SUMMARY
To define the mutation spectrum in non-Down syndrome acute megkaryoblastic leukemia (non-DS-AMKL), we performed transcriptome sequencing on diagnostic blasts from 14 pediatric patients and validated our findings in a recurrency/validation cohort consisting of 34 pediatric and 28 adult AMKL leukemia samples. Our analysis identified a cryptic chromosome 16 inversion [inv(16)(p13.3q24.3)] in 27% of pediatric cases, which encodes a CBFA2T3-GLIS2 fusion protein. Expression of CBFA2T3-GLIS2 in Drosophila and murine hematopoietic cells induced bone morphogenic protein (BMP) signaling, and resulted in a marked increase in the self-renewal capacity of hematopoietic progenitors. These data suggest that expression of CBFA2T3-GLIS2 directly contributes to leukemogenesis.
INTRODUCTION
Acute megakaryoblastic leukemia (AMKL) accounts for approximately 10% of pediatric acute myeloid leukemia (AML) and 1% of adult AML (Athale et al., 2001; Barnard et al., 2007; Oki et al., 2006; Tallman et al., 2000). AMKL is divided into two subgroups: AMKL arising in patients with Down syndrome (DS-AMKL), and leukemia arising in non-Down syndrome patients (non-DS-AMKL). Although DS-AMKL patients have an excellent prognosis with an ~80% survival, non-DS-AMKL patients do not fare as well with a reported survival of only 14-34% despite high intensity chemotherapy (Athale et al., 2001; Barnard et al., 2007; Creutzig et al., 2005). With the exception of the t(1;22) seen in infant non-DS-AMKL, little is known about the molecular lesions that underlie this leukemia subtype (Carroll et al., 1991; Lion et al., 1992; Ma et al., 2001; Mercher et al., 2001).
We recently reported data from a high resolution study of DNA copy number abnormalities (CNAs) and loss of heterozygosity on pediatric de novo AML (Radtke et al., 2009). These analyses demonstrated a very low burden of genomic alterations in all pediatric AML subtypes except AMKL. AMKL cases were characterized by complex chromosomal rearrangements and a high number of CNAs. To define the functional consequences of the identified chromosomal rearrangements in non-DS-AMKL, the St. Jude Children's Research Hospital – Washington University Pediatric Cancer Genome Project performed transcriptome and exome sequencing on diagnostic leukemia samples.
RESULTS
AMKL is Characterized by Chimeric Transcripts
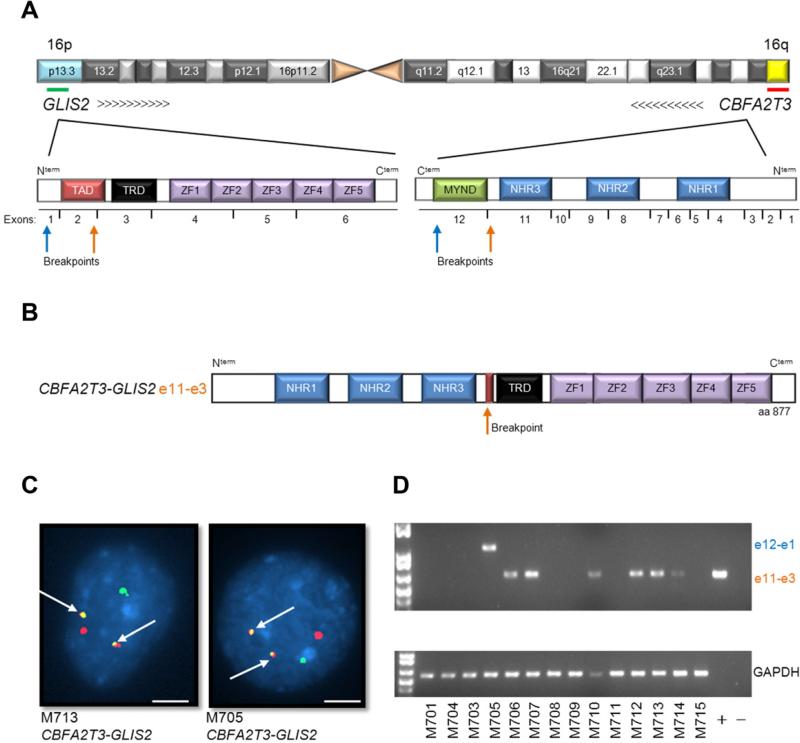
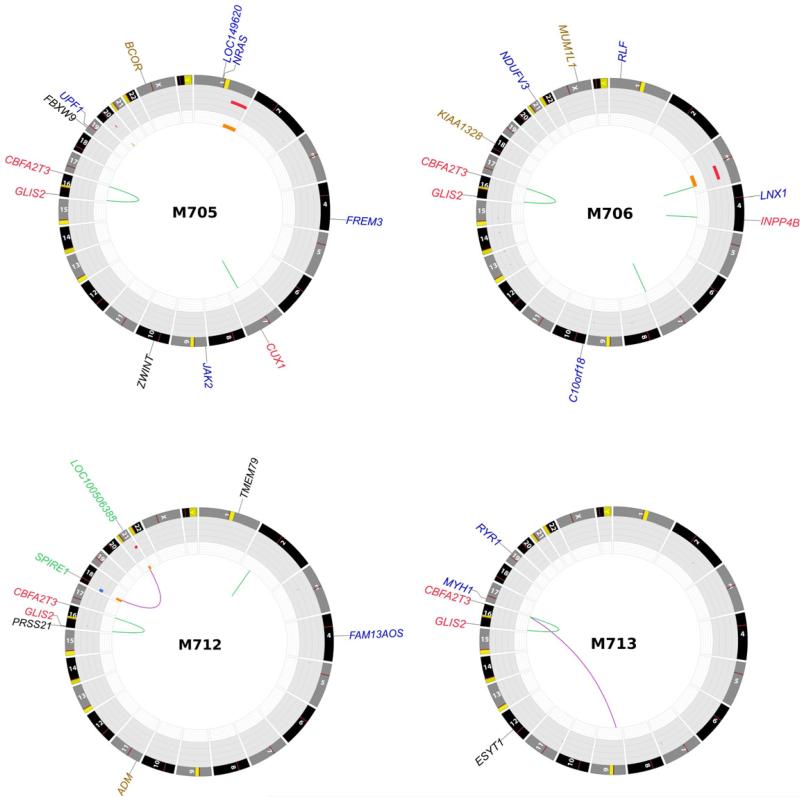
Transcriptome sequencing was performed on diagnostic leukemia cells from 14 pediatric non-DS-AMKL patients (discovery cohort) (Table S1 and S2). Our analysis identified structural variations (SVs) that resulted in the expression of chimeric transcripts encoding fusion proteins in 12 of 14 cases (Table S3). Remarkably, in 7 of 14 cases a cryptic inversion on chromosome 16 [inv(16)(p13.3q24.3)] was detected that resulted in the joining of CBFA2T3, a member of the ETO family of nuclear corepressors, to GLIS2, a member of the GLI family of transcription factors (Figures 1-2 and S1). In six of these cases exon 10 of CBFA2T3 was fused to exon 3 of GLIS2, while in the remaining one case exon 11 of CBFA2T3 was fused to exon 1 of GLIS2. Both encoded proteins retain the three CBFA2T3 N-terminal nervy homology regions that mediate protein interactions, and the five GLIS2 C-terminal zinc finger domains that bind the Glis DNA consensus sequence (Figure 1A and B). Whole genome sequence analysis of tumor and germ line DNA from four cases demonstrated that the CBFA2T3-GLIS2 chimeric gene resulted from simple balanced inversions in three cases and a complex rearrangement involving chromosomes 16 and 9 in the fourth case (Figure 2, and S1).
Figure 1. inv(16)(p13.3;q24.3) encodes a CBFA2T3-GLIS2 chimeric transcript.
(A) Schematic of chromosome 16 with locations of GLIS2 and CBFA2T3 shown. Arrows indicate orientation of the gene and the green and red lines the probes used for FISH. The protein structure of the genes is shown below chromosome 16 and is not drawn to scale. Breakpoints are indicated by arrows. TAD, transactivation domain; TRD, transcriptional regulatory domain; ZF, zinc finger; NHR, nervy homology region. (B) Schematic of CBFA2T3-GLIS2 chimeric protein. (C) Interphase FISH analysis of two representative patient samples carrying CBFA2T3-GLIS2. The GLIS2 probe is green, the CBFA2T3 probe is red. White arrows indicate the fusion event. Scale bars, 10 m. (D) RT-PCR for CBFA2T3-GLIS2 and GAPDH on the discovery cohort. See also Figure S1 and Tables S1-S6.
Figure 2. Somatic mutations in whole genome sequenced AMKL cases.
Plots depict structural genetic variants, including DNA copy number alterations, intraand inter-chromosomal translocations, and sequence alterations (Krzywinski et al., 2009). DNA copy number alterations: Loss of heterozygosity (LOH), orange; amplification, red; deletion, blue. Sequence mutations in Refseq genes: silent single nucleotide variants (SNVs), black; UTR, brown; non-silent SNVs, blue. Genes at structural variant breakpoints: genes involved in in-frame fusions, red; others, green.
Chimeric transcripts were also detected in 5 of 7 leukemia samples that lacked expression of CBFA2T3-GLIS2, including one case each expressing in-frame fusions of GATA2-HOXA9, MN1-FLI1, NIPBL-HOXB9, NUP98-KDM5A, GRB10-SDK1 and C8orf76-HOXA11AS (Figure 3 and Table S3). Importantly, several of the genes involved in these translocations play a direct role in normal megakaryocytic differentiation (GATA2 and FLI1), have been previously shown to be involved in leukemogenesis (HOXA9, MN1, HOXB9, NUP98, KDM5A), or are highly expressed in hematopoietic stem cells or myeloid/megakaryocytic progenitors (Figure S2) (Argiropoulos and Humphries, 2007; Buijs et al., 2000; Heuser et al., 2011; Kawada et al., 2001; Visvader et al., 1995; Wang et al., 2009). Analysis of a recurrency/validation cohort consisting of diagnostic leukemia cells from 62 AMKL cases (34 pediatric and 28 adult) revealed 6 additional pediatric samples carrying CBFA2T3-GLIS2 for an overall frequency of 27% (13/48) in pediatric AMKL (Table S1). None of the adult AMKL cases contained this chimeric transcript suggesting that this lesion is restricted to pediatric non-DS-AMKLs. NUP98-KDM5A was the only other chimeric transcript that was recurrent, being detected in 8.3% (4/48) of pediatric cases (Table S1). This chimeric transcript was also not detected in adult AMKLs.
Figure 3. Low frequency chimeric transcripts in pediatric AMKL.
Four chimeric transcripts were identified in one case each of the discovery cohort: GATA2-HOXA9, MN1-FLI1, NIPBL-HOXB9, and NUP98-KDM5A. (A) RT-PCR validation of the discovery cohort. Primers and conditions are described in supplementary experimental procedures. (B) Interphase FISH analysis of M703 carrying the MN1-FLI1 chimeric protein. The MN1 probe is red, the FLI1 probe is green. White arrows indicate the fusion event. Scale bar, 10 m. (C) Schematic of chimeric proteins. Exons and domains are not drawn to scale. TAD, transactivation domain; NRD, negative regulatory domain; ZNF, zinc finger; MIM, Meis interaction motif; HD, Hox domain; Ets, E-twenty six domain; FLS, Fli1 specific region; CTA, C-terminal transactivation domain; GLN, glutamine rich domain; NLS, nuclear localizing signal; HEAT, Huntingtin/EF3/PP2A/TOR1 domain; FG, phenylalanine-glycine repeats; JMJ, jumonji domain; ARID, AT-rich interaction domain; PHD, plant homeodomain. See also Figure S2.
Cooperating Lesions in AMKL
In addition to the described chimeric transcripts, exome sequence analysis on 10 of the 14 samples in the discovery cohort that had matched germ line DNA, coupled with CNAs detected by Affymetrix SNP6 microarrays, revealed an average of 5 (range 1-14) somatic non-silent sequence mutations, and 5 (range 0-11) CNAs involving annotated genes per case. (Tables S4-S6 and Figure S1). Despite the relative paucity of somatic mutations, recurrent lesions were identified in JAK kinase genes, MPL and GATA1, which have been previously shown to play a role in AMKL (Malinge et al., 2008). Sequence analysis of these genes in cases within the recurrency cohort that had available genomic DNA revealed activating mutations in JAK Kinases (9/51, 17.6%) and MPL (2/51, 3.9%), as well as inactivating mutations in GATA1 (5/51, 9.8%) (Tables S1 and S6). In addition, 7 of 14 cases with available copy number data contained amplification of chromosome 21 in the Down syndrome critical region (chr21q22, DSCR) that includes genes known to play a role in AML such as RUNX1, ETS2, and ERG (Table S4 and Figure S1). Three of these cases carry the CBFA2T3-GLIS2 chimeric gene. Importantly, the total burden of somatic mutations was significantly lower in the CBFA2T3-GLIS2 expressing cases (7.17±3.60 versus 16.60±5.13, p=0.009, Table S5).
CBFA2T3-GLIS2 AMKL is a Distinct Subtype of Pediatric AMKL with a Poor Prognosis
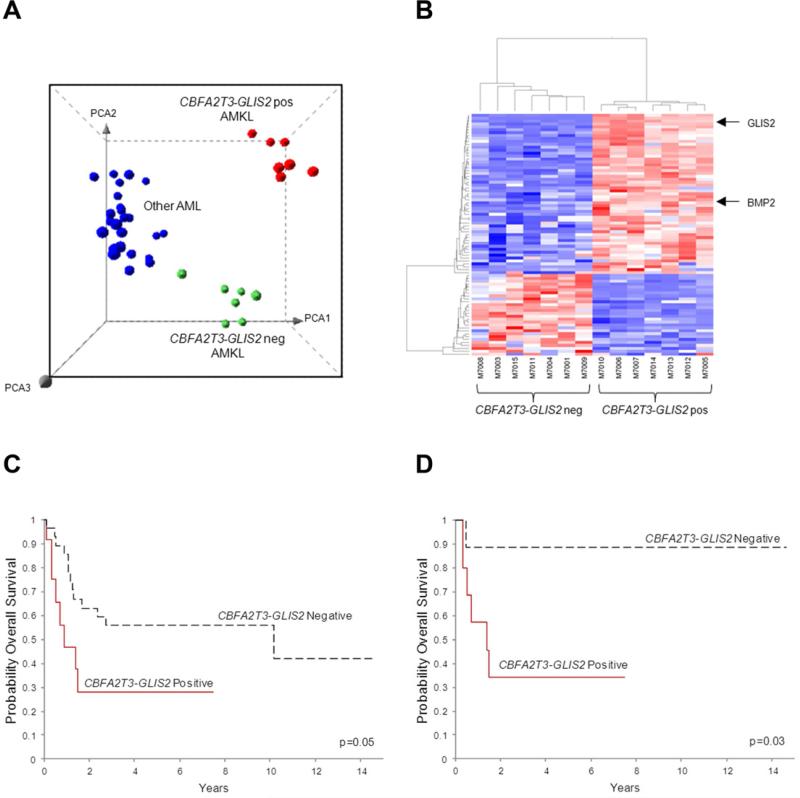
The gene expression profile of CBFA2T3-GLIS2 AMKL was distinct from that of AMKL cells lacking this chimeric transcript, and from other genetic subtypes of pediatric AML (Figure 4A-B). A detailed co-expression network analysis of the top 4000 differentially expressed genes suggest that expression of CBFA2T3-GLIS2 leads to marked upregulation of BMP2, a downstream target of Hedgehog signaling (Figure 4B, S3 and Table S7). Moreover, gene set enrichment analysis based on KEGG pathway annotation of the top scoring network module demonstrated Hedgehog and JAK-STAT pathways to be significantly upregulated in CBFA2T3-GLIS2 positive AMKL (Figure S3).
Figure 4. CBFA2T3-GLIS2 defines a unique subtype of AML with a distinct gene expression signature and poor outcomes.
(A) Principal component analysis of the gene expression profiles of the AMKL discovery cohort and 32 other non-AMKL AML samples representing all other known genetic subtypes of pediatric AML. Clusters were generated using 1000 genes selected by k-means algorithm. A detailed description of the samples included in this analysis can be found at NCBI gene expression omnibus, accession GSE35203. (B) Heat map of differentially expressed genes in the top scoring network module of CBFA2T3-GLIS2 positive and negative AMKL patient samples. For gene relationships please see Figure S3. For a detailed list of the top 500 differentially expressed the genes (not limited to this network), please see Table S7. (C) Overall survival of 40 pediatric non-DS AMKL cases treated at multiple institutions (CBFA2T3-GLIS2 negative cases n=28, and CBFA2T3-GLIS2 expressing cases, n=12). The curves for the two groups were tested by logrank method and exact test using permutation which yielded a p value of p=0.05. (D) Overall survival of 19 pediatric non-DS AMKL cases treated at St. Jude Children's Research Hospital (CBFA2T3-GLIS2 negative cases, n=9, and CBFA2T3-GLIS2 expressing cases, n=10). The curves for the two groups were tested by logrank method and exact test using permutation which yield a p value of p=0.03. See also Figure S3 and Table S7.
Given the historically poor outcomes seen in pediatric non-DS-AMKL, we next explored whether the presence of CBFA2T3-GLIS2 identified a clinically distinct subset of cases. Outcome data were available on 40 pediatric patients. Although these patients were treated at a number of different centers using a variety of different therapeutic approaches, the presence of CBFA2T3-GLIS2 identified a subgroup of patients with a significantly worse overall survival at 5 years as compared to AMKL patients that lacked this chimeric transcript (28.1% vs. 41.9%, p=0.05, Figure 4C). Moreover, when this analysis was limited to patients treated at a single institution (St. Jude, n=19) the adverse prognostic impact of CBFA2T3-GLIS2 on survival was maintained (34.3% vs. 88.9%, p=0.03, Figure 4D).
CBFA2T3-GLIS2 Modified Hematopoietic Cells Demonstrate Enhanced Self Renewal
CBFA2T3 (also known as MTG16) was initially identified as a fusion partner with RUNX1 in rare cases of therapy-related AML that contain a t(16;21)(q24;q22) (Gamou et al., 1998). More recently, CBFA2T3 has been implicated in the maintenance of hematopoietic stem cell quiescence (Chyla et al., 2008). By contrast, GLIS2 has not been previously implicated in leukemogenesis. GLIS2 is a member of GLI-similar (GLIS1-3) subfamily of Krüppel-like zinc finger transcription factors and is closely related to the GLI-family of transcription factors that function as critical elements of the hedgehog signaling pathway (Kim et al., 2007; Lamar et al., 2001). GLIS2 is expressed in the kidney and germ line inactivating mutations lead to nephronophthisis, an autosomal recessive cystic kidney disease (Attanasio et al., 2007). Although GLIS2 is not normally expressed in the hematopoietic system, its fusion to CBFA2T3 as a result of the inv(16)(p13.3q24.3) results in high level expression of the C-terminal portion of the protein including its DNA-binding domain (Figure S1).
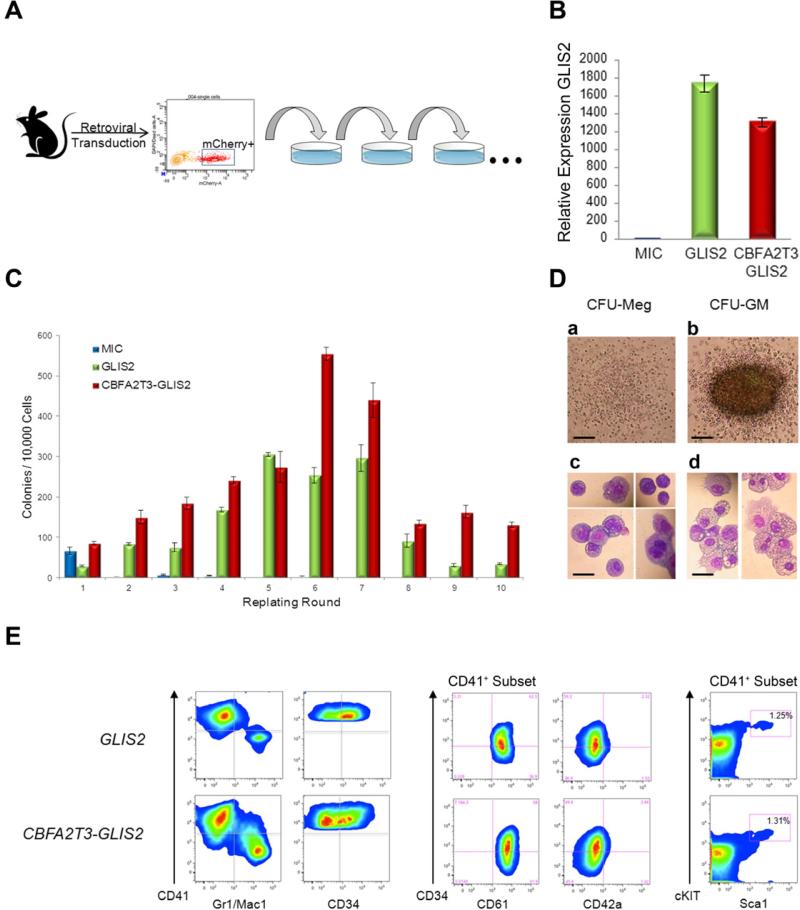
To explore the functional effects of the CBFA2T3-GLIS2 fusion protein, we transduced murine hematopoietic cells with a retrovirus expressing either CBFA2T3-GLIS2 or GLIS2 alone and assessed their effect on in vitro colony formation, differentiation, and replating efficiency as a surrogate measure of self-renewal (Figure 5A,B). On the initial plating, the expression of CBFA2T3-GLIS2 had no effect on colony numbers, size or overall myeloid/erythroid differentiation when cells were grown in the presence of IL3, IL6, SCF, and EPO. However, hematopoietic cells transduced with the empty retrovirus (MIC) failed to form colonies after the second replating, whereas expression of either CBFA2T3-GLIS2 or wild-type GLIS2 resulted in a marked increase in the self-renewal capacity, with colony formation persisting through ten replatings (Figure 5C). Upon serial replating, two colony types were detected, CFU-GM and CFU-Meg (Figure 5D). Immunophenotypic analysis at the 3rd replating also revealed evidence of megakaryocytic differentiation with CD41/CD61 dual expression and the absence of cKIT and Sca1 expression in the majority of cells (Figure 5E). Importantly,CBFA2T3-GLIS2 expressing cells remained growth factor dependent suggesting that cooperating mutations in growth factor signaling pathways are likely required for full leukemic transformation (data not shown). Moreover, transplantation of CBFA2T3-GLIS2 transduced bone marrow cells into syngeneic recipients failed to induce overt leukemia at day 365 as demonstrated by normal blood counts and low level reporter gene expression in peripheral blood (<5%) (data not shown), consistent with a requirement for cooperative mutations. Failure to induce leukemia in mice as a single lesion has been previously reported for other chimeric genes that confer the ability to serially replate in colony forming assays, including AML1-ETO (Higuchi et al., 2002).
Figure 5. CBFA2T3-GLIS2 leads to enhanced replating of hematopoietic cells.
(A) Experimental design. Murine bone marrow cells were transduced with retroviral vectors expressing mCherry alone (MSCV-IRES-mCherry, “MIC”), or mCherry along with GLIS2, or CBFA2T3-GLIS2. Transduced cells were purified by sorting mCherry positive cells and plated onto methylcellulose containing IL3, IL6, SCF, and EPO. Colonies were counted after 7 days of growth and replated serially. (B) Semi-quantitative RT-PCR of GLIS2 utilizing cells harvested from first round of plating. GLIS2 primers are specific for the 3’ half of the transcript and thus pick up both full length GLIS2 as well as CBFA2T3-GLIS2. Expression in MIC cells was defined as 1, and data is pooled from two separate experiments with similar results. p=<0.0001 as determined by one-way ANOVA. Error bars represent mean ± SEM of two independent experiments. (C) Number of colonies detected at 7 days following each plating. Error bars represent mean ± SEM of two independent experiments. (D) Colony morphology detected in GLIS2 and CBFA2T3-GLIS2 modified cells from the 2nd plating and beyond. a, CFU-Meg; b, CFU-GM. Scale bars, 500 m. Representative cytospins and morphology of each colony type are shown. c, CFU-Meg; d, CFU-GM. Scale bars, 50 m. (E) Cells harvested from colony forming assays after 3 or more replatings were subjected to flow cytometry. Cells were negative for acetylcholinesterase (data not shown).
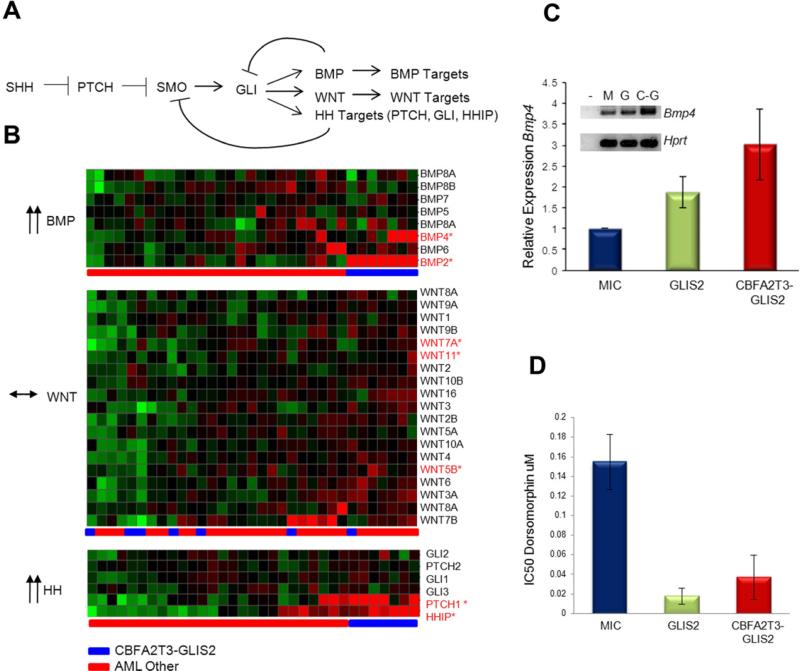
CBFA2T3-GLIS2 Induces BMP Signaling
GLIS2 can function as both a transcriptional activator and repressor depending on the cellular context and has been implicated in altered signaling through a number of pathways including sonic hedgehog-GLI1 (SHH) and WNT/ß-catenin (Attanasio et al., 2007; Kim et al., 2007). Analysis of the gene expression signatures of CBFA2T3-GLIS2 expressing AMKLs revealed altered expression of a number of genes in the SHH and WNT pathways, as well as genes in the bone morphogenic protein (BMP) pathway, which is directly influenced by SHH signaling (Figure 4B, 6A, and S3) (Dahn and Fallon, 2000; Ingham and McMahon, 2001; Vokes et al., 2007). When this analysis was limited to genes containing GLI consensus DNA-binding sites (Gli-BS) in their promoters, or to genes known to be transcriptional targets of GLIS2, marked over-expression of PTCH1, HHIP, BMP2 and BMP4 was observed (Figures 6B, S3, S4 and Table S7) (Attanasio et al., 2007). Consistent with this observation, although CBFA2T3-GLIS2 only weakly activated transcription of a reporter construct containing the Gli-BS (Figure S4), it strongly activated transcription of the Gli-BS-containing BMP4-promoter driven luciferase construct and induced expression of BMP4 in murine hematopoietic cells (Figure 6C and S4). Moreover, CBFA2T3-GLIS2 strongly activated a BMP response element (BRE) containing luciferase reporter construct and induced expression of the BMP downstream transcriptional target, inhibitor of differentiation 1 (ID1) (Korchynskyi and ten Dijke, 2002), consistent with the induced expression of BMP2/4 (Figure S4).
Figure 6. CBFA2T3-GLIS2 activates the BMP pathway.
(A) The Hedgehog (HH) signaling pathway. In addition to classic hedgehog targets such as PTCH and HHIP, WNT and BMP gene expression have been demonstrated to be affected by the GLI transcription factor in various models (Dahn and Fallon, 2000; Ingham and McMahon, 2001; Vokes et al., 2007). (B) Gene expression profiles from CBFA2T3-GLIS2 containing AMKL cases and other AML subtypes were evaluated for expression levels of BMP, WNT, and HH target genes. CBFA2T3-GLIS2 negative AMKL cases are not shown in this analysis. Significantly upregulated probe sets (FDR less than 0.05) are designated with red font: BMP2 FDR 1.06×10-17, BMP4 FDR 0.015976, PTCH1 FDR 2.05×10-6, and HHIP FDR 0.0038. (C) Murine bone marrow cells were transduced with retroviral vectors carrying mCherry alone (MSCV-IRES-mCherry, “MIC”), mCherry plus GLIS2, or CBFA2T3-GLIS2. mCherry positive cells were sorted and plated in methylcellulose containing IL3, IL6, SCF, and EPO. Following one week of growth, RNA was isolated, reverse transcribed, and amplified with Bmp4 or Hprt specific primers. Error bars represent mean ± SEM of four independent experiments. A representative gel is shown (-, neg; M, MIC; G, GLIS2; and C-G, CBFA2T3-GLIS2). p=0.047 as determined by one-way ANOVA. (D) GLIS2 and CBFA2T3-GLIS2 sensitize murine hematopoietic cells to BMP receptor type I inhibition. Colony formation assays were conducted in the presence or absence of dorsomorphin at the indicated concentrations (Yu et al., 2008). IC50 values were calculated as the amount of drug required to inhibit 50% of the colony formation as determined by colony counts. Error bars represent mean ± SEM of two independent experiments. p=0.036 as determined by one-way ANOVA. See also Figure S4.
BMP signaling plays a critical role in the specification of hematopoiesis in developing embryos and studies suggest that BMP4 stimulation can augment megakaryocytic output from CD34 progenitors (Jeanpierre et al., 2008; Soderberg et al., 2009). To determine if the observed CBFA2T3-GLIS2 induced BMP expression contributes to the enhanced replating capacity of murine hematopoietic cells, colony replating assays were repeated in the presence of dorsomorphin, a selective small molecule inhibitor of the BMP type I receptors that blocks BMP mediated phosphorylation of SMAD 1/5/8 (Yu et al., 2008). Importantly, CBFA2T3-GLIS2 as well as GLIS2 expressing hematopoietic cells were significantly more sensitive to dorsomorphin than wild type cells in the first plating (Figure 6D). Continuous exposure to dorsomorphin inhibited colony formation in a dose dependent manner on subsequent platings (data not shown). Interestingly, sublethal doses of dorsomorphin in CBFA2T3-GLIS2 positive cells led to an upregulation of Bmp4 and Id1 transcripts over time with colony counts returning to untreated levels, suggesting cells are able to overcome this inhibition by upregulating the BMP pathway (data not shown).
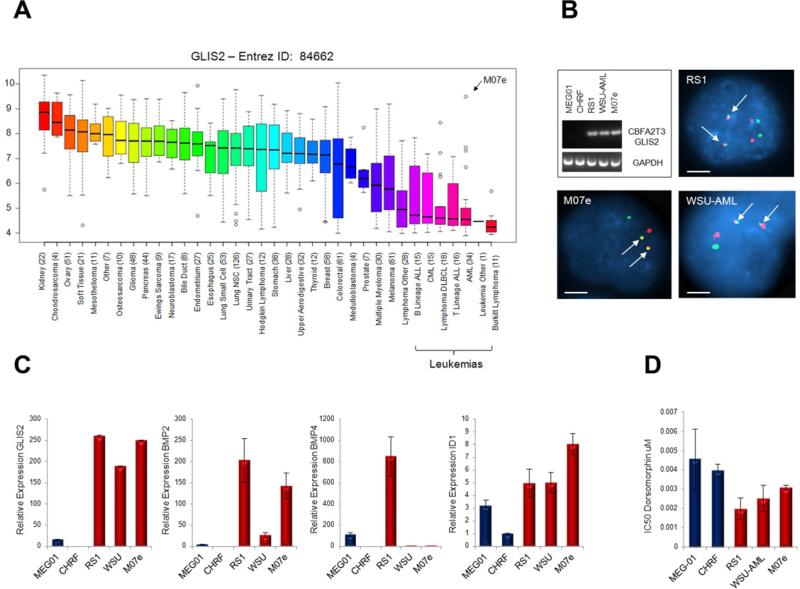
To further explore the downstream signaling of CBFA2T3-GLIS2 in human leukemia cell lines, we first assessed the expression level of GLIS2 in human cancer cell lines using the recently published Broad-Novartis Cancer Cell Line Encyclopedia (Figure 7A) (Barretina et al., 2012). Interestingly, this analysis showed that GLIS2 expression levels are lowest in leukemia cell lines. Moreover, within the leukemias the highest expressing cell line was the pediatric AMKL cell line M07e. To further explore AMKL cell lines, we performed RT-PCR for CBFA2T3-GLIS2 on 5 human AMKL cell lines. Three of the five cell lines (RS1, WSU-AML, and M07e) expressed CBFA2T3-GLIS2 (Figure 7B). The presence of the chimeric gene in these lines was validated by FISH analysis (Figure 7B). We went on to determine the relative expression of BMP genes by semi-quantitative RT-PCR and found a trend towards upregulation of these genes in the CBFA2T3-GLIS2 positive cells (Figure 7C). We also assessed our AMKL cell lines for dorsomorphin sensitivity and found a trend towards increased sensitivity in cell lines expressing CBFA2T3-GLIS2 as determined by a standard MTT assay (Figure 7D).
Figure 7. CBFA2T3-GLIS2 is present in AMKL cell lines.
(A) GLIS2 expression as determined by gene expression arrays in 991 human cancer cell lines. Log2 transformed expression levels are shown. Data obtained from the Broad-Novartis Cancer Cell Line Encyclopedia (http://www.broadinstitute.org/ccle/home). 34 AML cell lines are included, the extreme outlier of this subtype, M07e, is indicated. The GLIS2 probe set recognizes the end of the transcript and thus does not distinguish between wild type GLIS2 and CBFA2T3-GLIS2. (B) RT-PCR on 5 AMKL cell lines: MEG-01, CHRF-288-11, RS-1, WSU-AML, and M07e. The three cell lines carrying CBFA2T3-GLIS2 were validated by FISH. Scale bars, 10 μm. (C) Real time semi-quantitative RT-PCR of GLIS2, BMP2, BMP4, and ID1 on the 5 AMKL cell lines. Expression levels relative to Beta Actin are shown. CHRF-288-11 expression levels were set to 1 for comparison across cell lines. Error bars represent mean ± SEM of two independent experiments. (D) Dorsomorphin sensitivity in the cell lines as determined by MTT assay. Error bars represent mean ± SEM of two independent experiments. For cell line information and MTT assay, please see supplemental experimental procedures.
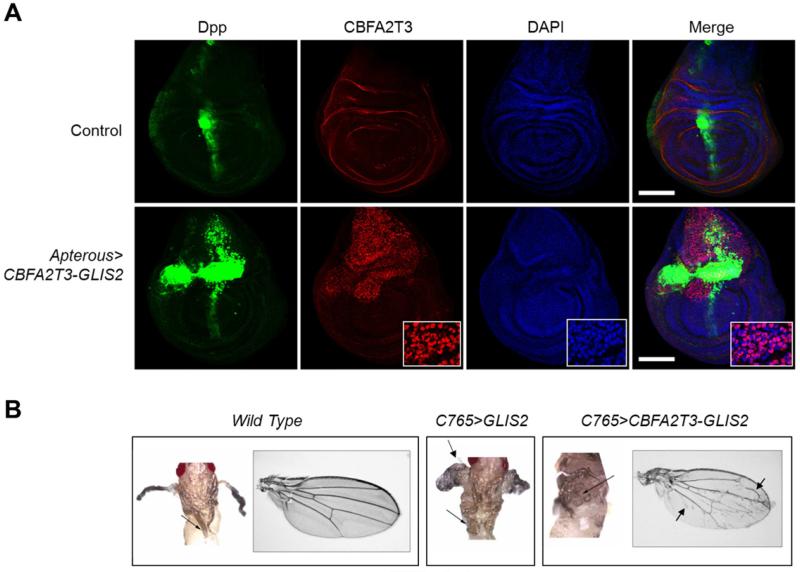
To determine if CBFA2T3-GLIS2 induces the upregulation of BMP signaling in vivo, we generated transgenic Drosophila expressing either CBFA2T3-GLIS2 or full length GLIS2 using an epithelial promoter, and examined their effect on fly development. During Drosophila development, the WNT, BMP, and SHH homologs (Wg, Dpp, and Hh, respectively) have distinct roles in patterning adult wing structures (Dahn and Fallon, 2000; Ingham and McMahon, 2001; Vokes et al., 2007). When altered these signaling pathways trigger characteristic loss and gain-of-function phenotypes (Tabata and Takei, 2004). Expression of CBFA2T3-GLIS2 and full length GLIS2 in Drosophila resulted in ectopic expression of endogenous dpp, the fly homolog of BMP4, in wing imaginal discs (Figure 8A and S5). Immunofluorescence confirmed the nuclear localization of CBFA2T3-GLIS2 (Figure 8A). Both CBFA2T3-GLIS2 and GLIS2 over-expression induced lethality; however, a small number of escapers developed to pharate adults and demonstrated a morphologic dpp gain-of-function phenotype; wing hinges were converted to notum and legs were shortened and broadened (Figure 8B) (Grieder et al., 2009). Rare CBFA2T3-GLIS2 transgenic flies developed to adulthood and demonstrated mild ectopic veination throughout the wing blade, as well as wing blistering consistent with a dpp gain-of-function phenotype (Figure 8B) (Sander et al., 2010).
Figure 8. Transgenic CBFA2T3-GLIS2 Drosophila ectopically express Dpp.
(A) CBFA2T3-GLIS2 was expressed under control of Apterous-Gal4 (strong epithelial dorsal driver). dpp-lacZ serves as a reporter for dpp induction. Wing imaginal discs were isolated at the late 3rd instar, stained for β-gal as a read out for dpp (green), CBFA2T3 (red), and DAPI (blue) followed by immunofluorescence analysis. Nuclear localization of CBFA2T3-GLIS2 can be seen by the pink signal (inset). Scale bars, 100 μm. (B) CBFA2T3-GLIS2 was expressed under control of C765, a weak epithelial driver. Pharate adults were dissected from pupal casings and imaged. Arrows indicate ectopic notum, broadened and shortened legs. No C765>GLIS2 Drosophila matured to adulthood. Arrows indicate ectopic veins in wings of rare C765>CBFA2T3-GLIS2 escapers. See also Figure S5.
DISCUSSION
Sequence analysis of pediatric non-DS-AMKLs revealed the expression of an inv(16)-encoded CBFA2T3-GLIS2 in almost 30% of pediatric non-DS-AMKL patients and its presence defined a distinct subgroup of patients that had an exceptionally poor outcome when compared to AMKL patients that lacked this lesion. In addition, five other chimeric transcripts (GATA2-HOXA9, MN1-FLI1, NIPBL-HOXB9, GRB10-SDK1 and C8orf76-HOXA11AS) were detected in single AMKL cases. Surprisingly, none of the identified chimeric transcripts were detected in adult AMKL cases highlighting the significant biological differences between pediatric and adult AMKL. Importantly, each of the detected chimeric transcripts is predicted to encode a fusion protein that would alter signaling pathways known to play a role in normal hematopoiesis suggesting that these lesions are “driver” mutations that directly contribute to the development of leukemia. In addition to these somatic structural alterations, a variety of other somatic mutations were detected including activating mutations in kinase signaling pathways in 21.6% of cases (JAK kinase family members and MPL), inactivating mutations in GATA1 in 9.8% of cases, and amplification of chromosome 21 in the Down syndrome critical region that includes genes known to play a role in AML such as RUNX1, ETS2, and ERG in 50% of the cases. How these mutations interact to not only induce overt leukemia but also to influence therapeutic responses remains to be determined.
As part of the St. Jude Children's Research Hospital - Washington University Pediatric Cancer Genome Project we have sequenced 260 cases of pediatric cancers across multiple tumor types (Downing et al., 2012). The CBFA2T3-GLIS2 fusion was limited to AMKL cases. This specificity may exist for several reasons. The N-terminal portion of the fusion, CBFA2T3 is primarily expressed in the hematopoietic compartment leading one to predict that expression of the inversion product, if it were to occur, would primarily be limited hematopoietic cells. While we do not know the exact target cell of transformation, induction of BMP4 signaling in human CD34+ progenitors has been demonstrated to increase the percentage of megakaryocyte and erythroid colonies in vitro (Fuchs et al., 2002; Jeanpierre et al., 2008). Thus enhanced BMP signaling as a result of the expression of the inv(16)-encoded CBFA2T3-GLIS2 may directly contribute to the megakaryocytic differentiation of the leukemia cells.
The inv(16)-encoded CBFA2T3-GLIS2 chimeric gene induced aberrant high level expression of the DNA-binding domain of GLIS2 in hematopoietic cells, along with the disruption of one allele of CBFA2T3, a gene whose encoded protein has been shown to play a role in maintaining normal hematopoietic stem cell quiescence (Chyla et al., 2008). GLIS2 is a distant member of the GLI superfamily of transcriptional factors that function as critical transcriptional targets of the SHH signaling pathway (Hui and Angers, 2011). Although alterations in the SHH pathway have been directly implicated in a range of cancers (Barakat et al., 2010), the role of SHH signaling in normal hematopoiesis and leukemia remains poorly defined (Lim and Matsui, 2010). Our data suggests that aberrant expression of GLIS2 results in upregulation of the classic SHH negative feedback inhibitors PTCH and HHIP, coupled with a marked increase in the expression of BMP 2 and 4, resulting in enhanced BMP signaling. These results indicate that CBFA2T3-GLIS2 functions, in part, as a gain-of-function GLIS2 allele. The exact mechanisms by which GLIS2 induced the upregulation of BMP2/4 remains incompletely defined, although our data suggest that a direct transcription effect of GLIS2 on the BMP4 promoter is likely, although an indirect mechanism may also contribute.
Interestingly, BMP4 has been shown to expand and maintain human cord blood hematopoietic stem cells in vitro both directly, as well as indirectly via SHH signaling (Bhardwaj et al., 2001; Bhatia et al., 1999). Furthermore, ID1, a downstream BMP target previously implicated in leukemogenesis was found to be upregulated in CBFA2T3-GLIS2 modified hematopoetic cells demonstrating that this pathway is activated (Wang et al., 2011b). Consistent with these findings, we demonstrated that activation of BMP signaling contributed to the marked increase in the replating capacity of myeloid/erythroid committed progenitors. Accordingly, we found that murine hematopoietic cells carrying either full length GLIS2, or CBFA2T3-GLIS2 demonstrated an increased sensitivity to BMP inhibition suggesting that upregulation of this pathway contributes to the observed phenotype. In addition, BMP4 signaling has been shown to induce the differentiation of human CD34+ progenitors into megakaryocytes (Jeanpierre et al., 2008), suggesting that the upregulation of this pathway is also contributing to the megakaryocyte differentiation phenotype of these leukemias. Lastly, BMP4, like thrombopoietin, appears to exert its effects on human megakaryopoiesis in part through the JAK/STAT pathways (Jeanpierre et al., 2008). Interestingly, functional pathway analysis of gene expression profiles in CBFA2T3-GLIS2 positive AMKL samples identified genes in the Jak-STAT signaling pathway to be significantly upregulated (p=0.0038, FDR 0.022978, Figure S4). Combined with the identification in some cases of activating mutations in either JAK family members or MPL in CBFA2T3-GLIS2 expressing leukemias, our data suggests that these lesions likely cooperate in leukemogenesis.
Taken together, these data define a poor prognostic subgroup of pediatric AMKL patients that are characterized by the inv(16)( p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein. Expression of CBFA2T3-GLIS2 induces an enhanced replating capacity of lineage committed myeloid progenitors, along with megakaryocytic differentiation, in part through enhanced BMP2/4 signaling. Whether altered SHH and CBFA2T3-induced signaling also contribute to leukemogenesis remains to be determined. Nevertheless, the presented data raises the important possibility that inhibition of the BMP pathway may have a therapeutic benefit in this aggressive form of pediatric AML.
EXPERIMENTAL PROCEDURES
Patients and Samples
Paired-end transcriptome sequencing on diagnostic leukemic blasts was performed on 14 Pediatric non-DS-AMKL cases using the illumina platform. Four of these cases underwent whole genome sequencing on diagnostic leukaemia blasts and matched germ line samples. All 14 cases underwent whole exome sequencing for which 10 had matching germ line samples. One additional diagnostic sample with matched germline DNA had whole exome sequencing done that did not undergo transcriptome sequencing. All 15 of these patients were treated at St Jude Children's Research Hospital from 1990-2008. The recurrence cohort consisted of 61 additional cases including 33 pediatric specimens and 28 adult specimens . All samples were obtained with patient or parent/guardian provided informed consent under protocols approved by the Institutional Review Board at each institution and St. Jude Children's Research Hospital.
Sequencing
RNA and DNA library construction for transcriptome and whole genome DNA sequencing, respectively has been described previously (Mardis et al., 2009; Zhang et al., 2012). Analysis of whole-genome sequencing data (WGS) and whole-exome sequencing data which include mapping, coverage and quality assessment, SNV/indel detection, tier annotation for sequence mutations, prediction of deleterious effects of missense mutations, and identification of loss-of-heterozygosity were described previously (Zhang et al., 2012). Please see supplementary experimental procedures for details.
Recurrencey screening for sequence variations and fusions
We performed recurrence screening on a cohort of 61 AMKL samples. All 61 were screened by RT-PCR (see supplementary experimental procedures for primers and conditions) for CBFA2T3-GLIS2, GATA2-HOXA9, MN1-FLI1, NIPBL-HOXB9, and NUP98-KDM5A. Whole genome amplified DNA (Qiagen) from 38 cases underwent PCR and Sanger sequencing by Beckman Coulter Genomics for JAK1, JAK2, JAK3, and MPL mutations. In 8/38 cases, a paired matched germline was available. Putative SNVs and indel variants were detected by SNPdetector (Zhang et al., 2005).
Overall Survival Probabilities
Outcome data was available for 40 Pediatric patients tested for CBFA2T3-GLIS2. CBFA2T3-GLIS2 was found in 13 patients. Overall survival was defined as the date of diagnosis or study enrollment to the date of death with surviving patients censored at the date of last follow-up. Survival curves were estimated using the Kaplan-Meier method and compared using the exact log-rank test based on 10,000 permutations.
Affymetrix SNP array
Affymetrix SNP 6.0 array genotyping was performed for 14 of 15 AMKL cases in the discovery cohort, and array normalization and DNA copy number alterations identified as previously described (Lin et al., 2004; Mullighan et al., 2007; Olshen et al., 2004; Pounds et al., 2009). To differentiate inherited copy number alterations from somatic events in leukaemia blasts from patient's lacking matched normal DNA, identified putative variants were filtered using public copy number polymorphism databases and a St. Jude database of SNP array data from several hundred samples (Iafrate et al., 2004; McCarroll et al., 2008). SNP array data are available at dbGaP accession number phs000413.v1.p1.
Gene Expression Profiling
Gene expression profiling was performed using Affymetrix Human Exon 1.0 ST Arrays (Affymetrix) according to manufacturer's instructions. This cohort comprised 39 Pediatric AML samples including AMKL (N=14), AML1-ETO (N=4), CBFB-MYH11 (N=2), MLL rearranged (N=3), PML-RARA (N=2), NUP98-NSD1 (N=2), HLXB9-ETV6 (N=1) and AML cases lacking chimeric genes (N=11). Please see supplementary experimental procedures for further details.
FISH
Dual-color FISH was performed on archived bone marrow cells and cell lines as described previously (Mullighan et al., 2007). Probes were derived from bacterial artificial chromosome (BAC) clones (Invitrogen). BACs used were RP11-830F9 (CBFA2T3), CTD-25555M20 (GLIS2), RP11-345E21 (MN1), and CTD-2542E23 (FLI1). BAC clone identity was verified by T7 and SP6 BAC-end sequencing and by hybridization of fluorescently labeled BAC DNA with normal human metaphase preparations.
Cloning of CBFA2T3-GLIS2 and GLIS2
Total RNA was extracted from leukaemia blasts and MEG01 cells using RNeasy (Qiagen) and reverse transcribed using Superscript III (Invitrogen) as per manufacturer's instructions. The coding region of CBFA2T3-GLIS2 was PCR amplified from patient M712 and M707 using primers CBFA2T3_119F and GLIS2_1685R (see supplementary experimental procedures for primers and conditions). GLIS2 was PCR amplified from the MEG01 cell line (Sato et al., 1989) cDNA using primers GLIS2_21F and GLIS2_1685R (see supplementary experimental procedures for primers and conditions). PCR products were subcloned into the pGEM-T Easy Vector and sequenced (Promega). Clones containing the correct sequence were then subcloned into the MSCV-IRES-mCherry retroviral backbone (Volanakis et al., 2009).
Murine Bone Marrow Transduction and Colony Forming Assays
All experiments involving mice were reviewed and approved by the Institutional Animal Care and Use Committee. Bone marrow from 4-6 week old female C57/BL6 was harvested and cultured in the presence of recombinant murine SCF (rmSCF, Peprotech, 50ng/ml), IL3 (rmIL3, Peprotech, 50ng/ml), and IL6 (rmIL6, Peprotech, 50ng/ml) for 24 hours prior to transduction on RetroNectin (Takara Bio Inc.) coated plates. Ecotropic envelope pseudotyped retroviral supernatant was produced by transient transfection of 293T cells as previously described (Soneoka et al., 1995). 48 hours following transduction cells were harvested, sorted for mCherry expression, and plated on methylcellulose containing IL3, IL6, SCF and EPO (Stem Cell technologies, Vancouver, BC) as per manufacturer's instructions. Colonies were counted after 7 days of growth at 37 C, harvested and replated. In a subset of experiments, Dorsomorphin (Sigma-Aldrich) was added to the methylcellulose at the indicated concentrations.
Flow Cytometry
Cells were resuspended in PBS and pre-incubated with anti-CD16/CD32 Fc-block (BD Pharmingen) if staining did not include conjugated anti-murine CD16/32. Aliquots were stained for 15 minutes at 4°C with conjugated antibodies. Cells were washed and resuspended in 4',6-diamidino-2-phenylindole containing solution (1ug/ml DAPI in PBS) for subsequent analysis using FACS LSR II D (BD Biosciences). For a list of antibodies used, please see supplementary experimental procedures.
Luciferase Assays
The human BMP4 promoter driven luciferase construct pSLA4.1EX (Van den Wijngaard et al., 1999) was kindly provided by E. Joop van Zoelen, Nijmegen, Netherlands. The murine BMP response element (pBRE) (Korchynskyi and ten Dijke, 2002) was kindly provided by Peter ten Dijke, Leiden, Netherlands. The 8x3'Gli-BS luciferase reporter (pGli-BS) (Sasaki et al., 1997) has been previously described. TOPFlash and FOPFlash (Korinek et al., 1997) constructs were obtained from Addgene. For details on luciferase reporter assays, please see supplementary experimental procedures.
ELISA
Bmp4 protein levels in the supernatants from transduced murine hematopoietic cells were determined by ELISA. Briefly, mCherry positive bone marrow cells transduced with empty (MIC), GLIS2, or CBFA2T3-GLIS2 contaiing retroviruses were placed in media containing IL3, IL6, and SCF for 48 hours and supernatant was then harvested and the level of murine BMP4 determines using an ELISA kit purchased from TSZ Elisa (www.Tszelisa.com). Measurements were done according to manufacturer's instructions.
Transgenic Drosophila
CBFA2T3-GLIS2 and GLIS2 cDNAs were subcloned into the pUAS-attB plasmid (Bischof et al., 2007). Transgenic UAS-CBFA2T3-GLIS2 and UAS-GLIS2 flies weregenerated using site-specific ϕC31 integration system (Bischof et al., 2007). Embryo injections were performed by Best Gene, Inc. UAS constructs were targeted to chromosome 2R-51D in order to avoid differential positional effects on transgene expression. For wing imaginal disc staining, relevant crosses were performed to generate flies carrying all three transgenes: Apterous-Gal4 (a strong epithelial dorsal compartment specific GAL4 driver), UAS-CBFA2T3-GLIS2, and a dpp-lacZ enhancer trap reporter. Gal4 driver and dpp-lacZ reporter stocks were obtained from the Bloomington Stock Center. Wing imaginal discs were dissected from wandering 3rd instar larvae, fixed and immunostained using anti-β-gal (Promega, Z378), anti-CBFA2T3 (Abcam, ab33072), and DAPI (Invitrogen, D3571) as previously described (Carroll et al.). To assess the phenotypic effects of CBFA2T3-GLIS2 and GLIS2, UAS transgenes were expressed under control of the epithelial driver C765-Gal4 and progeny were observed. Pharate adults were dissected from pupal casings and imaged.
Supplementary Material
Significance.
Acute megakaryoblastic leukemia (AMKL) accounts for 10% of childhood acute myeloid leukemia (AML). Although AMKL patients with Down syndrome (DS-AMKL) have an excellent survival, non-DS-AMKL patients have an extremely poor outcome with a 3 year survival of less than 40%. With the exception of the t(1;22) seen in the majority of infants with non-DS-AMKL, little is known about the molecular lesions that underlie this leukemia subtype. Our results identified a fusion gene, CBFA2T3-GLIS2 that functions as a driver mutation in a subset of these patients. Importantly, pediatric patients with CBFA2T3-GLIS2 expressing AMKL had inferior outcomes (5-year survival 34.3% vs. 88.9%; p=0.03) demonstrating that this lesion is a prognostic factor in this leukemia population.
Highlights.
CBFA2T3-GLIS2 is a novel recurrent fusion gene in pediatric AMKL
CBFA2T3-GLIS2 AMKL has a distinct expression profile and an inferior outcome
CBFA2T3-GLIS2 induces BMP signaling and enhanced self-renewal of hematopoietic progenitors
ACKNOWLEDGEMENTS
The authors would like to specifically thank Joy Nakitandwe for critical input and discussions, Susana Raimondi for review of cytogenetics, Matt Stine for assistance with data deposition, Bill Pappas and Scott Malone for support of the information technology infrastructure, and the staff of Tissue Resources Laboratory, Flow Cytometry and Cell Sorting Core, the Hartwell Center for Biotechnology and Bioinformatics of St Jude Children's Research Hospital, and Emily Dolezale for assistance in sample procurement at Memorial Sloan Kettering Cancer Center. This work was supported by grants from the National Institutes of Health (Cancer Center Support Grant P30 CA021765), the Eric Trump Foundation, a Leukemia & Lymphoma Society Specialized Center of Research grant LLS7015, and the American Lebanese Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The sequence data and single nucleotide polymorphism microarray data have been deposited in the dbGaP database (http://www.ncbi.nlm.nih.gov/gap) under the accession number phs000413.v1.p1. Affymetrix gene expression data have been deposited in the NCBI gene expression omnibus ((http://www.ncbi.nlm.nih.gov/geo/) under GSE35203.
REFERENCES
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Athale UH, Razzouk BI, Raimondi SC, Tong X, Behm FG, Head DR, Srivastava DK, Rubnitz JE, Bowman L, Pui CH, Ribeiro RC. Biology and outcome of childhood acute megakaryoblastic leukemia: a single institution's experience. Blood. 2001;97:3727–3732. doi: 10.1182/blood.v97.12.3727. [DOI] [PubMed] [Google Scholar]
- Attanasio M, Uhlenhaut NH, Sousa VH, O'Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nature genetics. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends in molecular medicine. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DR, Alonzo TA, Gerbing RB, Lange B, Woods WG. Comparison of childhood myelodysplastic syndrome, AML FAB M6 or M7, CCG 2891: report from the Children's Oncology Group. Pediatr Blood Cancer. 2007;49:17–22. doi: 10.1002/pbc.20951. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, Chadwick K, Ling LE, Karanu FN, Bhatia M. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol. 2001;2:172–180. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- Bhatia M, Bonnet D, Wu D, Murdoch B, Wrana J, Gallacher L, Dick JE. Bone morphogenetic proteins regulate the developmental program of human hematopoietic stem cells. The Journal of experimental medicine. 1999;189:1139–1148. doi: 10.1084/jem.189.7.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs A, van Rompaey L, Molijn AC, Davis JN, Vertegaal AC, Potter MD, Adams C, van Baal S, Zwarthoff EC, Roussel MF, Grosveld GC. The MN1-TEL fusion protein, encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity. Molecular and cellular biology. 2000;20:9281–9293. doi: 10.1128/mcb.20.24.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Civin C, Schneider N, Dahl G, Pappo A, Bowman P, Emami A, Gross S, Alvarado C, Phillips C, et al. The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia: a Pediatric Oncology Group Study. Blood. 1991;78:748–752. [PubMed] [Google Scholar]
- Carroll CE, Marada S, Stewart DP, Ouyang JX, Ogden SK. The extracellular loops of Smoothened play a regulatory role in control of Hedgehog pathway activation. Development. 139:612–621. doi: 10.1242/dev.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyla BJ, Moreno-Miralles I, Steapleton MA, Thompson MA, Bhaskara S, Engel M, Hiebert SW. Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Molecular and cellular biology. 2008;28:6234–6247. doi: 10.1128/MCB.00404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutzig U, Reinhardt D, Diekamp S, Dworzak M, Stary J, Zimmermann M. AML patients with Down syndrome have a high cure rate with AML-BFM therapy with reduced dose intensity. Leukemia. 2005;19:1355–1360. doi: 10.1038/sj.leu.2403814. [DOI] [PubMed] [Google Scholar]
- Dahn RD, Fallon JF. Interdigital regulation of digit identity and homeotic transformation by modulated BMP signaling. Science. 2000;289:438–441. doi: 10.1126/science.289.5478.438. [DOI] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui C-H, Ding L, Ley TJ, Evans WE. The Pediatric Cancer Genome Project. Nature genetics. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs O, Simakova O, Klener P, Cmejlova J, Zivny J, Zavadil J, Stopka T. Inhibition of Smad5 in human hematopoietic progenitors blocks erythroid differentiation induced by BMP4. Blood cells, molecules & diseases. 2002;28:221–233. doi: 10.1006/bcmd.2002.0487. [DOI] [PubMed] [Google Scholar]
- Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder NC, Morata G, Affolter M, Gehring WJ. Spalt major controls the development of the notum and of wing hinge primordia of the Drosophila melanogaster wing imaginal disc. Dev Biol. 2009;329:315–326. doi: 10.1016/j.ydbio.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Heuser M, Yun H, Berg T, Yung E, Argiropoulos B, Kuchenbauer F, Park G, Hamwi I, Palmqvist L, Lai CK, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer cell. 2011;20:39–52. doi: 10.1016/j.ccr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annual review of cell and developmental biology. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nature genetics. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jeanpierre S, Nicolini FE, Kaniewski B, Dumontet C, Rimokh R, Puisieux A, Maguer-Satta V. BMP4 regulation of human megakaryocytic differentiation is involved in thrombopoietin signaling. Blood. 2008;112:3154–3163. doi: 10.1182/blood-2008-03-145326. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kang HS, ZeRuth G, Lichti-Kaiser K, Vasanth S, Yin Z, Kim YS, Jetten AM. Gli-similar (Glis) Kruppel-like zinc finger proteins: insights into their physiological functions and critical roles in neonatal diabetes and cystic renal disease. Histology and histopathology. 2010;25:1481–1496. doi: 10.14670/hh-25.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada H, Ito T, Pharr PN, Spyropoulos DD, Watson DK, Ogawa M. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. International journal of hematology. 2001;73:463–468. doi: 10.1007/BF02994008. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kang HS, Jetten AM. The Kruppel-like zinc finger protein Glis2 functions as a negative modulator of the Wnt/beta-catenin signaling pathway. FEBS letters. 2007;581:858–864. doi: 10.1016/j.febslet.2007.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. The Journal of biological chemistry. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar E, Kintner C, Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- Lim Y, Matsui W. Hedgehog signaling in hematopoiesis. Critical reviews in eukaryotic gene expression. 2010;20:129–139. doi: 10.1615/critreveukargeneexpr.v20.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- Lion T, Haas OA, Harbott J, Bannier E, Ritterbach J, Jankovic M, Fink FM, Stojimirovic A, Herrmann J, Riehm HJ, et al. The translocation t(1;22)(p13;q13) is a nonrandom marker specifically associated with acute megakaryocytic leukemia in young children. Blood. 1992;79:3325–3330. [PubMed] [Google Scholar]
- Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nature genetics. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- Malinge S, Ragu C, Della-Valle V, Pisani D, Constantinescu SN, Perez C, Villeval JL, Reinhardt D, Landman-Parker J, Michaux L, et al. Activating mutations in human acute megakaryoblastic leukemia. Blood. 2008;112:4220–4226. doi: 10.1182/blood-2008-01-136366. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature genetics. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Mercher T, Coniat MB, Monni R, Mauchauffe M, Nguyen Khac F, Gressin L, Mugneret F, Leblanc T, Dastugue N, Berger R, Bernard OA. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki Y, Kantarjian HM, Zhou X, Cortes J, Faderl S, Verstovsek S, O'Brien S, Koller C, Beran M, Bekele BN, et al. Adult acute megakaryocytic leukemia: an analysis of 37 patients treated at M.D. Anderson Cancer Center. Blood. 2006;107:880–884. doi: 10.1182/blood-2005-06-2450. [DOI] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Pounds S, Cheng C, Mullighan C, Raimondi SC, Shurtleff S, Downing JR. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics. 2009;25:315–321. doi: 10.1093/bioinformatics/btn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J, Ganti R, Cai Z, Goorha S, Pounds SB, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander V, Eivers E, Choi RH, De Robertis EM. Drosophila Smad2 opposes Mad signaling during wing vein development. PloS one. 2010;5:e10383. doi: 10.1371/journal.pone.0010383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Sato T, Fuse A, Eguchi M, Hayashi Y, Ryo R, Adachi M, Kishimoto Y, Teramura M, Mizoguchi H, Shima Y, et al. Establishment of a human leukaemic cell line (CMK) with megakaryocytic characteristics from a Down's syndrome patient with acute megakaryoblastic leukaemia. Br J Haematol. 1989;72:184–190. doi: 10.1111/j.1365-2141.1989.tb07681.x. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Soderberg SS, Karlsson G, Karlsson S. Complex and context dependent regulation of hematopoiesis by TGF-beta superfamily signaling. Ann N Y Acad Sci. 2009;1176:55–69. doi: 10.1111/j.1749-6632.2009.04569.x. [DOI] [PubMed] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic acids research. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Takei Y. Morphogens, their identification and regulation. Development. 2004;131:703–712. doi: 10.1242/dev.01043. [DOI] [PubMed] [Google Scholar]
- Tallman MS, Neuberg D, Bennett JM, Francois CJ, Paietta E, Wiernik PH, Dewald G, Cassileth PA, Oken MM, Rowe JM. Acute megakaryocytic leukemia: the Eastern Cooperative Oncology Group experience. Blood. 2000;96:2405–2411. [PubMed] [Google Scholar]
- Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic acids research. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Wijngaard A, Pijpers MA, Joosten PH, Roelofs JM, Van zoelen EJ, Olijve W. Functional characterization of two promoters in the human bone morphogenetic protein-4 gene. J Bone Miner Res. 1999;14:1432–1441. doi: 10.1359/jbmr.1999.14.8.1432. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Crossley M, Hill J, Orkin SH, Adams JM. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Molecular and cellular biology. 1995;15:634–641. doi: 10.1128/mcb.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Volanakis EJ, Williams RT, Sherr CJ. Stage-specific Arf tumor suppression in Notch1-induced T-cell acute lymphoblastic leukemia. Blood. 2009;114:4451–4459. doi: 10.1182/blood-2009-07-233346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma J, Rusch MC, Chen K, Harris CC, Ding L, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nature methods. 2011a;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, Hatlen MA, Vu L, Liu F, Xu H, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011b;333:765–769. doi: 10.1126/science.1201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wheeler DA, Yakub I, Wei S, Sood R, Rowe W, Liu PP, Gibbs RA, Buetow KH. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.