Abstract
Background
Regulatory T cells (Tregs) are a subset of T cells that play an important role in modulating T effector responses during infectious challenges. The aim of this study was to evaluate possible associations between regulatory gene polymorphisms and the risk of uncomplicated malaria and the control of Plasmodium falciparum parasite density levels.
Methods
Twelve regulatory single nucleotide polymorphisms (SNPs) in the promoter regions of FOXP3 (ss270137548, rs11091253), IL10RA (rs56356146, rs7925112), IL10RB (rs8178433, rs8178435, rs999788), STAT6 (rs3024941, rs3024943, rs3024944) and TNFRSF18 (ss2080581728, rs3753344) were genotyped in a cohort of Congolese children. Studied subjects were followed up (passively) during one year. The children who experienced one or several clinical episodes were genotyped as “uncomplicated malaria” group (n=179) and those children who did not experience any episode were genotyped as “asymptomatic children” group (n=138).
Results
The prevalence of rs3024944CC genotype of STAT6 was significantly higher in the group of asymptomatic children compared to that of uncomplicated malaria (P=0.003). Similarly, the minor allele rs3024944C was more prevalent in the group of asymptomatic children (P=0.019). Two novel SNPs were observed including -163T/G (ss491228441) in IL10RA gene and -163C/T (ss491228440) in TNFRSF18 gene. The genotype ss491228441TT and the minor allele ss491228441G of the IL10RA were more frequent in the group of asymptomatic children (P=0.006 and P=0.007, respectively). The genotype rs11091253CT of the FOXP3 was associated with high parasite density levels. In addition, a new promoter IL10RA variant (ss491228441) contributes to shield against mild malaria.
Conclusion
The study indicated that the STAT6 promoter polymorphism rs3024944 was associated with uncomplicated malaria, whereas the FOXP3 promoter variant rs11091253 was associated with significant P. falciparum parasitaemia levels. These genetic data may contribute to the understanding of molecular mechanisms that regulate immune response to P. falciparum infections.
Keywords: Plasmodium falciparum, Tregs, FOXP3, IL10RA, STAT6
Background
Malaria is one of the major causes of morbidity and mortality in tropical and sub-tropical areas. Approximately 216 million clinical cases of malaria occurred worldwide in 2010, resulting in almost 655,000 deaths [1]. Sub-Saharan Africa remains the region with the highest burden of the disease, accounting for nearly 91% of global malaria deaths. Malaria is caused by protozoan parasites of the genus Plasmodium and Plasmodium falciparum being the most virulent species. Plasmodium falciparum-infected individuals show different clinical phenotypes, which range from asymptomatic infections to severe forms of malaria [1]. Several investigations demonstrated that immune gene polymorphisms in human host, such as those having an impact on expression of T regulatory cells (Treg), contribute to the variability of malaria phenotypes [2,3]. The generation of Tregs has been demonstrated to modulate the immune response in malaria infection as well as in other infectious diseases [4-8]. A few of the Tregs are induced in response to infectious challenge and the others are considered as natural regulators [4]. Expression of Tregs is well known to be regulated by number of human host genes, such as the forkhead box P3 (FOXP3), interleukin 10 receptors alpha and beta (IL10RA and IL10RB), the signal transducer and activator of transcription 6 (STAT6), and the tumour necrosis factor receptor superfamily member 18 (TNFRSF18) [9].
FOXP3 belongs to the family of transcription factors that play a role in various cellular processes and is described as an important regulator of natural Tregs development and function [10-12]. In humans, mutations in the FOXP3 gene are known to be associated with immune dysregulation polyendocrinophathy and enterophathy X-linked syndrome (IPEX) caused by lack of Tregs [13]. IL10RA and IL10RB are two receptor chains forming the human interleukin 10-receptor (IL10R) which belongs to the class II cytokine receptors [14]. The major role of IL10RA precursor is to mediate high affinity and ligand binding and subsequent signal transduction, whereas the IL10RB precursor is believed to contribute only in signalling process [15]. Reports have demonstrated that mutations in the genes encoding IL10RA and IL10RB precursors of the IL10R annul the IL10 mediated immunomodulatory signalling and are strongly associated with inflammatory bowel disorders [16]. Recently combined promoter haplotypes of the IL10RA and IL10RB genes have been shown to be associated with protection against severe malaria in Gabonese children [14]. The STAT6 gene, which belongs to the STAT family of transcription factors, has been reported to mediate various cytokine-induced responses. Recent reports have shown that STAT6 gene polymorphisms play crucial roles in controlling helminth infection. More precisely, a promoter variant rs324013 was found to be associated with the control of Schistosoma haematobium in a Dogon population (where S. haematobium is endemic) in Mali [17]. Similarly, the variant rs324015 was associated to the control of infection levels in Chinese children from an endemic area for Ascaris lumbricoides[18]. TNFRSF18, also known as glucocorticoid-induced TNF receptor, is a member of TNF superfamily, which plays a key role in the co-stimulatory process for T cell activation for both conventional and CD25+CD4+ T cells. In addition, TNFRSF18 expressed on Tregs was demonstrated to play a crucial role in the maintenance of peripheral tolerance [9]. Therefore, all these findings suggest that mutations in FOXP3, IL10RA, IL10RB, STAT6 and TNFRSF18 may lead to various immunopathological reactions in auto-immune disorders and/or parasitic infections.
In a previous investigation, regulatory SNPs in the promoter region of FOXP3, IL10RA, IL10RB, STAT6 and TNFRSF18 genes were identified in the Gabonese population in Central Africa. These identified SNPs were further validated for their allelic gene expression using transient transfection assays [14,19-21]. Against this background, in the present study, the possible relationship between the reported SNP variants and risk of having uncomplicated P. falciparum malaria was investigated. For this purpose, this study utilized a case-control cohort of Congolese children, who were followed up (passively) during one year for surveillance of clinical malaria cases, comprising a group of children who experienced one or several uncomplicated malaria episodes and that of children who had no clinical episode during the follow up. As a second objective, the impact of these SNPs carriage on the P.falciparum parasite density levels was assessed.
Methods
Study area, study population and ethical consideration
The study was carried out in three districts (Kinsana, Mbouono and Ngoko) of Makélékélé health division in the southern area of Brazzaville. These districts are semi-urban with about 6,000 inhabitants and located along the Congo River. Malaria is transmitted throughout the year with P. falciparum being the predominant species and Anopheles gambiae the main mosquito vector [9]. This study, as part of a cohort study for malaria surveillance in the study area (Ndounga et al, unpublished), is a community-based case-control study. The study population consisted of children aged one to nine years permanently living in the study area. All the study individuals belong to Bantu ethnicity and had equivalent risk for infection. Children were recruited between April and June 2010 and passively followed up during one year. At the time of enrolment 4 ml of whole blood were collected from all children and a thick and thin blood smears were performed. The inclusion criteria were: absence of clinical malaria in the previous two weeks and one week after enrolment (an axillary temperature of <37.5°C, no disease symptoms such as muscular pain and headaches), negative thick blood smear or positive blood smear with less than 5,000 parasites per microlitre of blood (p/μl), and not being homozygous for sickle cell trait. During the follow up, at any time, when a child was suspected to have fever or any malaria-related symptoms, parents or guardians were invited to present the child at the health centre for clinical examination. In case of malaria (positive thick and thin blood smears and axillary temperature ≥37.5°C), blood sample was collected, parasite density determined and children were treated with artesunate-amodiaquine or artemether-lumefantrine. No data were available about the present or past history of helminth infection.
The ethical approval was given by the Institutional Ethics Committee for Research on Health Sciences of the Republic of Congo. Written informed consent was obtained from parents or guardians of children.
Microscopic examination
Thick and thin blood films were stained with 10% Giemsa for 15 min and read by two independent, competent microscopists to determine the parasite density and Plasmodium species. Asexual parasites were counted against 200 leucocytes and expressed as the number of asexual parasites per μl, assuming the leucocytes count of 8,000/μl of blood.
DNA extraction
Genomic DNA was extracted from 200 μl of peripheral whole blood samples obtained from patients and asymptomatic children using the commercially available QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions and stored at -20°C until use.
SNP genotyping
The promoter region of each of the five targeted genes in all the samples was genotyped for the promoter SNP variants that revealed altered gene expression as reported in earlier investigations. Those SNPs are: -794C/G (ss270137548) and -738C/T (rs11091253), for the FOXP3 gene; -185C/T (rs56356146), and -116T/C (rs7925112) for the IL10RA gene; -978T/G (rs8178433), -754A/G (rs8178435), and -740C/T (rs999788) for the IL10RB gene; -3728C/G (rs3024941), -3430G/A (rs3024943), and -3387G/C (rs3024944) for the STAT6 gene; and -265C/T (ss2080581728), and -199C/T (rs3753344) for the TNFRSF18 gene. Polymerase chain reaction (PCR) amplifications were carried out in 20 μl reaction volumes with 5 ng of genomic DNA, 1X PCR buffer (20 mM of Tris-HCL pH 8.4, 50 mM of KCL, 1.5 mM of MgCl2), 0.2 mM of dNTPs, 0.5 mM of each primer and 1 U of Taq DNA polymerase (Qiagen, Hilden, Germany). Thermal cycling parameters are mentioned in Table 1. All the PCR products were purified with the PCR DNA Purification Kit (GE Healthcare Europe GmbH) and 1 μl of the purified product was directly used as template for sequencing using the BigDye terminatorv.2.0 cycle sequencing Kit (Applied Biosystems, USA) on an ABI 3130 DNA sequencer, according to the manufacturer’s instructions. Sequence-specific primers designed and used for amplifications and sequencing are listed in Table 1. Polymorphisms were identified by assembling the sequences with respective reference sequences obtained from SNPper database using Codon code Aligner 4.0 software and were reconfirmed visually from their respective electropherograms.
Table 1.
Primers used for genotyping and sequencing
| Gene ID | Chr. | Primer sequences (Forward/reverse) | Size | Thermal Conditions |
|---|---|---|---|---|
|
FOXP3 |
X |
5′-AGGCTGAGGGCCTCAGAAGCATC-3′* |
429 bp |
94°C(5mn), [94°C(15S), 61°C(1mn) 72°C(1mn)]x35, 72°C(10mn) |
| 5′-CTCCAGGCCTCAGTTTCCCTATAG-3′ | ||||
|
IL10RA |
11 |
5′-GGGAAGGAAAGGGAGGGGTGGC-3′* |
241 bp |
94°C(2mn), [94°C(30S) 61°C(30S),72°C(30S)]x35, 72°C(2mn) |
| 5′-GCGCGCCTCCAGCTACCCTTG-3′ | ||||
|
IL10RB |
21 |
5′-AAGGATGAGGCTGTGAGGAGG-3′* |
401 bp |
94°C(2mn), [94°C(30S) 61°C(30S),72°C(1mn)]x35, 72°C(2mn) |
| 5′-CAAGCAGAGGGAAGTGAATGCG-3′ | ||||
|
STAT6 |
12 |
5′-GGCGTGTCTCAGTGTTTACCCC-3′* |
530 bp |
94°C(2mn), [94°C(30S) 61°C(30S),72°C(1mn)]x30, 72°C(2mn) |
| 5′-GTCCCCCTCAAAAGCATCAGC-3′ | ||||
| TNFRSF18 | 1 | 5′-TCCATGGTTGAGGCTCTCTGG-3′* |
293 bp | 94°C(2mn), [94°C(30S) 61°C(30S),72°C(1mn)]x35, 72°C(2mn) |
| 5′-GTGTGAGGAGGGGGTGTAGAC-3′ |
Chr: Chromosome, *: Sequencing primers, bp: Base pairs, mn: minutes, S: seconds.
Statistical analysis
In this study, children without any reported malaria episode during the follow-up period were considered as asymptomatic and were classified in a group as asymptomatic children (as they could possibly be parasite carriers). Similarly, those who experienced at least one clinical malaria attack were classified in another group as uncomplicated malaria. Clinical and parasitological data for each child who had uncomplicated malaria were obtained from the longitudinal cohort study for malaria surveillance (Ndounga et al, unpublished). Those data included number of malaria attacks or episodes, parasite density and axillary temperature values.
Statistical analysis was performed using binary logistic regression model with Intercooled Stata v. 9.1 (STATA Corporation, Texas, USA) to determine the differences in allele frequencies and genotype distributions between uncomplicated malaria and asymptomatic children adjusted for age and gender. The level of significance was set at P<0.05. In addition, genetic models such as allelic, dominant and recessive were examined to evaluate the contributions of the genotype and allele frequencies using STATA. Kruskal-Wallis tests were employed to analyse the relationship between genotype distributions and the number of episodes as well as the risk of parasite density levels in the group of children with uncomplicated malaria. Genotype or haplotype frequencies were determined by simple gene counting and by using the expectation-maximum (EM) algorithm. The significance of deviation from Hardy-Weinberg equilibrium was tested using the random-permutation procedure as implemented in the Arlequin v. 3.5.1.2 software [22]. Linkage disequilibrium (LD) analysis was performed using Haploview V. 3.2 program.
Results
Characteristics of the study participants
Of the 323 children enrolled in the cohort study for malaria surveillance (Ndounga et al, personal communication), samples from 317 children were available for the present study. All the 317 children were successfully followed up during one year, and among them, 179 had at least one clinical malaria episode (forming the group of uncomplicated malaria children) whereas 138 did not experience any clinical malaria (forming the group of asymptomatic children). A total of 297 episodes were recorded without any case of recrudescence. No case of severe malaria was recorded and P. falciparum was the only Plasmodium species detected in children with uncomplicated malaria. Characteristics of recruited children are summarized in Table 2.
Table 2.
Baseline characteristics of recruited children
| Characteristics | Mild malaria (n=179) | Asym (n=138) | P value |
|---|---|---|---|
| Mean age (year) |
4.73 ± 2.51 |
3.84 ± 2.56 |
NS |
| Sex ratio (Male/Female) |
1.16 (96/83) |
1.16 (74/64) |
NS |
| Parasite carriers (%) |
21 (11%) |
10 (7.24%) |
NS |
| Hb AA carries (%) |
148 (82.69) |
109 (79%) |
NS |
| Hb SS carriers (%) | 31 (17.31%) | 29 (21%) | NS |
Hb: haemoglobin; NS: non-significant; Unc malaria: uncomplicated malaria;
Asym: asymptomatic.
Contribution of regulatory gene polymorphisms to uncomplicated clinical malaria
All the analysed SNPs did not deviate from Hardy-Weinberg equilibrium in each group. Genotype distributions and allele frequencies of these SNP variants are summarized in Table 3. Two novel SNPs were identified in the promoter region of IL10RA at the position -163T/G (ss491228441) and at the position -163C/T (ss491228440) in the promoter region of TNFRSF18 gene, respectively. The identified novel SNPS were submitted to the SNPper database and appropriate SNP #ss IDs were received. The new SNP IDs for -163T/G of IL10RA and -163C/T variants are rs199832262 and rs150621809 respectively. Association analysis revealed that genotype rs3024944CC of STAT6 gene was observed more in children who did not have clinical malaria episode during the follow up (asymptomatic group) compared to uncomplicated malaria group (Adjusted OR: 0.17, 95%CI: 0.05-0.55, P=0.003). The minor allele rs3024944C of STAT6 gene was associated with decreased risk of clinical uncomplicated malaria (Adjusted OR: 0.57, 95%CI: 0.35-0.91, P=0.019). Interestingly, the heterozygous genotype ss491228441TG of the IL10RA was also found to be more frequent in asymptomatic children and the homozygous ss491228441TT genotypes were observed to be a risk factor (Adjusted OR: 6.35, 95%CI: 1.68-23.9, P=0.006). Also it was revealed that the minor allele ss491228441G of the IL10RA gene, despite it low overall frequency in Congolese children, was significantly more frequent in asymptomatic children compared to uncomplicated malaria group (Adjusted OR: 0.16, 95%CI: 0.04-0.61, P=0.007). The other regulatory polymorphisms in the FOXP3, IL10RB and TNFRSF18 loci do not reveal any significant associations either at allele, genotype or at haplotype levels.
Table 3.
Distribution of gene variants in children from the group of uncomplicated malaria and that of asymptomatic children
| Gene Variant | Unc malar (n=179) | Asym (n=138 ) | OR (95%CI) | P value | |
|---|---|---|---|---|---|
|
IL10RA ss491228441 T/G |
Genotypes |
|
|
|
|
| TT |
176 (0.98) |
125 (0.91) |
|
|
|
| TG |
3 (0.02) |
13 (0.09) |
|
|
|
| GG |
0 (0) |
0 (0) |
|
|
|
| Allele |
|
|
|
|
|
| T |
355 (0.99) |
263 (0.95) |
Reference |
||
| G |
3 (0.01) |
13 (0.05) |
0.16 (0.04-0.61) |
0.007 |
|
| Dominant |
|
|
|
|
|
| TT |
176 (0.98) |
125 (0.91) |
6.35 (1.68-23.9) |
0.006 |
|
| TG+GG |
3 (0.02) |
13 (0.09) |
|
|
|
| Recessive |
|
|
|
|
|
| GG |
0 (0) |
0 (0) |
NA |
||
| TT+TG |
179 (1.0) |
138 (1.0) |
|||
| STAT6 rs3024944 G/C | Genotypes |
|
|
|
|
| GG |
145 (0.81) |
106 (0.77) |
|
|
|
| GC |
30 (0.17) |
17 (0.12) |
|
|
|
| CC |
4 (0.02) |
15 (0.11) |
|
|
|
| Allele |
|
|
|
|
|
| G |
320 (0.89) |
229 (0.82) |
Reference |
||
| C |
38 (0.11) |
47 (0.18) |
0.57 (0.35-0.91) |
0.019 |
|
| Dominant |
|
|
|
|
|
| GG |
145 (0.81) |
106 (0.77) |
NS |
||
| GC+CC |
34 (0.19) |
32 (0.23) |
|||
| Recessive |
|
|
|
|
|
| CC |
4 (0.02) |
15 (0.11) |
0.17 (0.05-0.55) |
0.003 |
|
| GG+GC | 175 (0.98) | 123 (0.89) | |||
NA: not applicable; NS: not significant; Unc malar: uncomplicated malaria; Asym: asymptomatic, Odds ratio and P value were calculated by using binary logistic regression model and adjusted for age and gender.
The reconstructed haplotypes of STAT6 (rs3024941/rs30240943/rs3024944) and IL10RA (rs56356146/rs7925112/ss491228441) were observed as a susceptibility factor between both studied groups and are presented in Table 4. In total, five different haplotypes were observed for STAT6 locus, of which the major haplotype CGG was marginally significant with more prevalence among children who presented at least one clinical episode (uncomplicated malaria) (Adjusted OR: 1.4, 95%CI: 1.0-2.0, P=0.05). Similarly, five haplotypes were observed for the IL10RA, and the minor haplotype CTG was found to be associated with protection against the disease (Adjusted OR: 0. 16, 95%CI: 0.04-0.61, P=0.007). Different haplotypes of the other genes were not found to be significantly distributed between both groups of children. The linkage disequilibrium pattern of studied loci in promoter regions of STAT6 and IL10RA are presented in Figure 1.
Table 4.
Distribution of STAT6 and IL10RA haplotypes
| Gene ID | Haplotypes | Unc. Malaria n=358(%) | Asymptomatic n=276(%) | OR (95%CI) | P value |
|---|---|---|---|---|---|
|
STAT6(rs3024941/ rs3024943 /rs3024944) |
CGG |
276 (0.771) |
193 (0.699) |
1.4 (1.0-2.0) |
0.054 |
| GAG |
43 (0.121 |
36 (0.130) |
NA |
NS |
|
| CGC |
38 (0.106) |
43 (0.156) |
NA |
NS |
|
| CAG |
1 (0.002) |
0 (0) |
NA |
NS |
|
| GAC |
0 (0) |
3 (0.011) |
NA |
NS |
|
| CAC |
0 (0) |
1 (0.004) |
NA |
NS |
|
| IL10RA(rs56356146 /rs7925112 /ss491228441) | CTT |
284 (0.8) |
209 (0.758) |
NA |
NS |
| TTT |
44 (0.12) |
40 (0.145) |
NA |
NS |
|
| CCT |
26 (0.07) |
14 (0.05) |
NA |
NS |
|
| CTG |
3 (0.008) |
13 (0.047) |
0.16 (0.04-0.61) |
0.007 |
|
| TCT | 1 (0.002) | 0 (0) | NA | NS |
NA: not applicable; NS: not significant; Unc malar: uncomplicated malaria; Asym: asymptomatic, Odds ratio and P value were calculated by using binary logistic regression model and adjusted for age and gender.
Figure 1.
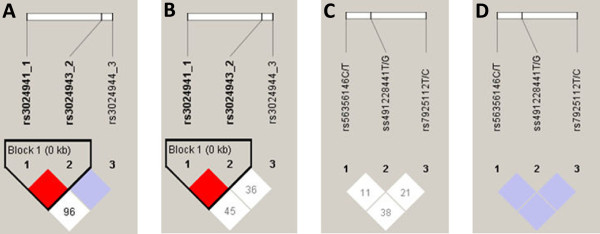
Linkage disequilibrium of studied variants in asymptomatic and uncomplicated malaria. Linkage disequilibrium pattern of studied SNPs in promoter regions of STAT6 in uncomplicated malaria (A); STAT6 in asymptomatic children (B); IL10RA in uncomplicated malaria (C); and, IL10RA in asymptomatic children (D). At the top, the SNPs are shown according to their succession from the promoter region of studied gene. Empty squares indicate a high degree of LD (LD coefficient D’ = 1) between pairs of markers, numbers indicate the D’ value, red squares indicate strong LD, purple and white squares indicate lower degree of LD with LOD <2. The haplotype block is outlined by a solid line.
Contribution of regulatory gene polymorphisms to clinical episodes and parasite density
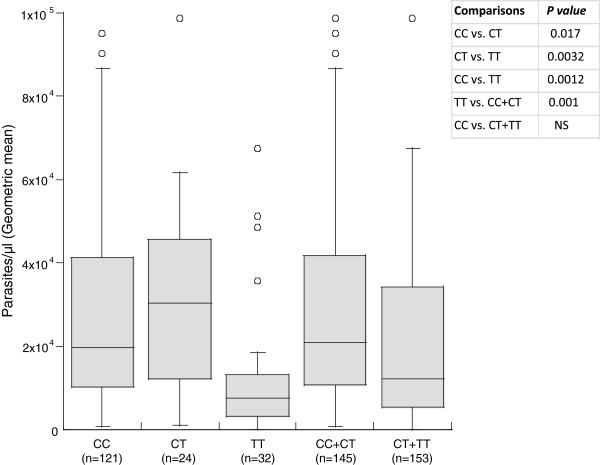
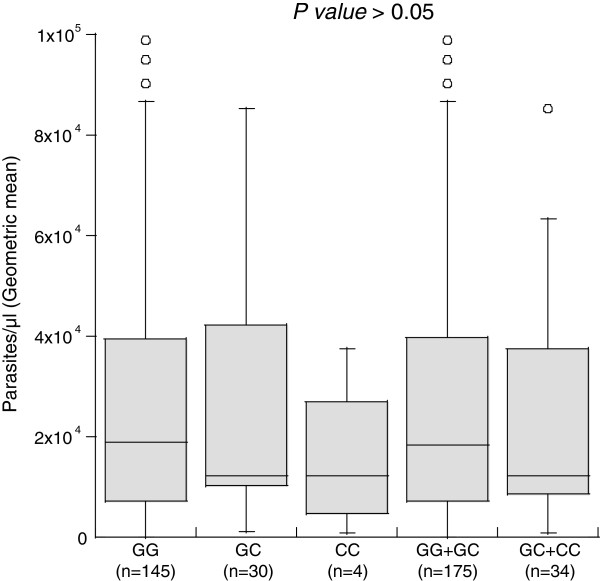
The effects of each single SNP variant on the number of clinical malaria episodes were investigated by simple comparison of genotype and allele distributions between individuals who experienced one malaria attack and those who experienced two or more malaria attacks. No association was found between the investigated SNPs and the number of malaria attacks. However analyses of the effect of individual single nucleotide polymorphisms on parasite density levels (Kruskal-wallis tests) showed an association between the FOXP3 rs11091253 variant and P. falciparum parasitaemia. More precisely, patients carrying the rs11091253CT genotype harboured higher parasite densities compared to those carrying the rs11091253CC or rs11091253TT genotype (Figure 2). These results also showed that patients having the STAT6 rs3024944CC genotype, which was correlated with protection against uncomplicated malaria, were found to have low levels of infection than those with G/G (not significant) (Figure 3).
Figure 2.
Association of the FOXP3 rs11091253 SNP with parasite density levels. Note. P values were calculated by Kruskal-Wallis test. Shown P values are corrected for multiple comparisons.
Figure 3.
Association of rs3024944 of STAT6 and parasite density levels. Note. P values were calculated by Kruskal-Wallis test.
Discussion
Malaria has exerted selective pressure on the human genome that has led to changes, such as haemoglobin S which confers some resistance against severe malaria [23]. Understanding the mechanisms leading to increased susceptibility or resistance against the disease would assist in the elaboration of better tools to fight the disease. In this search, immune responses that are on the front of the defence are particularly investigated. Tregs are suggested to be central in the control of P. falciparum infection. Therefore, investigation of human polymorphisms in genes associated with Treg activity may underlie both susceptibility to infection and level of Treg expression.
The present study evaluated the contribution of promoter SNPs of FOXP3, IL10RA, IL10RB, STAT6 and TNFRSF18 genes in P. falciparum infection outcome among Congolese children. Earlier studies have identified some regulatory SNPs on the promoter loci of these five genes from 40 unrelated Gabonese individuals who had earlier infection episodes with various parasites, including P. falciparum[14,19-21]. These SNPs were then validated for their allelic gene expression using transient transfection assays. Some of the identified were already pre-described, while others were newly discovered. All the SNPs selected for the current investigation were found to alter expression of their respective gene [14,19-21]. Therefore any possible correlations of these SNP variants with resistance or susceptibility to mild malaria attacks and/or the control of P. falciparum infection levels were evaluated.
The results obtained in this study demonstrated that the STAT6 rs3024944C/C genotype was significantly more prevalent in asymptomatic children. This suggests the contribution of this genotype to protection of Congolese children against mild clinical malaria. The SNP rs3024944 is located in a transcription factor-binding site TCF-1 which is a member of family of genes with homology to high mobility group I (HMG) proteins [24]. TCF-1, in part with β-catenin, plays a pivotal role in T cell activation and is specifically expressed in T lymphocytes [25,26]. And the fact that in the uncomplicated malaria group, children with the minor allele rs3024944C have low parasite densities compared to those having the major allele rs3024944G, that speculates that the rs3024944C/C variant directs activation of T cells in response to P. falciparum infection, thus contributing to protection against the disease through parasite elimination. In haplotype level, the major haplotype CGG was found to be correlated with the marginal risk of uncomplicated malaria. This emphasizes the role of the SNP rs3024944 on susceptibly/resistance to uncomplicated malaria among Congolese children. This is the first study that shows the correlation between a promoter SNP STAT6 rs3024944 and protection against uncomplicated malaria. However, in previous investigations, the promoter SNP rs324013 was found to be associated with the control of S. haematobium infection levels in a Dogon population in Mali [17]. Similarly, the variant rs324015 was associated to the control of A. lumbricoides infection levels in Chinese children [18].
Two novel promoter SNPs were identified including, ss491228441 and ss491228440 of IL10RA and TNFRSF18 respectively. In both cases the frequency of the minor allele was low. Meanwhile the minor allele ss491228441G of IL10RA appeared to contribute to protection against uncomplicated malaria. Interestingly, the haplotype CTG involving this minor allele ss491228441G was significantly more present in asymptomatic children. This provides an additional evidence of the involvement of this allele in protection against uncomplicated clinical malaria. However, larger association studies in different malaria endemic areas including children and adults as well as functional studies of this ss491228441 SNP variant would be of great interest to verify the current observations.
The data also showed the association between the carriage of the FOXP3 rs11091253 SNP variant and P. falciparum parasite density levels. In particular, the rs11091253CT genotype was found to be associated with high infection levels, whereas the homozygous for the minor allele, the rs11091253TT genotype, was associated with lower infection levels. This suggests that FOXP3 contributes to the modulation of the immune response against P. falciparum infection. The SNP rs11091253 is located in a putative binding site for the transcription factors nuclear factor kappa B (NF-κB) and c-Rel, which is also a member of NF-κB family. These transcription factors have been demonstrated to be critical for regulation and transcription of FOXP3[27,28]. Therefore the differential binding properties of the promoter carrying the rs11091253CT genotype and that of the promoter carrying the rs11091253TT genotype may explain the associations observed in this study. One possible explanation may be that the binding of NF-κB and/or c-Rel with the promoter carrying the rs11091253C/T genotype results in upregulation of FOXP3 which in turn leads to increased number of fully functional Tregs. Consequently, parasite multiplication is facilitated by suppression of either P. falciparum-responsive effector cells or pro-inflammatory cytokine response. This is in line with what was reported by previous study showing that upregulation of FOXP3, and CD4+CD25+ regulatory cells correlates with more rapid parasite growth in human malaria infection [7].
Conclusion
To summarize, selected regulatory SNPs in the promoter region of FOXP3, IL10RA, IL10RB, STAT6 and TNFRSF18 genes have been investigated for possible association with uncomplicated malaria and high P. falciparum parasite density among Congolese children. These findings indicate that the STAT6 promoter polymorphism rs3024944 is highly associated with protection against uncomplicated malaria, whereas the FOXP3 promoter variant rs11091253 is more likely to be involved in the control of P. falciparum parasite density levels. In addition, a new SNP detected in the promoter region of IL10RA (ss491228441) contributes to protection against clinical malaria. These findings deserve to be further investigated in a larger population of children in the Republic of Congo but also in other malaria endemic areas.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
MN designed and performed the field study. VTP designed and supervised the experiments. FKK performed the experiments. FKK, HVT, and VTP analysed data. VTP, FN, and AAA contributed materials/analysis tools. FKK, FN, and VTP wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Felix Koukouikila-Koussounda, Email: felixkoukouikila@yahoo.fr.
Francine Ntoumi, Email: ffntoumi@hotmail.com.
Mathieu Ndounga, Email: ngoualandounga@yahoo.fr.
Hoang V Tong, Email: tong.van-hoang@uni-tuebingen.de.
Ange-Antoine Abena, Email: abena_cg@yahoo.fr.
Thirumalaisamy P Velavan, Email: velavan@medizin.uni-tuebingen.de.
Acknowledgements
We thank the team of CERVE for its contribution in the field study. We are grateful to Prof Pascal Rihet for his helpful advices on results analysis. We also thank Silvelia Grummes and Viola Galinat for the excellent technical help for sequencing. FKK was a recipient of a training grant from CANTAM network. This work has been supported through the CANTAM project. CANTAM (Central Africa Network on Tuberculosis, HIV/AIDs and Malaria) is a network of excellence supported by EDCTP and NACCAP.
References
- World Health Organization. World malaria report 2011. Geneva: World Health Organization; 2011. [Google Scholar]
- Maizels RM. Parasite immunomodulation and polymorphisms of the immune system. J Biol. 2009;8:62. doi: 10.1186/jbiol166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall DJ, Clegg JB. Genetic variability in response to infection: malaria and after. Genes Immun. 2002;3:331–337. doi: 10.1038/sj.gene.6363878. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol. 2005;6:353–360. doi: 10.1038/ni1181. [DOI] [PubMed] [Google Scholar]
- Hisaeda H, Maekawa Y, Iwakawa D, Okada H, Himeno K, Kishihara K, Tsukumo S, Yasutomo K. Escape of malaria parasites from host immunity requires CD4+ CD25+ regulatory T cells. Nat Med. 2004;10:29–30. doi: 10.1038/nm975. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Sakaguchi N. Regulatory T cells in immunologic self-tolerance and autoimmune disease. Int Rev Immunol. 2005;24:211–226. doi: 10.1080/08830180590934976. [DOI] [PubMed] [Google Scholar]
- Walther M, Tongren JE, Andrews L, Korbel D, King E, Fletcher H, Andersen RF, Bejon P, Thompson F, Dunachie SJ, Edele F, de Souza JB, Sinden RE, Gilbert SC, Riley EM, Hill AV. Upregulation of TGF-beta, FOXP3, and CD4+CD25+ regulatory T cells correlates with more rapid parasite growth in human malaria infection. Immunity. 2005;23:287–296. doi: 10.1016/j.immuni.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Velavan TP, Ojurongbe O. Regulatory T cells and parasites. J Biomed Biotechnol. 2011;2011:520940. doi: 10.1155/2011/520940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoratti FM, Farias L, Alves FP, Suarez-Mutis MC, Coura JR, Kalil J, Camargo EP, Moraes SL, Ramasawmy R. Variants in the toll-like receptor signaling pathway and clinical outcomes of malaria. J Infect Dis. 2008;198:772–780. doi: 10.1086/590440. [DOI] [PubMed] [Google Scholar]
- Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A, Ocheltree EL, Greenberg PD, Ochs HD, Rudensky AY. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci USA. 2006;103:6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Wing K, Miyara M. Regulatory T cells - a brief history and perspective. Eur J Immunol. 2007;37(Suppl 1):S116–S123. doi: 10.1002/eji.200737593. [DOI] [PubMed] [Google Scholar]
- Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Powrie F. Emerging challenges in regulatory T cell function and biology. Science. 2007;317:627–629. doi: 10.1126/science.1142331. [DOI] [PubMed] [Google Scholar]
- Velavan TP, Buyukyazici B, Kremsner PG, Kun JF. Combined promoter haplotypes of the IL10R genes are associated with protection against severe malaria in Gabonese children. Immunogenetics. 2012;64:87–95. doi: 10.1007/s00251-011-0563-3. [DOI] [PubMed] [Google Scholar]
- Gasche C, Grundtner P, Zwirn P, Reinisch W, Shaw SH, Zdanov A, Sarma U, Williams LM, Foxwell BM, Gangl A. Novel variants of the IL-10 receptor 1 affect inhibition of monocyte TNF-alpha production. J Immunol. 2003;170:5578–5582. doi: 10.4049/jimmunol.170.11.5578. [DOI] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hätscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Isnard A, Kouriba B, Cabantous S, Dessein A, Doumbo O, Chevillard C. A STAT6 gene polymorphism is associated with high infection levels in urinary schistosomiasis. Genes Immun. 2008;9:195–206. doi: 10.1038/gene.2008.2. [DOI] [PubMed] [Google Scholar]
- Peisong G, Yamasaki A, Mao XQ, Enomoto T, Feng Z, Gloria-Bottini F, Bottini E, Shirakawa T, Sun D, Hopkin JM. An asthma-associated genetic variant of STAT6 predicts low burden of ascaris worm infestation. Genes Immun. 2004;5:58–62. doi: 10.1038/sj.gene.6364030. [DOI] [PubMed] [Google Scholar]
- Hanel SA, Velavan TP, Kremsner PG, Kun JF. Novel and functional regulatory SNPs in the promoter region of FOXP3 gene in a Gabonese population. Immunogenetics. 2011;63:409–415. doi: 10.1007/s00251-011-0524-x. [DOI] [PubMed] [Google Scholar]
- Velavan TP, Bechlars S, Huang X, Kremsner PG, Kun JF. Novel regulatory SNPs in the promoter region of the TNFRSF18 gene in a Gabonese population. Braz J Med Biol Res. 2011;44:418–420. doi: 10.1590/S0100-879X2011007500036. [DOI] [PubMed] [Google Scholar]
- Velavan TP, Bechlars S, Tomiuk J, Kremsner PG, Kun JF. Molecular characterization of regulatory polymorphisms in the promoter region of the STAT6 gene in a Gabonese population. Mem Inst Oswaldo Cruz. 2011;106:65–69. doi: 10.1590/S0074-02762011000100011. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fortin A, Stevenson MM, Gros P. Susceptibility to malaria as a complex trait: big pressure from a tiny creature. Hum Mol Genet. 2002;11:2469–2478. doi: 10.1093/hmg/11.20.2469. [DOI] [PubMed] [Google Scholar]
- Castrop J, van WD, Koomans-Bitter M, van de Wetering M, de WR, van DJ, Clevers H. The human TCF-1 gene encodes a nuclear DNA-binding protein uniquely expressed in normal and neoplastic T-lineage lymphocytes. Blood. 1995;86:3050–3059. [PubMed] [Google Scholar]
- Staal FJ, Burgering BM, van de Wetering M, Clevers HC. Tcf-1-mediated transcription in T lymphocytes: differential role for glycogen synthase kinase-3 in fibroblasts and T cells. Int Immunol. 1999;11:317–323. doi: 10.1093/intimm/11.3.317. [DOI] [PubMed] [Google Scholar]
- Yu Q, Sharma A, Sen JM. TCF1 and beta-catenin regulate T cell development and function. Immunol Res. 2010;47:45–55. doi: 10.1007/s12026-009-8137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S. c-Rel: a pioneer in directing regulatory T-cell lineage commitment? Eur J Immunol. 2010;40:664–667. doi: 10.1002/eji.201040372. [DOI] [PubMed] [Google Scholar]
- Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]