Abstract
There is a critical need to identify molecular markers that can reliably aid in stratifying esophageal adenocarcinoma (EAC) risk in patients with Barrett's esophagus. MicroRNAs (miRNA/miR) are one such class of biomolecules. In the present cross-sectional study, we characterized miRNA alterations in progressive stages of neoplastic development, i.e., metaplasia–dysplasia–adenocarcinoma, with an aim to identify candidate miRNAs potentially associated with progression. Using next generation sequencing (NGS) as an agnostic discovery platform, followed by quantitative real-time PCR (qPCR) validation in a total of 20 EACs, we identified 26 miRNAs that are highly and frequently deregulated in EACs (≥4-fold in >50% of cases) when compared to paired normal esophageal squamous (nSQ) tissue. We then assessed the 26 EAC-derived miRNAs in laser microdissected biopsy pairs of Barrett's metaplasia (BM)/nSQ (n = 15), and high-grade dysplasia (HGD)/nSQ (n = 14) by qPCR, to map the timing of deregulation during progression from BM to HGD and to EAC. We found that 23 of the 26 candidate miRNAs were deregulated at the earliest step, BM, and therefore noninformative as molecular markers of progression. Two miRNAs, miR-31 and –31*, however, showed frequent downregulation only in HGD and EAC cases suggesting association with transition from BM to HGD. A third miRNA, miR-375, showed marked downregulation exclusively in EACs and in none of the BM or HGD lesions, suggesting its association with progression to invasive carcinoma. Taken together, we propose miR-31 and –375 as novel candidate microRNAs specifically associated with early- and late-stage malignant progression, respectively, in Barrett's esophagus.
INTRODUCTION
Despite advances in multimodality therapy over the last three decades, esophageal adenocarcinoma (EAC) carries a higher annual ratio of mortality-to-incidence than any other malignancy, including hepatobiliary and pancreatic cancer. Over the same period, the incidence of EAC has risen at rates far exceeding all other cancers, for reasons that remain largely unclear (Jemal et al., 2010; Pohl et al., 2010). Most EACs are initiated in metaplastic esophageal epithelium (Barrett's esophagus) where they develop progressively through dysplastic stages into invasive adenocarcinomas, the “M-D-A” sequence (Prasad et al., 2010; Reid et al., 2010). Although Barrett's esophagus is the only known precursor of EAC, the rate of neoplastic progression is only 1 per 200 patient-years (Prasad et al., 2010; Reid et al., 2010). As a consequence, it is extremely challenging to distinguish the small number of Barrett's individuals who will progress to EAC, from the majority who will not. The current clinical standard for assessing cancer risk in Barrett's patients relies on interval endoscopic biopsies and histopathologic grading for dysplasia (Prasad et al., 2010; Reid et al., 2010). This approach has not demonstrably decreased mortality and is hampered by well-recognized limitations including sampling error and intra- and inter-observer diagnostic variability (Prasad et al., 2010; Reid et al., 2010). There is therefore an ongoing need to develop molecular markers for stratifying cancer risk in Barrett's esophagus.
MicroRNAs (miRNA/miR) are a class of small noncoding regulatory RNAs that play a major role in the pathogenesis of various cancers, and are being pursued as both predictive and prognostic biomarkers. However, the utility of miRNAs as classifiers of cancer risk in the Barrett's disease model remains largely unknown. In the present cross-sectional study, we employed next generation sequencing (NGS) and laser capture micro-dissection (LCM) techniques, together with a rigorous analytical approach to identify reliable candidate miRNA markers associated with neoplastic progression in Barrett's esophagus. Based on our findings, we propose that the majority of the deregulated miRNAs in EACs are metaplasia-derived, and therefore unsuitable as markers of progression. However, we have identified two specific miRNAs, miR-31 and –375, as unique and promising markers of early- and late-stage neoplastic progression, respectively.
MATERIALS AND METHODS
Detailed methods for NGS-based miRNA detection and quantitation, LCM, and quantitative real-time PCR are provided in the Supporting Information Methods section.
Tissue Samples
Patient characteristics, number and type of lesions utilized in this study are summarized in Table 1. Paired, normal (uninvolved) esophageal squamous tissue (nSQ) from each of the Barrett's metaplasia (BM), high-grade dysplasia (HGD), and EAC cases was obtained a minimum of 3 cm proximal to the squamo-columnar junction, and a minimum of 3 cm away from the lesional biopsy at the time of endoscopy. In order to ensure preservation of high quality RNA, all tissue samples were frozen immediately following the collection and stored at –80°C. The majority of patients included in the study are Caucasian males (Table 1), which is in accordance with the well-recognized white male predominance in patients susceptible to Barrett's esophagus. To minimize the heterogeneity among our cancer cases, tumors of the esophago-gastric junction were excluded, and only intestinal-type EACs were included in this study. Additionally, Barrett's with low-grade dysplasia (LGD) lesions were not included in our study due to the nonreproducibility and marked inter-observer variability in the histopathological diagnosis of LGD (Prasad et al., 2010; Reid et al., 2010).
TABLE 1.
Patient Characteristics
| Barrett's metaplasia (BM) | High-grade dysplasia (HGD) | Esophageal adenocarcinoma (EAC) | |
|---|---|---|---|
| No. matched pairs of lesion/nSQ | 15 | 14 | 20 |
| Age at endoscopy (range) | 65 (18–86) | 69 (33–89) | 67 (45–83) |
| Male White cases | 13 | 10 | 18 |
| Female White cases | 1 | 4 | 2 |
| Female African American cases | 1 | 0 | 0 |
miRNA Expression Profiling in EACs Using Next Generation Sequencing
Global miRNA expression levels were assessed in a panel of paired EAC/nSQ biopsy tissues (n = 9 pairs) using NGS. The raw miRNA expression counts were normalized to the geometric mean expression counts of miR-103 and miR-191, two miRNAs that we identified as house-keeping miRNAs in the esophageal tissue, and were previously shown to be highly consistent in their expression across a wide variety of normal and tumor tissues (Peltier and Latham, 2008). A ≥4-fold variation in expression levels between EAC and paired nSQ tissues was considered significant.
Laser-Capture Microdissection and RNA Extraction from BM and HGD Lesions
Frozen tissue sections from paired biopsies of BM/nSQ (n = 15) and HGD/nSQ (n = 14) were laser microdissected to enrich for pure populations of BM, HGD, and nSQ epithelia. Total RNA, including the miRNA fraction, was extracted according to the Ambion RNAqueous Micro protocol (Applied Biosystems, Carlsbad, CA), including DNAse treatment to remove DNA contamination.
Quantitative Real-Time PCR of miRNAs
For quantitative real-time PCR (qPCR), 250 ng of total RNA from paired biopsies of EAC/nSQ (n = 11), laser microdissected BM/nSQ (n = 15), and HGD/nSQ (n = 14) was first reverse-transcribed using miRCURY locked nucleic acid (LNA) universal cDNA synthesis kit, followed PCR using miRCURY LNA Universal SYBR Green system with pre- or custom-designed LNA miRNA primer sets (Exiqon, Woburn, MA). Each qPCR reaction was carried out in triplicate, and miRNA expression levels were normalized to the geometric mean expression values of miR-103 and miR-191. A ≥4-fold variation in miRNA expression levels between lesion and paired nSQ tissue was considered significant.
Statistical Analysis
To determine which miRNAs are associated with progression from BM to HGD to EAC, the proportion of cases with ≥4-fold alteration in miRNA expression relative to paired nSQ tissues in each of the BM, HGD, and EAC cohorts were first estimated. Then, using a one-sided two-group exact proportional test, pairwise comparisons were performed to estimate for significant differences in deregulation frequencies for each of the miRNAs between BM vs. HGD, BM vs. EAC, and HGD vs. EAC groups. A P-value of <0.05 was considered significant. Survival analysis was performed between the patients with loss of both miR-31 and miR-375 and the patients with loss of one of the two miRs or no loss of any of the two miRs. Survival time was calculated in days. The Kaplan–Meier method was used to estimate the overall survival difference between the two groups. Cox proportional hazards model was also performed by adjusting age at diagnosis, gender, and stage. All variables of interest were tested for agreement with the proportional hazards assumption of the Cox model. Analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC) and R version 2.13.2. R package “survival” was employed for the survival analysis.
RESULTS AND DISCUSSION
Identifying miRNAs Deregulated in Esophageal Adenocarcinomas
The main objective of this study is to identify candidate miRNA markers associated with tumor progression in Barrett's esophagus. A primary requirement for such miRNA markers is that they accurately represent the malignant endpoint of disease progression. Accordingly, we first determined genome-wide alterations in miRNA expression levels in a discovery panel of 9 EAC/nSQ paired tissue samples using the NGS platform (Supporting Information Table S1). This yielded a total of 152 highly deregulated miRNAs (110 upand 42 downregulated) based on selection criteria of 4-fold alteration in 50% of EAC lesions as compared to paired nSQ tissue (Supporting Information Table S2). We then filtered these 152 miRs based on a minimum requirement of 500 raw NGS expression read counts in at least one member of the pair, anticipating a minimum qPCR threshold (Supporting Information Methods section). This identified 36 miRNAs that were carried forward for further validation in an independent set of 11 EAC/nSQ pairs tested by qPCR. Real-time assays for 3 of the 36 miRNAs did not pass established qPCR quality guidelines (Supporting Information Methods), and therefore were eliminated from further analysis. Of the remaining 33 miRs, 26 were confirmed to be highly altered (≥4-fold) in at least 50% of the combined NGS discovery and qPCR validation EAC cohorts (n = 20) and carried forward for analysis (Supporting Information Table S3).
The Majority of Highly Deregulated miRNAs in EAC Lesions are Metaplasia-Derived
Next, we mapped the timing along the M–D–A progression sequence at which each of the 26 EAC derived miRNAs become deregulated. Accordingly, we characterized the expression status of the 26 miRNAs by qPCR in a panel of BM/nSQ (n = 15) and HGD/nSQ (n = 14) biopsy pairs. To accurately quantify for the levels of miRNAs in these early-stage lesions, pure cell populations of BM, HGD, and paired nSQ epithelial biopsies were first enriched by laser microdissection prior to qPCR analysis. Given that only a small number of Barrett's patients develop cancer (Prasad et al., 2010; Reid et al., 2010), we were particularly interested in miRNAs that are altered in a significantly lower proportion of BM cases compared to HGD and/or EAC, and might therefore be associated with neoplastic progression. However, for 23 of the 26 EAC derived miRs, deregulation clearly mapped to the BM stage (Table 2), with these 23 miRs demonstrating 4-fold or greater change from normal squamous mucosa in 50%–100% of all BM samples (proportions that were not significantly different from HGD or from EAC). To address the possibility that our 4-fold cutoff was too stringent, we also tested a relaxed 2-fold cutoff and observed that stage-wise comparisons were not significantly altered (Supporting Information Table S4). Overall, these findings strongly suggest that the majority of miRNAs commonly deregulated in HGD and EAC lesions represent a metaplasia-derived signal and are not suitable as candidate markers of neoplastic progression.
TABLE 2.
Comparison of miRNA Deregulation Frequencies in Barrett's Associated Premalignant and Malignant Lesions
| Proportion of cases with ≥4-fold alteration in expression within each group |
Two-group proportion test P-value |
|||||
|---|---|---|---|---|---|---|
| miRNA | % BM (n = 15) | %HGD (n = 14) | %EAC (n = 20) | BM vs. HGD | BM vs. EAC | HGD vs. EAC |
| Up-regulated | ||||||
| miR-196b | 60.00 | 92.86 | 70.00 | 0.049 | 0.397 | 0.986 |
| miR-424a | 66.67 | 57.14 | 70.00 | 0.819 | 0.560 | 0.340 |
| miR-196aa | 66.67 | 92.86 | 90.00 | 0.099 | 0.101 | 0.810 |
| miR-146aa | 73.33 | 57.14 | 65.00 | 0.905 | 0.813 | 0.456 |
| miR-199b-5pa | 80.00 | 78.57 | 80.00 | 0.709 | 0.660 | 0.623 |
| miR-145a | 86.67 | 85.71 | 70.00 | 0.728 | 0.945 | 0.933 |
| miR-199a-5pa | 86.67 | 100.00 | 95.00 | 0.259 | 0.390 | 1.000 |
| miR-30a* | 93.33 | 71.43 | 50.00 | 0.983 | 1.000 | 0.947 |
| miR-30aa | 93.33 | 85.71 | 70.00 | 0 900 | 0.989 | 0.933 |
| miR-7a | 93.33 | 85.71 | 80.00 | 0 900 | 0.952 | 0.810 |
| miR-143a | 93.33 | 71.43 | 85.00 | 0 983 | 0.908 | 0.295 |
| miR-192a | 100.00 | 100.00 | 95.00 | 1.000 | 1.000 | 1.000 |
| miR-194a | 100.00 | 100.00 | 100.00 | 1.000 | 1.000 | 1.000 |
| miR-215a | 100.00 | 100.00 | 100.00 | 1.000 | 1.000 | 1.000 |
| Down-regulated | ||||||
| miR-375a | 0.00 | 0.00 | 55.00 | 1.000 | <0.001 | <0.001 |
| miR-31a | 13.33 | 57.14 | 60.00 | 0.017 | 0.006 | 0.573 |
| miR-31* | 13.33 | 42.86 | 65.00 | 0.086 | 0.003 | 0.177 |
| miR-23ba | 46.67 | 50.00 | 55.00 | 0.576 | 0.442 | 0.524 |
| miR-99aa | 53.33 | 71.43 | 75.00 | 0.268 | 0.165 | 0.560 |
| miR-210a | 60.00 | 85.71 | 85.00 | 0.129 | 0.100 | 0.701 |
| miR-452 | 86.67 | 85.71 | 70.00 | 0.728 | 0.945 | 0.933 |
| miR-944 | 93.33 | 92.86 | 95.00 | 0.776 | 0.681 | 0.661 |
| miR-224a | 100.00 | 92.86 | 80.00 | 1.000 | 1.000 | 0.944 |
| miR-205a | 100.00 | 92.86 | 85.00 | 1.000 | 1.000 | 0.896 |
| miR-149a | 100.00 | 92.86 | 95.00 | 1.000 | 1.000 | 0.661 |
| miR-203a | 100.00 | 92.86 | 95.00 | 1.000 | 1.000 | 0.661 |
miRNA fold changes indicate the relative expression differences between lesions and paired nSQ tissue. Shown are the deregulation frequencies of the 26 miRNAs in BM, HGD, and EAC lesions. Of these, miR-31 was the only miRNA altered in a significantly lower proportion of BM cases when compared to HGD or EAC lesions. A similar trend in deregulation frequency was observed for its complementary miRNA, miR-31*, among the BM, HGD, and EAC cases. Down-regulation of miR-375 was restricted to EAC lesions. P-values <0.05 are indicated in bold-italic font.
Represents miRNAs that have been previously shown to be similarly altered in Barrett's associated lesions in at least one of the published studies (Bansal et al., 2011; Fassan et al., 2011; Feber et al., 2008; Kan et al., 2009; Maru et al., 2009; Wijnhoven et al., 2010; Yang et al., 2009). The 20 EAC cases include both the NGS discovery (n = 9), and qPCR validation (n = 11) EAC cohorts. miR-103 and –191 were used as endogenous controls for normalizing miRNA expression levels in each of the samples in NGS and qPCR assays as described in Supporting Information Methods.
miR-31 and miR-375 are Candidate Markers of Neoplastic Progression
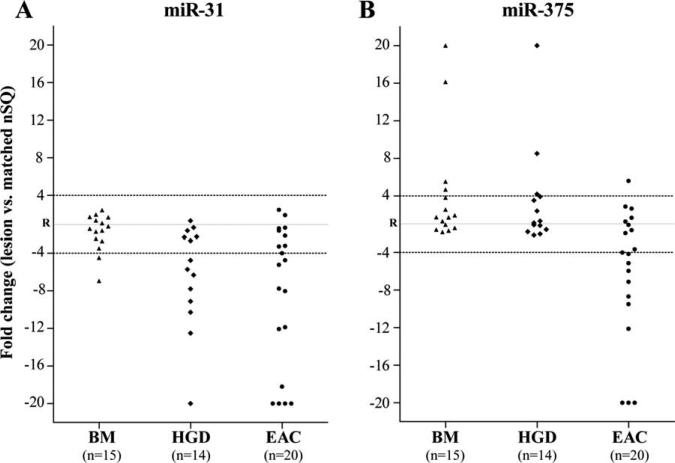
Comparison of BM vs. HGD and vs. EAC lesions identified two miRNAs, miR-31 and –375, as potential markers of neoplastic progression. We first noted a marked reduction (>4-fold) in miR-31 expression in >55% of HGD and EAC lesions (as compared to paired nSQ tissues); whereas only 13% of BM cases showed such a downregulation (P < 0.02) (Table 2, Fig. 1A). Moreover, the majority of lesions with miR-31 downregulation also showed a significant reduction in the expression of its complementary miRNA, miR-31*, thereby indicating a generalized deregulation in the precursor transcript from which these mature miRNAs are derived (Table 2). Interestingly, miR-31 has been previously suggested to have tumor suppressor function in epithelial malignancies (Valastyan et al., 2009; Creighton et al., 2010; Stuelten and Salomon, 2010). Moreover, miR-31, along with CDKN2A, is located on chromosome 9p21.3, a genomic locus whose loss of heterozygosity in Barrett's patients is associated with significantly increased cancer risk (Galipeau et al., 2007; Paulson et al., 2009). Of note, among the 15 BM cases utilized in the study, five cases showed at least a 2-fold reduction in miR-31 expression (Supporting Information Table S4), of which, three cases progressed to HGD or EAC at 1- to 2-years follow-up, while none of the remaining 10 BM cases with normal expression of miR-31 showed disease progression (data not shown). These findings suggest that loss of miR-31 may play an important functional role in the metaplasia to dysplasia transition, and that loss of miR-31 may have potential value as an early molecular marker of dysplastic progression.
Figure 1.
Alterations in miR-31 and –375 expression levels in Barrett's carcinogenesis. MiRNA expression levels in each of the BM, HGD, and EAC lesions were shown as linear fold changes relative to respective paired nSQ tissues. The solid gray reference line in the middle of the Y-axis, represented by the letter “R,” corresponds to a miRNA expression ratio value of 1.0 between lesion and paired nSQ tissues, thus indicating no change. The horizontal dotted lines at +4 and –4 correspond to a 4-fold up- and downregulation in miRNA expression levels between lesion and paired nSQ tissues, respectively. For simplicity, lesions with greater than 20-fold up- and downregulation in miRNA expression were plotted on the Y-axis at 20 and –20, respectively. miRNA expression levels in BM and HGD cases were determined using qPCR. For EACs, data derived from the NGS discovery set (n = 9), and qPCR validation cohort (n = 11) were combined in the figure. Both miR-103 and –191 were used as endogenous controls for normalizing miRNA expression levels in each of the samples in NGS and qPCR assays (described in Supporting Information Methods section).
We next noted that downregulation of miR-375 was exclusively and specifically a feature of the EAC cohort (Table 2, Fig. 1B). Whereas >50% of the EACs showed a > 4-fold reduction in miR-375 expression, none of the BM or HGD lesions exhibited a comparable loss (Table 2, Fig. 1B). miR-375 has been shown to have functional activity in suppressing growth of gastroesophageal malignant cells (Ding et al., 2010; Liu et al., 2010; Li et al., 2011), and reduced expression of miR-375 in EACs has been associated with poor prognosis (Mathe et al., 2009; Nguyen et al., 2010). Our finding that loss of miR-375 occurs exclusively in EAC provides additional evidence supporting an important role for miR-375 late in EAC tumorigenesis, and as a potential molecular marker of late steps in neoplastic transformation.
Down-Regulation of miR-31 and miR-375 is Associated with Poor Prognosis in EAC Patients
In addition to having an association with malignant progression (Table 2), we investigated whether reduced expression of miR-31 and –375 also correlates with poor survival in patients with EAC. Survival data were available for 19 of the 20 EAC cases included in our analysis (one case was lost to clinical follow-up). Basic clinicopathologic characteristics including, age, gender, stage at diagnosis, and survival interval were obtained from the medical record. Survival analysis showed a significantly (P < 0.0002) poorer median survival for combined downregulation of miR-31 and –375 in EACs (1,028 days, n = 11), compared to cases with downregulation of only one or neither of the two miRs, (1,298 days, n = 8), (Supporting Information Table S5). Survival difference was independent of age, stage, and gender by Cox proportional hazards model, thus suggesting the potential value of miR-31 and –375 as prognostic classifiers in this disease.
There remains a critical need for developing molecular markers to stratify cancer risk in Barrett's esophagus patients. The ideal biomarker for clinical decision-making should: (1) first become positive at that stage of a disease process for which clinical intervention is most meaningful; and (2) remain positive through subsequent stages for purposes of surveillance. In the esophageal cancer progression model, HGD is arguably the most actionable stage, since both Barrett's esophagus and LGD carry too low a risk of progression to warrant treatment (~0.5% and ~1% per patient year, respectively), while EAC is generally refractory to treatment once diagnosed (~85% mortality) (Prasad et al., 2010; Reid et al., 2010). Accordingly, a marker that predicts progression to HGD in Barrett's esophagus would be of high value in identifying those who need close surveillance, while a marker that predicts progression from dysplasia to cancer would be useful in formulating rational therapeutic interventions in patients with HGD. In addition, such markers are more likely to play a fundamental role in tumor development, and therefore will be attractive targets for cancer chemoprevention.
Although prior reports have examined miRNA alterations in the Barrett's cancer model (Feber et al., 2008; Yang et al., 2009; Wijnhoven et al., 2010; Bansal et al., 2011), only a few studies have proposed specific miRNAs as indicators of neoplastic progression (Kan et al., 2009; Maru et al., 2009; Fassan et al., 2011). For example, Maru et al. proposed miR-196a as a progression marker based on their observation of an incremental upregulation in expression at successive stages of progression (Maru et al., 2009). Likewise, Kan et al. identified a modest but incremental upregulation (1.5- to 2.0-fold) in the expression of the polycistronic miR-106b, –25, and –93 complex between metaplasia and carcinoma lesions, and suggested their involvement in driving tumor evolution (Kan et al., 2009). More recently, Fassan et al., proposed specific miRNAs including, let-7c, miR-203, –205, –192, and –215, as being directly involved in neoplastic progression based on their consistent deregulation in the majority of metaplasia, dysplasia, and carcinoma lesions (Fassan et al., 2011). Similarly, our study detected these miRs as deregulated in esophageal neoplasia, but mapped the timing of that deregulation to the BM stage (Table 2). Hence, contrary to prior assertions (Kan et al., 2009; Maru et al., 2009; Fassan et al., 2011), these miRs appear to be metaplasia-derived rather than markers of progression. Further, we note that the modest incremental deregulation of some miRs previously attributed to progression (Kan et al., 2009; Maru et al., 2009) may be influenced by the increased neoplastic cellular content in more advanced lesions, an effect we obviated by use of LCM. Our data did not confirm previous reports of let-7c and miR-106b polycistron being commonly deregulated in EACs, with let-7c altered in only a minor proportion of EACs (Supporting Information Table S3), and miR-106b polycistron showing no significant alterations in EACs by NGS (data not shown). Although the present study is limited by the relatively small sample size, our primary intent was to identify a reliable set of candidate miRNA markers that can subsequently be incorporated into a much larger longitudinal study powered to ascertain progression risk in Barrett's esophagus patients. In conclusion, by adopting a stringent methodology to determine the timing of miRNA alterations in esophageal neoplasia, we have identified miR-31 and 375 as novel candidate miRNAs whose deregulation is specifically associated with multistep malignant progression in Barrett's esophagus.
Supplementary Material
ACKNOWLEDGMENTS
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the awarding organizations.
Supported by: NCRR-CTSA, Grant number: UL1 RR024989; NCI, Grant number: CA135692; NCI, Grant number: CA152756; NCI, Grant number: CA163060; NIDDK, Grant number: K23 DK068149; Case Comprehensive Cancer Center Core Grant, Grant number: NCI-P30 CA043703; American Society of Clinical Oncology Young Investigator Award; NCI K12 Clinical Oncology Research Career Development Award through the Case Comprehensive Cancer Center, Grant number: CA076917; K24 grant from the NIDDK, Grant number: DK002800; NCI K08 Mentored Clinical Scientist Research Career Development Award, Grant number: CA148980; Case Comprehensive Cancer Center Specialized Program of Research Excellence (SPORE) Career Development Program Scholar Award, Grant number: NCI-CA150964.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Bansal A, Lee IH, Hong X, Anand V, Mathur SC, Gaddam S, Rastogi A, Wani SB, Gupta N, Visvanathan M, Sharma P, Christenson LK. Feasibility of microRNAs as biomarkers for Barrett's esophagus progression: A pilot cross-sectional, phase 2 biomarker study. Am J Gastroenterol. 2011;106:1055–1063. doi: 10.1038/ajg.2011.37. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM, Anderson ML. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- Fassan M, Volinia S, Palatini J, Pizzi M, Baffa R, De Bernard M, Battaglia G, Parente P, Croce CM, Zaninotto G, Ancona E, Rugge M. MicroRNA expression profiling in human Barrett's carcinogenesis. Int J Cancer. 2011;129:1661–1670. doi: 10.1002/ijc.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipeau PC, Li X, Blount PL, Maley CC, Sanchez CA, Odze RD, Ayub K, Rabinovitch PS, Vaughan TL, Reid BJ. NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, Selaru FM, Hamilton JP, Yang J, Abraham JM, Mori Y, Meltzer SJ. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci. 2011;56:2849–2856. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, Rashid A, Luthra R. Micro-RNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, Altorki NK, Casson AG, Liu CG, Wang XW, Yanaihara N, Hagiwara N, Dannenberg AJ, Miyashita M, Croce CM, Harris CC. MicroRNA expression in squamous cell carcinoma and adeno-carcinoma of the esophagus: Associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen GH, Schetter AJ, Chou DB, Bowman ED, Zhao R, Hawkes JE, Mathe EA, Kumamoto K, Zhao Y, Budhu A, Hagiwara N, Wang XW, Miyashita M, Casson AG, Harris CC. Inflammatory and microRNA gene expression as prognostic classifier of Barrett's-associated esophageal adenocarcinoma. Clin Cancer Res. 2010;16:5824–5834. doi: 10.1158/1078-0432.CCR-10-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson TG, Maley CC, Li X, Li H, Sanchez CA, Chao DL, Odze RD, Vaughan TL, Blount PL, Reid BJ. Chromosomal instability and copy number alterations in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–3314. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl H, Sirovich B, Welch HG. Esophageal adenocarcinoma incidence: Are we reaching the peak? Cancer Epidemiol Bio-markers Prev. 2010;19:1468–1470. doi: 10.1158/1055-9965.EPI-10-0012. [DOI] [PubMed] [Google Scholar]
- Prasad GA, Bansal A, Sharma P, Wang KK. Predictors of progression in Barrett's esophagus: Current knowledge and future directions. Am J Gastroenterol. 2010;105:1490–1502. doi: 10.1038/ajg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett's oesophagus and oesophageal adenocarcinoma: Time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuelten CH, Salomon DS. miR-31 in cancer: location matters. Cell Cycle. 2010;9:4608–4609. doi: 10.4161/cc.9.23.13928. [DOI] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- Yang H, Gu J, Wang KK, Zhang W, Xing J, Chen Z, Ajani JA, Wu X. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.