT-image experiment using a standard two-offset acquisition scheme (+3.5 ppm for label and -3.5 ppm for reference) and eight signal averages. One image without RF saturation is acquired for normalization.
Calculate the APT image (see Note 2) and examine the effect of the B0 inhomogeneity on the APT images.
Acquire APT-image data using six frequency offsets (±3, ±3.5, ±4 ppm) and eight signal averages. One image without RF saturation is acquired for normalization. In this practical acquisition scheme, four extra offsets around ±3.5 ppm are acquired in the high SNR scan, and it is possible to correct for the artifacts on the APT image caused by B0 inhomogeneity.
Acquire a z-spectrum (33 offsets from 8 to -8 ppm with intervals of 0.5 ppm, one average) as an extra scan (see Note 7). One image without RF saturation is acquired for normalization.
Calculate the corrected APT image (see Section 3.6). The APT image is shown in color. For the healthy subject, a typical APT image is quite homogenous over the whole slice.
3.5. Scanning on Patients
The optimized z-spectrum and APT-image scans are added to the conventional MRI protocol. The total scan time for each subject should be limited to one hour to maximize patient comfort.
Complete the initial screening and the consent form.
Put the subject into the center of the magnet. The head of the subject should be restrained to avoid motion artifacts. This is accomplished using standard configurations with a very comfortable foam head holder and a headband.
Acquire localizer images and a SENSE reference scan.
Acquire T2-weighted images using the dual-echo TSE sequence.
Acquire fluid attenuated inversion recovery (FLAIR) images.
Identify the location of the lesion from the T2-weighted and FLAIR images, and put the APT-image slice(s) on the lesion.
Acquire the six-offset APT-image scan.
Turn off the prescan to avoid changes in shim and frequency offset settings.
Acquire the z-spectrum scan. The same slice localization as that used for the APT-image scan should be used.
Acquire the T1-weighted images using the magnetization-prepared rapid gradient-echo (MPRAGE) sequence.
Inject the gadolinium contrast agent into the patient (0.2 mL/kg, i.v.).
Acquire the gadolinium-enhanced T1-weighted images using the MPRAGE sequence.
Move the subject out of the magnet.
Perform the B0 inhomogeneity correction on a voxel-by-voxel basis for each z-spectrum experiment (see Section 3.6). Plot the z-spectra and MTRasym-spectra (see Note 2) from the lesion and other ROIs.
Calculate the corrected APT image (see Section 3.6). The APT image is displayed in color. An example of the results produced is shown in Fig. 3.
Fig. 3.
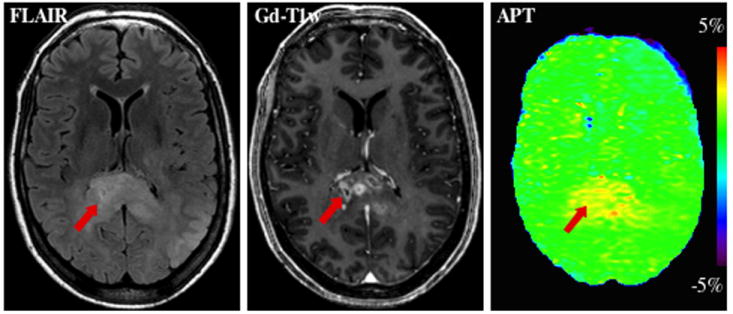
MR images of a patient with an anaplastic astrocytoma. Elevated APT signal can be seen in Gd-enhanced tumor core (red arrow), potentially providing unique information about the presence and grade of brain tumors, without the injection of exogenous contrast agents. The hyperintense APT area is comparable in size to the lesion identified on FLAIR, but larger than that on the Gd-T1w image. (Reproduced from ref. 9 with permission from Wiley-Liss, Inc.)
3.6. Data Processing
These instructions assume the use of IDL. They are easily adaptable to other languages, such as MATLAB.
Fit the full z-spectrum through all 33 offsets using a 12th-order polynomial (the maximum order available with IDL) on a voxel-by-voxel basis (see Fig. 2).
Interpolate the fitted curve using an offset resolution of 1 Hz (2049 points).
The actual water resonance should be at the frequency with the lowest signal intensity. The deviation of the water frequency in Hz forms a map of water-center-frequency shifts.
To correct for the field inhomogeneity effects on z-spectra, the measured z-spectrum for each voxel is interpolated to 2049 points and shifted along the direction of the offset axis to correspond to 0 ppm at its lowest intensity.
The realigned z-spectra are interpolated back to 33 points for visual purposes.
Plot the z-spectrum and MTRasym-spectrum for a small ROI. The outermost points of ±7.5 and ±8 ppm are excluded in the display.
To correct for the field inhomogeneity effects on APT images (see Fig. 2), the acquired APT data for offsets +4, +3.5, and +3 ppm (or +512, +448, +384 Hz) for each voxel are interpolated to 257 points over the range from +4.5 to +2.5 ppm (or +576, +575, …, +320 Hz).
Realign the APT-image data using the fitted z-spectrum central frequency shift for the same voxel.
Perform the B0 inhomogeneity correction on a voxel-by-voxel basis for the negative-offset data (-3, -3.5, -4 ppm).
Calculate the corrected APT image using the shift-corrected data at the two offsets ±3.5 ppm.
The calculated APT image is thresholded based on the signal intensity of the S0 image to remove voxels outside the brain, and displayed in color.
Acknowledgments
The author thanks the team at the F.M. Kirby Research Center for Functional Brain Imaging for helpful discussions and technical assistance. This work was supported in part by grants from NIH (EB002634, EB005252, EB009112, EB009731, and RR015241), and the Dana Foundation.
Footnotes
Many polymers, such as poly-L-lysine, dendrimers, and histone (12, 13), have a strong APT effect. However, these chemicals are very expensive; thus, they are not cost-effective for making large phantoms for use in human scanners. Egg whites in a bottle (protein 10%) from the supermarket are a straightforward protein phantom for APT imaging studies. In addition, some fruits, such as cantaloupe, are very useful phantoms with which to test imaging pulse sequences. Using a cantaloupe, up to about 25% of the CEST effect can be observed around a 1-2 ppm offset, which is due to various sugars in fruits.
In MT-type imaging (1, 2), water saturation is often measured as a function of transmitter frequency, producing the “z-spectra (14).” Such spectra are dominated by large direct water saturation around the water frequency at about 4.7 ppm and other saturation effects, such as conventional MT based on semi-solid tissue structures. The CEST effect is generally identified by asymmetry analysis with respect to this water signal (3), which is generally assigned to a reference frequency of 0 ppm.
Under higher-order slice shimming, as seen in the shift in the center of the z-spectrum, the B0 inhomogeneity is typically less than 20 Hz over most of the slice, but it could be as large as 60-80 Hz in the sinus and ear areas. A small B0 inhomogeneity can easily cause a few percentage point change in the MT asymmetry data, thus resulting in large artifacts on APT images.
The magnitude of the APT effect increases exponentially with the RF saturation time, and several seconds of saturation time are required to maximize the measurements. The use of weak CW saturation pulses is feasible on animal MRI scanners. It may also be feasible on human MRI systems if a transmit/receive (T/R) head coil is used. When body coil excitation with a phased-array coil receive is used, the RF saturation pulse is restricted. In the Philips 3T MRI scanner with the tube amplifier, as used in this study, the saturation pulse duration is limited to 500 ms.
The clinical application of APT imaging to data remains limited to single-slice. Multi-slice or whole-brain imaging would be feasible, but many technical issues, such as long scan times, APT contrast loss between slices, magnetic field inhomogeneity, and SAR must be resolved properly.
As the applied RF saturation power increases, MTRasym(3.5ppm) for gray matter and white matter increases initially, and then decreases back to zero, while that for CSF remains at around zero. At the characteristic RF saturation power, MTRasym(3.5ppm) is approximately zero for both normal brain tissue and CSF. In contrast, MTRasym(3.5ppm) would be hyperintensive in tumors and hypointensive in stroke regions.
If the z-spectrum measurement has the same saturation parameters as the APT-image scan, the z-spectrum data can be used for both identification of the APT effect and fitting of the B0 inhomogeneity map. If the z-spectrum is used to fit the B0 inhomogeneity map only, lower saturation power and shorter saturation time may be used. Such a scan, called water saturation shift referencing (WASSR) (15), would provide a narrower z-spectrum; thus, the B0 inhomogeneity map can be fitted more accurately.
References
- 1.Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn Reson Med. 1989;10:135–44. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- 2.Henkelman RM, Stanisz GJ, Graham SJ. Magnetization transfer in MRI: a review. NMR Biomed. 2001;14:57–64. doi: 10.1002/nbm.683. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Med. 2003;9:1085–90. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 4.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 5.Sun PZ, Zhou J, Sun W, Huang J, van Zijl PCM. Delineating the Boundary Between the Ischemic Penumbra and Regions of Oligaemia Using pH-weighted Magnetic Resonance Imaging (pHWI) J Cereb Blood Flow Metab. 2007;27:1129–36. doi: 10.1038/sj.jcbfm.9600424. [DOI] [PubMed] [Google Scholar]
- 6.Jokivarsi KT, Grohn HI, Grohn OH, Kauppinen RA. Proton transfer ratio, lactate, and intracellular pH in acute cerebral ischemia. Magn Reson Med. 2007;57:647–53. doi: 10.1002/mrm.21181. [DOI] [PubMed] [Google Scholar]
- 7.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50:1120–6. doi: 10.1002/mrm.10651. [DOI] [PubMed] [Google Scholar]
- 8.Jones CK, Schlosser MJ, van Zijl PC, Pomper MG, Golay X, Zhou J. Amide proton transfer imaging of human brain tumors at 3T. Magn Reson Med. 2006;56:585–92. doi: 10.1002/mrm.20989. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60:842–9. doi: 10.1002/mrm.21712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobbs SK, Shi G, Homer R, Harsh G, Altlas SW, Bednarski MD. Magnetic resonance imaging-guided proteomics of human glioblastoma multiforme. J Magn Reson Imag. 2003;18:530–6. doi: 10.1002/jmri.10395. [DOI] [PubMed] [Google Scholar]
- 11.Howe FA, Barton SJ, Cudlip SA, et al. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–32. doi: 10.1002/mrm.10367. [DOI] [PubMed] [Google Scholar]
- 12.Goffeney N, Bulte JWM, Duyn J, Bryant LH, van Zijl PCM. Sensitive NMR detection of cationic-polymer-based gene delivery systems using saturation transfer via proton exchange. J Am Chem Soc. 2001;123:8628–9. doi: 10.1021/ja0158455. [DOI] [PubMed] [Google Scholar]
- 13.McMahon MT, Gilad AA, Zhou J, Sun PZ, Bulte JWM, van Zijl PCM. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly-L-lysine and a starburst dendrimer. Magn Reson Med. 2006;55:836–47. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant RG. The dynamics of water-protein interactions. Annu Rev Biophys Biomol Struct. 1996;25:29–53. doi: 10.1146/annurev.bb.25.060196.000333. [DOI] [PubMed] [Google Scholar]
- 15.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PCM. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61:1441–50. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]


