Abstract
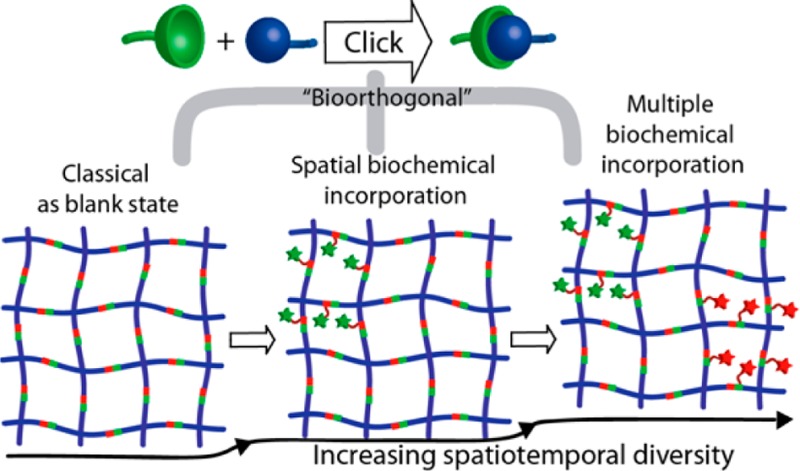
Over the past decade, bioorthogonal click chemistry has led the field of biomaterial science into a new era of diversity and complexity by its extremely selective, versatile, and biocompatible nature. In this viewpoint, we seek to emphasize recent endeavors of exploiting this versatile chemistry toward the development of poly(ethylene glycol) hydrogels as cell culture scaffolds. In these cell-laden materials, the orthogonality of these reactions has played an effective role in allowing the creation of diverse biochemical patterns in complex biological environments that provide new found opportunities for researchers to delineate and control cellular phenotypes more precisely than ever.
Since the realization that chemical conjugations are promising tools not only to interrogate biomolecules in their native environment,1,2 but also to build materials for biomedical applications,3−5 there has been a growing demand for engineering fast, selective, and high yielding organic reactions that can be conducted in a complex biological milieu at physiological conditions. Nonetheless, it is a daunting challenge to develop such distinctive reactions as, traditionally, most chemical reactions require longer reaction times, strict exclusion of water, protection of other competing functionalities, and vigorous heating/cooling.
A little over a decade ago, the notion of performing organic reactions under such restricted and controlled environments has, however, been challenged by the advent of an intriguing chemical strategy called “click chemistry”; the concept coined for chemical conjugations that are quick, selective, and high yielding.6,7 Up-to-date, there are a number of reactions (Figure 1) evolved to satisfy these criteria of efficiency in chemical conjugations.2−5,8 While most of these click reactions are convenient to perform in water and enable us to produce diverse and complex molecular architectures, executing these chemical reactions in complex biological media, for example, in the presence of cells, demand an even more stringent set of conditions: (i) the reagents used must be nontoxic to cells and (ii) fidelity of the reaction should not be affected by the plethora of endogenous functionalities that are present in cellular media. The pursuit for such characteristic reactions has led to the emergence of bioorthogonal click chemistry,2,8−10 an area that is rapidly expanding its applications, including labeling of biomolecules and imaging,11,12 cell surface modifications,13 protein engineering,14 and drug development.15 Toward these recent developments, bioorthogonal click reactions are now seeing widespread use in the engineering of biomaterials for cell culture applications.3,5,8,10,16,17 In this viewpoint, we focus on (i) the role of various bioorthogonal reactions in fabricating poly(ethylene glycol) (PEG) hydrogels as cell culture scaffold, for which we first seek to provide a brief introduction to hydrogels and their prospective cross-linking chemistries, and (ii) the exploitation of orthogonal functional groups to introduce spatiotemporally complex, and yet well-defined, biochemical cues in synthetic cell-laden hydrogels.
Figure 1.
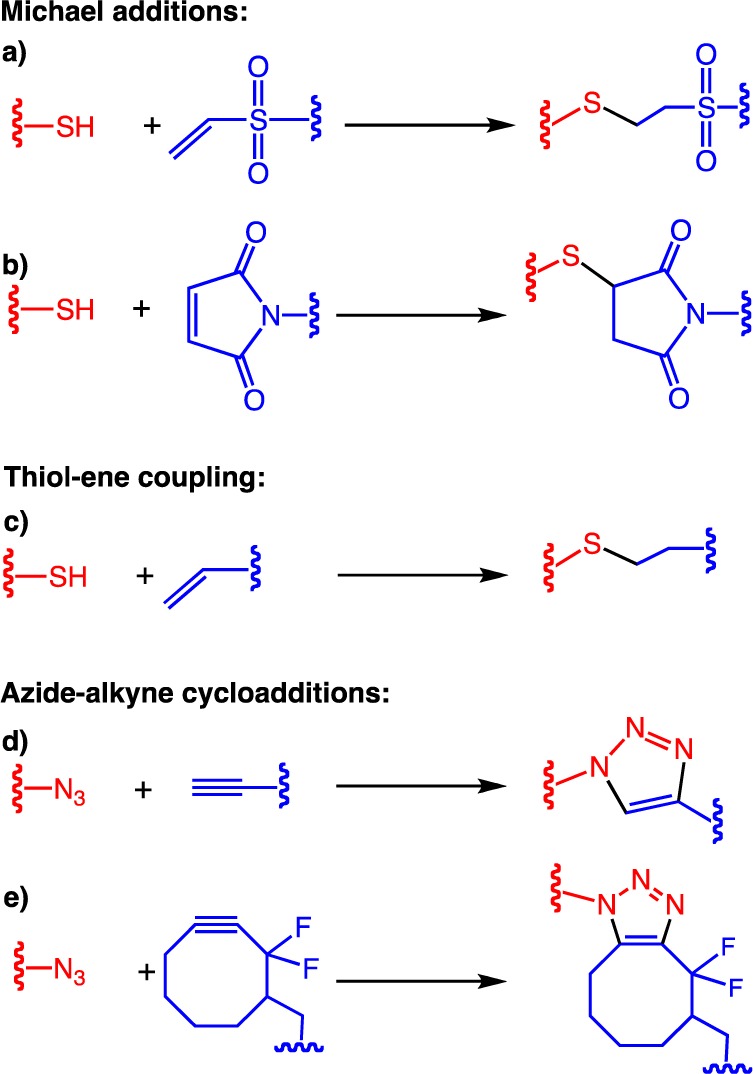
Examples of various click reactions that are commonly used in bioconjugation or hydrogel cross-linking: (a) copper-catalyzed Huisgen cycloaddition, (b) strain-promoted azide–alkyne cycloaddition (SPAAC), (c) base-catalyzed thiol-vinyl sulfone, (d) base-catalyzed thiol-maleimide Michael addition, (e) photoinitiated thiol–ene photocoupling.
As cell phenotype has been shown to vary greatly between cells that are cultured on 2D surfaces and in 3D matrices,18,19 fabrication of robust and biocompatible 3D material scaffolds that better mimic extracellular environment of natural tissues has become of growing interest to the fields of tissue engineering, regenerative medicine, and stem cell biology.20 Here, we focus on one very common 3D matrix, hydrogels or hydrated polymeric networks that have emerged as one of the promising synthetic extracellular matrices (ECM) for culturing cells in both 2D and 3D environments.21−23 Hydrogel networks are commonly fabricated from fully natural, synthetic polymers or a combination of both.24 Hydrogels of natural polymers (e.g., collagen and elastin)25,26 are inherently endowed with several fundamental biological features (e.g., cell adhesion moieties, proteolytic degradation sites, growth factor binding sites), but their batch-to-batch variation often fail to reproduce their mechanical and biochemical properties and, as a result, can limit the possibility to achieve matrices of well-defined properties.27,28 Alternatively, synthetic hydrogels enable one to precisely tune material properties, but the lack of biologically relevant chemistries necessitates the introduction of specific features found in natural ECMs in a highly controlled manner.24,27−30 Among the available synthetic repository, PEG hydrogels have been widely used to culture cells of different types in 2D and 3D architectures.24,27,31 Their hydrophilic nature renders PEG gels with elasticity, transport properties, and high water content, similar to many soft tissues, and the inherent minimal biological interactions of PEGs offer a blank environment that allows researchers not only to incorporate various biological signals, but to better understand how such signals influence host cells.
PEG polymer gelation can be achieved by ionic, physical, or covalent cross-linking of individual polymeric chains under aqueous conditions. However, ionic and physical (e.g., Pluronics)32 cross-linking leads to structurally weaker gels with a limited range of mechanical properties. To complement these approaches, covalent cross-linking provides hydrogels of higher and well-defined mechanical properties, but necessitates careful selection of cytocompatible, cross-linking chemistries. To date, covalently cross-linked hydrogels for cell encapsulation have been traditionally synthesized by radically initiated chain-growth polymerization of end-functionalized PEGs (e.g., PEG (di)acrylates and methacrylates); however, the indiscriminate choice of monomers by the growing chain results in a heterogeneous network structure, in which the cross-linking points are randomly distributed throughout the polydispersed kinetic chains,24 and potential damage to delicate primary cells33 and proteins.34 Alternatively, step-growth polymerization has emerged as an attractive method for constructing PEG hydrogels because (i) the method produces networks that are structurally uniform and yet mechanically superior and (ii) gelation can be achieved by reacting polymers containing any complementary reactive functional groups.24,29 In a standard setup, multifunctional molecular frames, (i.e., with a minimum of three functionalities) are reacted with bifunctional cross-linkers in a stoichiometric ratio to produce step-growth hydrogels (Figure 2), and to achieve hydrogels of different cross-linking densities, any of these molecular systems can be formulated from PEG-derived polymers of varying molecular weight and functionality. While step-growth polymerization paved the way for simplified procedures to achieve highly organized network structures, mild and cytocompatible cross-linking chemistries that enable gelation without compromising their fidelity and rate at physiological conditions, are the critical factors for hydrogels intended for cell delivery and regenerative medicine applications. In such stringent circumstances, bioorthogonal click chemistry reactions have emerged as superior and versatile chemical tools to construct hydrogels for studies involving cell encapsulation and culture in 3D.
Figure 2.
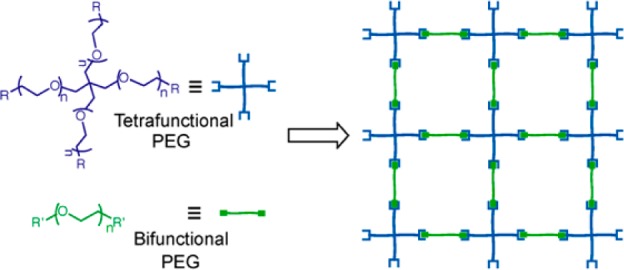
(left) Structures of multiarm and linear PEG precursors and (right) schematic of an idealistic step-growth hydrogel. The molecular weight of the precursors, their functionality, and their stoichiometric ratio can all influence the final network cross-linking density and ultimate material properties, including equilibrium water content, elasticity, and diffusion coefficients.
Among the bioorthogonal reaction tools, Michael additions (Figure 1a,b) have been broadly exploited as cross-linking modes for developing step-growth hydrogels, due to their virtue of simplicity, milder reaction conditions, and wider availability of functional precursors.3,8 Typically Michael-type addition involves a base-catalyzed addition of a Michael donor (e.g., thiols and amines) to an electrophilic carbon–carbon double bond conjugated with a carbonyl group, also called Michael acceptors. While a wide variety of Michael acceptors including acrylates, acrylamides, vinyl sulfones, and maleimides are investigated for hydrogel formulations, thiol-based Michael donors are largely utilized due to their higher nucleophilicity and selectivity at physiological pH and temperature.
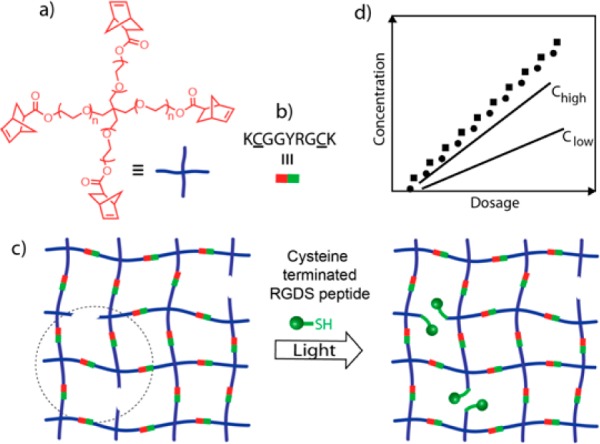
Hubbell and co-workers were the first to construct step-growth hydrogels using Michael additions as cross-linking chemistries to create peptide-functionalized biomaterial matrices.35 While their initial fabrications were based on acrylates, later they shifted their interest to more hydrolytically stable vinyl sulfones, especially for cell culture systems,36−39 in which thiol reactive vinyl groups were cleverly exploited as handles to install any cysteine-containing peptides, especially those that mimic ECM proteins, without the need for any postsynthetic modifications. In one such case, 4-armed tetravinyl sulfones were cross-linked, using cysteine-flanked matrix metalloproteinase (MMP) degradable peptides, to create cellularly remodeled gels and simultaneously introduced integrin binding, pendant peptide sequences (Figure 3).36,38 This pioneering work taught the field new strategies to create synthetic ECM analogs through peptide click reactions. While the base-catalyzed Michael reactions were managed by the addition of buffering agents, such as triethanolamine (TEA) or HEPES,39 these basic buffers are toxic to certain cell types (e.g., cells in ovarian follicles, pancreatic islets).40 However, a recent study by Garcia and co-workers on a set of hydrogels formulated from different Michael acceptors (e.g., acrylate, vinyl sulfone, maleimide) revealed that maleimide-based hydrogel formation require 2 orders of magnitude lower TEA as compared to its other counterparts, thereby significantly improving postencapsulation cell survival.41
Figure 3.

Schematic of a Michael addition driven step-growth hydrogel formed using thiol-reactive 4-arm PEG tetravinyl sulfone, cysteine-flanked MMP degradable peptides (↓ shows cleavage site), and simultaneous tethering of cysteine containing RGDS peptides.
In contrast to Michael additions, thiol–ene reactions (Figure 1c) are a radically mediated step growth polymerization, which requires creation of initial radicals either thermally or photochemically. However, photochemically driven reactions provide additional benefits when fabricating tissue culture matrices, because of their (i) lack of oxygen inhibition and rapid reaction rate at low initiating radical doses and (ii) the ability to spatiotemporally control the chemistry, thereby allowing site-specific incorporation of various biochemical or biomechanical cues.5,10,42 Our group has devised photoinitiated thiol–ene based fabrication of step-growth hydrogels employing 4-armed PEG tetra-norbornene and dicysteine-terminated (e.g., MMP and chymotrypsin cleavable) degradable peptides (Figure 4).43 This stepwise network can be formed in seconds to minutes using a water-soluble photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), and 365–420 nm single photon light or 720 nm multiphoton light, all in the presence of cells.17
Figure 4.
Thiol–ene hydrogel chemistry: (a) structure of 4-arm PEG tetranorbornene; (b) dicysteine-terminated chymotrypsin cleavable peptide; (c) schematic of spatial photopatterning throughout hydrogel networks created via thiol–ene by off-stoichiometrically reacting hydrogel precursors; (d) predictable relationship between photopatterning concentration and dosage of exposed light (● = constant intensity, varied exposure; ■ = constant exposure, varied intensity). The graph qualitatively shows that the extent of photopatterning can be varied by the alteration of light dosage, which in turn can be varied by exposure time/intensity. The graph also depicts the effect of photoinitiator concentration (Clow–Chigh) on photopatterning.
While supporting the facile incorporation of cysteine-containing peptides into PEG hydrogels, that is, similar to Michael additions, off-stoichiometrically performed photoinitiated thiol–ene polymerization affords additional opportunities for the precise and site-specific incorporation of peptide sequences or even thiolated proteins (Figure 4c), such as cell adhesion sites to control motility or inclusion of cytokines for regulating intracellular signaling, enabling researchers to dictate important cellular functions spatiotemporally. For example, when cell-adhesive RGDS peptides are photopatterned at specific locations in cell-laden gels (postencapsulation), cells residing in regions of high levels of RGDS exhibit a spread morphology and proliferate faster, while others in nonpatterned regions or at low concentrations remain spherical. The extent of such spreading is highly dependent on the local density of the patterned adhesive cues for many cell types.43 Furthermore, a simple variation of time of exposure, light intensity, initiator concentration and stoichiometric ratio of reactive functional groups offered exquisite control over the physical properties of gel network (e.g., stiffness) and concentration of patterned functionalities (Figure 4d), rendering this hydrogel system a simple yet powerful synthetic extra cellular matrix mimic.
Of all the click reactions, copper-driven azide–alkyne cycloaddition (Figure 1d) was not only the first one to set the standard for click reactions, but it has also emerged as one of the best utilized in both material and biomaterial sciences.6,44,45 Although the use of copper as a catalyst was critical for the revolutionary achievement of this cycloaddition reaction, the toxicity of copper has hampered its utility in cellular applications. Recently, Bertozzi and co-workers overcome this limitation upon employing strained alkynes: cyclooctynes that rapidly react with azides without the need for copper catalyst (Figure 1e).46 The nontoxic, copper free nature has raised this strain-promoted azide–alkyne cycloaddition (SPAAC) as one of the top choices of bioorthogonal reactions, in addition to the fact that none of the reactive functionalities of this cycloaddition reaction are found in or reactive toward biological systems.47 Driven by the superior bioorthogonality of SPAAC, DeForest et al. fabricated PEG hydrogels utilizing 4-arm PEG tetra azides and dicyclooctyne-flanked MMP-degradable peptides (Figure 5a).17,48 In this approach, a gem-difluoro cyclooctyne (DIFO) was adopted for its faster reaction kinetics due to the presence of stronger electron withdrawing fluorines along with the seminal ring strain.49 While gelation occurs in a few minutes (5 min) and complete network formation within an hour in the case of DIFO, a number of cyclooctynes with a wide range of reactivities have been developed in the past few years and, thus, offer the possibility of tuning gelation kinetics, as necessary.2 Note that, unlike other click reactions discussed above, SPAAC-based gel formation requires no additional reagents (e.g., initiator or buffering agents) and proceeds under physiological conditions with time scales that are reasonably appropriate for facile cellular encapsulations.
Figure 5.
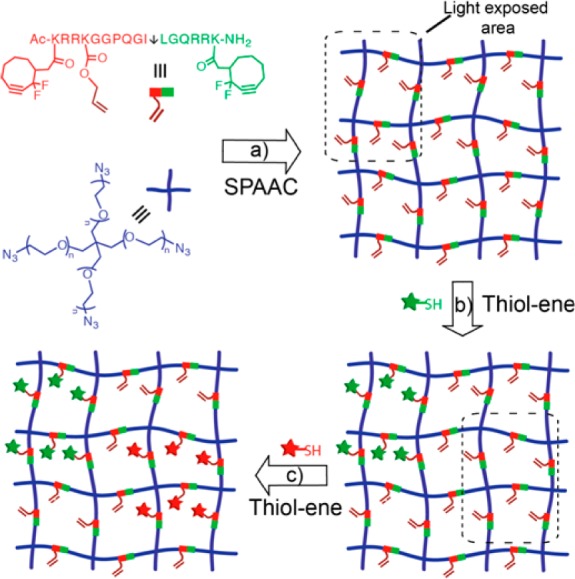
Sequential click approach for dynamically tuning extracellular microenvironments: (a) step-growth network formation via SPAAC that enables thiol–ene photopatterning without altering the original network structure by employing 4-arm PEG tetra azide and dicyclooctyne MMP cleavable (↓ shows cleavage site) peptide; (b) spatial thiol–ene photopatterning of a first biochemical unit; (c) patterning of a second biochemical unit at a different time point and location.
Despite various techniques employed to fabricate hydrogels of different network structures, as well as to immobilize several functional cues (e.g., integrin-binding peptides or proteins for cell survival, MMP-degradable peptides for cellular remodeling and migration, cytokines for regulating cell functions, such as proliferation, differentiation, and secretory properties) within a cellular scaffold, techniques that would enable one to introduce these signals at different time points in specified locations is critical for future biological studies aimed to better understand and produce complex extracellular features of living tissues.50,51 To achieve such spatiotemporal incorporation of biological cues, examples are now appearing in the literature demonstrating the utilization of two or more orthogonal click reactions in a sequential fashion.17,52 Often times the first reaction is used to form the hydrogel network, while the second, third, and so on reactions are used to introduce specific biochemical functionalities (Figure 5).17,48 In this regard, the light-driven thiol–ene photopatterning has become a unique and powerful orthogonal click chemistry because of its amenability for spatiotemporal manipulation.43 Motivated by the orthogonal nature of SPAAC and light-driven thiol–ene reactions, our group has demonstrated the formation of a cell-laden PEG hydrogel using SPAAC, comprising 4-arm PEG azides and dicyclooctyne-peptide with a pendant allyl functionality to enable postgelation photopatterning.17,48 These materials were used to create unique cell-culture niches with spatiotemporally regulated ligands (Figure 5).17,48 The generated biochemical photopatterns of this sequential click approach were not only well-defined, but more importantly displayed remarkable impact on dynamic cellular behaviors of encapsulated cells. For example, photopatterns of cell adhesive RGDS regions had definitive control over local cellular behavior, such as adhesion and morphology.
Click chemistry’s evolution to bioorthogonal materials development has the potential to broadly impact the field of biomaterials for applications ranging from the engineering of stem cell niches to the regeneration of complex tissue structures. As illustrated above, over the past decade, these flawless reactions have become powerful new tools for biomedical scientists, not only to build materials that are cell compatible, highly functional and organized in structure, but more importantly the power of their orthogonality in concert with one another enables the generation of highly complex patterns of biochemical cues (more than ever possible) within a single cellular scaffold. Such opportunities will enable numerous possibilities through spatiotemporally dictating cellular signals in an ever-precise manner and all in 3D.
Despite all these recent advancements, bioorthogonal click chemistry applied to biomaterials development is still an open field that is ripe with opportunities for researchers interested in biomaterials and their applications. Inverse demand Diels–Alder reaction of tetrazine and norbornene/trans-cyclooctene would be one of those new opportunities and perhaps beneficial as orthogonal to SPAAC and other click reactions.12,53 Also, the significant differences in the reactivity of tetrazines toward norbornene and trans-cyclooctene offer excellent possibilities to alter gelation time.54 Similarly, thiol-yne photoclick reactions are yet to be explored for biomaterial applications and could be useful, especially because of their capabilities to introduce dual thiol containing functionalities and ultimately provide materials of high cross-linking densities.55,56 Alternative to light-based chemistries (e.g., thiol–ene and thiol-yne), biomaterial scaffolds may also benefit from other biocompatible external energy sources such as ultrasound (e.g., ultrasound promoted Diels–Alder reactions).57 Toward that, ultrasound-mediated reversible click chemistries, such as cyclo-reversion of Diels–Alder adducts and 1,2,3-triazoles,58,59 have also emerged in recent years and should hold potential to alter gel stiffness or to release biochemical cues onsite. Further, recent reports of retro and exchange reactions of succinimide thioethers with free thiols might also be exploited for controlled release/exchange of biochemical signals.60
Advancing the scope of bioorthogonal micropatternings toward innovative cell culture studies to decipher and manipulate the complex extracellular microenvironments is the other critical future direction of biomedical scientists. For example, most of biochemical manipulations discussed above are limited to regulating and studying basic cellular features, such as adhesion, morphology, and motility; however, spatiotemporally fine-tuning (i) the presentation of certain crucial biochemical cues (e.g., growth and morphogenetic factors),61 (ii) local extracellular geometries (e.g., different geometrical patterning of cell adhesion proteins) in 3D for more efficient stem cell growth and differentiation would be challenging future goals.61,62 Complementarily, such real-time monitoring of cellular activities upon real-time altering of extracellular chemistry offers a fourth dimension, that is, time for researchers to study biology in a three-dimensional space, thanks to the optically clear gel matrix in visualizing cells and their extracellular surroundings.
Acknowledgments
The authors would like to thank the National Science Foundation (DM 1006711D) and the Howard Hughes Medical Institute for their financial support.
The authors declare no competing financial interest.
References
- Griffin B. A.; Adams S. R.; Tsien R. Y. Science 1998, 281, 269–272. [DOI] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Acc. Chem. Res. 2011, 44, 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo C. M.; Shoichet M. S. Bioconjugate Chem. 2011, 22, 2199–2209. [DOI] [PubMed] [Google Scholar]
- Golas P. L.; Matyjaszewski K. Chem. Soc. Rev. 2010, 39, 1338–1354. [DOI] [PubMed] [Google Scholar]
- Lowe A. B.; Hoyle C. E.; Bowman C. N. J. Mater. Chem. 2010, 20, 4745–4750. [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. Angew. Chem., Int. Ed. 2001, 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- Barner-Kowollik C.; Du Prez F. E.; Espeel P.; Hawker C. J.; Junkers T.; Schlaad H.; Van Camp W. Angew. Chem., Int. Ed. 2011, 50, 60–62. [DOI] [PubMed] [Google Scholar]
- Mather B. D.; Viswanathan K.; Miller K. M.; Long T. E. Prog. Polym. Sci. 2006, 31, 487–531. [Google Scholar]
- Jewett J. C.; Bertozzi C. R. Chem. Soc. Rev. 2010, 39, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondoni A. Angew. Chem., Int. Ed. 2008, 47, 8995–8997. [DOI] [PubMed] [Google Scholar]
- Best M. D. Biochemistry 2009, 48, 6571–6584. [DOI] [PubMed] [Google Scholar]
- Devaraj N. K.; Weissleder R.; Hilderbrand S. A. Bioconjugate Chem. 2008, 19, 2297–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J. M.; Prescher J. A.; Laughlin S. T.; Agard N. J.; Chang P. V.; Miller I. A.; Lo A.; Codelli J. A.; Bertozzi C. R. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A. J.; Mock M. L.; Tirrell D. A. Curr. Opin. Biotechnol. 2003, 14, 603–609. [DOI] [PubMed] [Google Scholar]
- Kolb H. C.; Sharpless K. B. Drug Discovery Today 2003, 8, 1128–1137. [DOI] [PubMed] [Google Scholar]
- Nimmo C. M.; Owen S. C.; Shoichet M. S. Biomacromolecules 2011, 12, 824–830. [DOI] [PubMed] [Google Scholar]
- DeForest C. A.; Polizzotti B. D.; Anseth K. S. Nat. Mater. 2009, 8, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Roy K. Tissue Eng. 2005, 11, 319–330. [DOI] [PubMed] [Google Scholar]
- Petersen O. W.; Ronnovjessen L.; Howlett A. R.; Bissell M. J. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 9064–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer R.; Vacanti J. P. Science 1993, 260, 920–926. [DOI] [PubMed] [Google Scholar]
- Peppas N. A.; Hilt J. Z.; Khademhosseini A.; Langer R. Adv. Mater. 2006, 18, 1345–1360. [Google Scholar]
- Tibbitt M. W.; Anseth K. S. Biotechnol. Bioeng. 2009, 103, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y.; Mooney D. J. Chem. Rev. 2001, 101, 1869–1879. [DOI] [PubMed] [Google Scholar]
- Kloxin A. M.; Kloxin C. J.; Bowman C. N.; Anseth K. S. Adv. Mater. 2010, 22, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacki J.; Mizuno S. Biopolymers 2008, 89, 338–344. [DOI] [PubMed] [Google Scholar]
- Daamen W. F.; Veerkamp J. H.; van Hest J. C. M.; van Kuppevelt T. H. Biomaterials 2007, 28, 4378–4398. [DOI] [PubMed] [Google Scholar]
- Nuttelman C. R.; Rice M. A.; Rydholm A. E.; Salinas C. N.; Shah D. N.; Anseth K. S. Prog. Polym. Sci. 2008, 33, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl F.; Sommer F.; Goepferich A. Biomaterials 2007, 28, 134–146. [DOI] [PubMed] [Google Scholar]
- Zhu J. M. Biomaterials 2010, 31, 4639–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. M.; Marchant R. E. Expert Rev. Med. Dev. 2011, 8, 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloxin A. M.; Kasko A. M.; Salinas C. N.; Anseth K. S. Science 2009, 324, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong B.; Kim S. W.; Bae Y. H. Adv. Drug Delivery Rev. 2002, 54, 37–51. [DOI] [PubMed] [Google Scholar]
- Lin C. C.; Raza A.; Shih H. Biomaterials 2011, 32, 9685–9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall J. D.; Anseth K. S. Biomacromolecules 2012, 13, 2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert D. L.; Pratt A. B.; Lutolf M. P.; Halstenberg S.; Hubbell J. A. J. Controlled Release 2001, 76, 11–25. [DOI] [PubMed] [Google Scholar]
- Lutolf M. P.; Hubbell J. A. Biomacromolecules 2003, 4, 713–722. [DOI] [PubMed] [Google Scholar]
- Lutolf M. P.; Lauer-Fields J. L.; Schmoekel H. G.; Metters A. T.; Weber F. E.; Fields G. B.; Hubbell J. A. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 5413–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeber G. P.; Lutolf M. P.; Hubbell J. A. Biophys. J. 2005, 89, 1374–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt A. B.; Weber F. E.; Schmoekel H. G.; Muller R.; Hubbell J. A. Biotechnol. Bioeng. 2004, 86, 27–36. [DOI] [PubMed] [Google Scholar]
- Shikanov A.; Smith R. M.; Xu M.; Woodruff T. K.; Shea L. D. Biomaterials 2011, 32, 2524–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps E. A.; Enemchukwu N. O.; Fiore V. F.; Sy J. C.; Murthy N.; Sulchek T. A.; Barker T. H.; Garcia A. J. Adv. Mater. 2012, 24, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.; Lin B. F.; Campos L.; Dimitriou M. D.; Hikita S. T.; Treat N. D.; Tirrell M. V.; Clegg D. O.; Kramer E. J.; Hawker C. J. Nat. Chem. 2010, 2, 138–145. [DOI] [PubMed] [Google Scholar]
- Fairbanks B. D.; Schwartz M. P.; Halevi A. E.; Nuttelman C. R.; Bowman C. N.; Anseth K. S. Adv. Mater. 2009, 21, 5005–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses J. E.; Moorhouse A. D. Chem. Soc. Rev. 2007, 36, 1249–1262. [DOI] [PubMed] [Google Scholar]
- Adzima B. J.; Tao Y. H.; Kloxin C. J.; DeForest C. A.; Anseth K. S.; Bowman C. N. Nat. Chem. 2011, 3, 256–259. [DOI] [PubMed] [Google Scholar]
- Agard N. J.; Prescher J. A.; Bertozzi C. R. J. Am. Chem. Soc. 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C. A.; Sims E. A.; Anseth K. S. Chem. Mater. 2010, 22, 4783–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codelli J. A.; Baskin J. M.; Agard N. J.; Berozzi C. R. J. Am. Chem. Soc. 2008, 130, 11486–11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. S.; Miller J. S.; West J. L. Adv. Mater. 2006, 18, 2679–2684. [Google Scholar]
- Khetan S.; Burdick J. A. Biomaterials 2010, 31, 8228–8234. [DOI] [PubMed] [Google Scholar]
- Wylie R. G.; Ahsan S.; Aizawa Y.; Maxwell K. L.; Morshead C. M.; Shoichet M. S. Nat. Mater. 2011, 10, 799–806. [DOI] [PubMed] [Google Scholar]
- Seitchik J. L.; Peeler J. C.; Taylor M. T.; Blackman M. L.; Rhoads T. W.; Cooley R. B.; Refakis C.; Fox J. M.; Mehl R. A. J. Am. Chem. Soc. 2012, 134, 2898–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman M. L.; Royzen M.; Fox J. M. J. Am. Chem. Soc. 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks B. D.; Scott T. F.; Kloxin C. J.; Anseth K. S.; Bowman C. N. Macromolecules 2009, 42, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimetti A. A.; Feaver K. R.; Anseth K. S. Chem. Commun. 2010, 46, 5781–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Snyder J. K. J. Am. Chem. Soc. 1989, 111, 1522–1524. [Google Scholar]
- Brantley J. N.; Wiggins K. M.; Bielawski C. W. Science 2011, 333, 1606–1609. [DOI] [PubMed] [Google Scholar]
- Wiggins K. M.; Brantley J. N.; Bielawski C. W. ACS Macro Lett. 2012, 1, 623–626. [DOI] [PubMed] [Google Scholar]
- Baldwin A. D.; Kiick K. L. Bioconjugate Chem. 2011, 22, 1946–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery M. J. Cell Sci. 2010, 123, 4201–4213. [DOI] [PubMed] [Google Scholar]
- DeForest C. A.; Anseth K. S. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 421–444. [DOI] [PubMed] [Google Scholar]


