Abstract
Homeobox genes represent a family of highly conserved transcription factors originally discovered to regulate organ patterning during development. More recently, several homeobox genes were shown to affect processes in adult tissue, including angiogenesis and wound healing. Whereas a subset of members of the Hox-family of homeobox genes activate growth and migration to promote angiogenesis or wound healing, other Hox genes function to restore or maintain quiescent, differentiated tissue function. Pathological tissue remodeling is linked to differential expression of activating or stabilizing Hox genes and dysregulation of Hox expression can contribute to disease progression. Studies aimed at understanding the role and regulation of Hox genes have provided insight into how these potent morphoregulatory genes can be applied to enhance tissue engineering or limit cancer progression.
Keywords: Hox genes, angiogenesis, endothelial cells, tumor vascularization, inflammation
Introduction
Delivery of nutrients and the removal of anabolites by the vascular network are critical for the viability of biological tissues. For this reason, nearly every cell in the human body lies at a distance no further than 100–200 µm from a blood vessel, which correlates with the mathematically-calculated limit at which oxygen tension reaches a critical level for cell survival.1,2 Not surprisingly, regulation of vascular development is closely tied to tissue architecture, and changes in tissue homeostasis during development and in pathological conditions trigger changes in the supporting vasculature.
Vascularization typically occurs via two processes: vasculogenesis and angiogenesis. Vasculogenesis is the de novo formation of blood vessels that arises from the coalescence of angioblasts to form the primary capillary plexus.3 Vasculogenesis occurs largely during embryonic development; however, the presence of a population of circulating endothelial progenitor cells (EPCs) originating from the bone marrow, suggests that it may also occur in the postnatal stages of life.4,5 Conversely, angiogenesis is the sprouting of vessels from existing vasculature and occurs during development and throughout life. This process involves endothelial cells (ECs) degrading the basement membrane of the parent vessel, collectively migrating through the provisional extracellular matrix (ECM) and subsequently establishing mature neovessels stabilized by pericytes.6 This requires the coordinated expression of cytokines, proteases, adhesion molecules and other cell cycle regulatory factors. During embryonic development, angiogenesis is the means by which vessels expand from the primary capillary plexus into an extensive, closed circulatory network. Excluding corpus luteum formation and endometrium redevelopment, angiogenesis generally does not occur in adults, but is initiated during wound healing or in response to tumor growth.
Activation of angiogenesis under pathological conditions is controlled by what Judah Folkman termed the “angiogenic switch,”7,8 which was originally coined to describe how tumors generate an angiogenic stimulus to support their growth and viability. This concept states that at homeostasis, there exists a balance between pro-angiogenic and anti-angiogenic factors, but under pathological conditions, expression of pro-angiogenic factors tilts the balance toward vessel formation, and conversely, when the process is complete the anti-angiogenic factors prevail, resulting in vessel pruning and regression. Tumors were shown to mimic features of healing wounds by producing copious amounts of angiogenic factors (e.g., VEGFA and FGF2) and macrophage chemoattractants (e.g., CCL2 and CSF1).8-10 Macrophage infiltration at the tumor site augments the pro-angiogenic environment by increasing the local availability of pro-angiogenic cytokines.11-17 However, exogenous addition of anti-angiogenic factors, such as thrombospondin 1, causes vessel regression by reversing the tilt of the angiogenic switch.18 Restoration of tumor suppressor genes that limit tumor growth and subsequent metabolic demand can also shift the balance toward an anti-angiogenic environment.19,20
Understanding the mechanisms that modulate the angiogenic switch during normal wound healing and pathological vascularization has been the subject of intense investigation by researchers with the aim of developing novel approaches to either drive therapeutic angiogenesis or impair this process as a means of limiting tumor growth. Much attention has focused on a few well-established factors, particularly VEGFA, in driving angiogenesis, but inconsistent results achieved when applying anti-VEGFA agents to different tumor types have underscored the need to investigate the contribution of numerous other factors that drive pathological angiogenesis.21 In this review we will focus on the role of homeobox genes during wound and tumor angiogenesis. These morphoregulatory genes, which are intimately linked to tissue architecture and homeostasis, have shown tremendous therapeutic potential to activate angiogenesis in poorly healing wounds or limit neovascularization in aggressive tumors.
Homeobox Genes
Homeobox genes encode a family of highly conserved transcription factors that were originally discovered to be essential for segmental identity in Drosophila.22 Further research showed that homeobox genes were highly conserved throughout the animal kingdom and acted as master regulators of tissue and organ patterning.23
Homeobox genes are divided into families and subfamilies based on the similarity of their DNA-binding homeodomains. Most families contain between two to nine genes; these include the Msx, En, Nkx, Pax and Dlx families. However, the largest, and most extensively studied, family of homeobox genes is the class I Hox family, which includes 39 members in mammals. The Hox genes are arranged in clusters—with eight genes clustered on a single chromosome in Drosophila, while mammals have 39 genes grouped in clusters of 9–13 genes on each of four chromosomes. During vertebrate development, there exists a strict temporospatial control of Hox gene expression in a manner such that 3′ Hox genes are expressed prior to 5′ genes within a given cluster.24 This expression pattern is responsible for controlling segmental identity and morphology on the anteroposterior axis; 3′ Hox genes are associated with the development of anterior or rostral tissues and 5′ genes are associated with posterior or caudal tissues. Mutations or deletions that lead to misexpression of Hox genes have shown to result in homeotic transformations in Drosophila and mammals, in which anterior features develop in posterior regions and vice versa.22,25,26 The finding that Hox genes are arranged into highly conserved chromosomal clusters, with the lowest density of interspersed repeats in the entire human genome,27 is most likely due to the need to preserve cis-regulatory elements to maintain this temporospatial control.
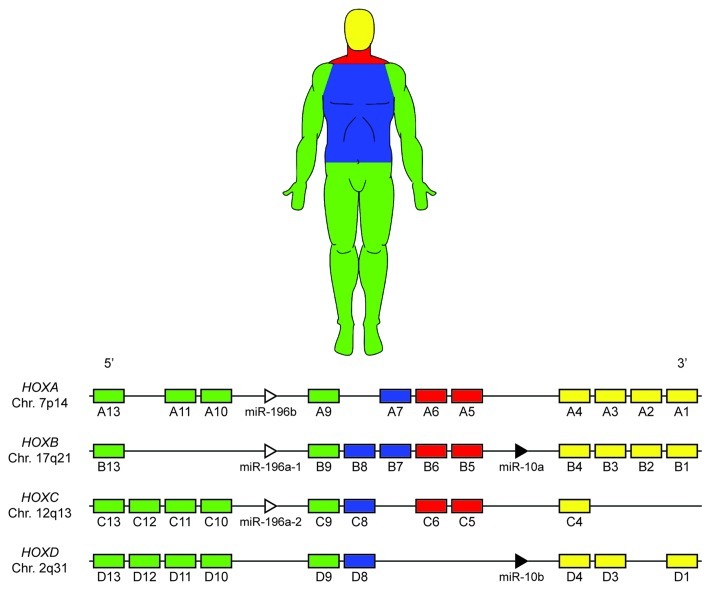
In humans, the 39 mammalian Hox genes are arranged into four chromosomal clusters; designated HOXA, HOXB, HOXC and HOXD, located on chromosomes 7p14, 17q21, 12q13 and 2q31, respectively. Each of these 39 genes is assigned to one of 13 paralog groups, with no single cluster containing a gene from each group (Fig. 1). Hox paralogs, defined by their homeodomain sequence, occupy the same relative positions on a given chromosomal cluster (e.g., HOXA3, HOXB3 and HOXD3).28 Paralogous group members have been shown to exhibit some level of functional redundancy, as they are able to perform compensatory functions during development upon mutation or deletion of a fellow group member.29-34 In these experiments, mice that are double or triple mutants for paralogous Hox group members display more complete and severe phenotypes than single mutants of any of these paralogs. Functional redundancy between non-paralogous, neighboring Hox genes,35-37 as well as non-paralogous genes in separate clusters has also been documented.38
Figure 1. Mammalian Hox clusters. Thirty-nine Hox genes are arranged into four distinct chromosomal loci. During human development Hox genes are activated in a 3′ to 5′ manner within each cluster, concomitant with the development of the anteroposterior axis; Hox gene activation is color-coded based approximately on anatomical location during development. Note the presence of two miRNA families (MIR10 and MIR196) within the Hox clusters, which target many Hox genes and may contribute to the posterior prevalence phenomenon.
Pro-Angiogenic Homeobox Genes
Studies over the last decade have clearly defined a role for the homeobox genes in regulating neovascularization during both embryonic vasculogenesis and pathological angiogenesis in adults.
The initial phases of embryonic vascular development consist of the formation of blood islands through the proliferation and differentiation of mesodermal hemangioblasts into angioblasts and hematopoietic progenitor cells.3 Fusion of these blood islands results in the formation of the primary capillary plexus and the primitive heart tube.
Early studies by Chisaka and Capecchi linked Hox function to embryonic vascular development in vertebrates.39 They observed that Hoxa3 (originally referred to as Hox1.5)-knockout mice died at or shortly after birth, with abnormalities including an absent right carotid artery and irregularly sized heart compartments. These effects could be ameliorated by inserting Hoxd3 within the Hoxa3 locus, suggesting that the paralogs are somewhat interchangeable if expressed “properly” in space and time.34
In addition, the non-Hox homeobox genes Prx1 and Prx2 are normally expressed in the vessel wall during development. Prx1−/− mice displayed an irregularly curved aortic arch along with a misdirected and elongated ductus arteriosus.40 In contrast, Prx2−/− mice did not display any vascular deformities. However, Prx1−/− Prx2−/− double mutant mice presented similar, but more pronounced, abnormalities as the Prx1−/− mice. Interestingly, although the great arteries were malformed in the double mutants, the heart developed normally compared with wild-type mice. Additionally, vascular smooth muscle cells (VSMCs), which represent the major cellular component of the vessel wall, were found to express HOXA5, HOXA11, HOXB1, HOXB7 and HOXC9 in fetal tissue, while Hox gene expression was absent in their adult counterparts, suggesting their importance during early vascular development.41
Subsequent studies revealed that homeobox genes, at least partially, direct the differentiation of EPCs into ECs. Transactivation of Vegfr2 occurs via cis-acting Hoxb5 in embryoid bodies, resulting in their differentiation into angioblasts. These angioblasts expand and further differentiate into an endothelial phenotype.42 We have also profiled the dynamic expression of several Hox genes during the differentiation of embryonic stem cells into ECs in culture.43 Early expression of HOXA3 and HOXD3 was concomitant with the emergence of an EC-phenotype and upregulation of pro-angiogenic factors such as CCL2, MMP14 and the β3 integrin subunit. Furthermore, Hoxa3 is critical in maintaining the endothelial phenotype in embryonic vasculature and preventing its hematopoietic differentiation.44 At later time points, HOXA3 and HOXD3 are downregulated while the anti-angiogenic HOXA5 and HOXD10 are upregulated. This corresponds with the dowregulation of the aforementioned pro-angiogenic factors and the expression of the potent angiogenesis inhibitor thrombospondin 2. A separate study identified HOXA9 as a master regulator of EC differentiation from progenitor cells by directly enhancing transcription of VEGFR2 and eNOS.45
The non-Hox homeobox gene Hex plays a complex role in vascularization. During embryonic vasculogenesis, Hex is expressed in the blood islands, and is required for endothelial and hematopoietic differentiation.46 Hex-knockout mice displayed embryonic lethality typified with several defects, including impaired vasculogenesis.47 However, Hex inhibits EC tube formation in culture through the downregulation of angiogenic factors such as VEGFR1, VEGFR2, TIE1, TIE2 and the αv integrin subunit,48,49 suggesting that Hex promotes vasculogenesis rather than angiogenesis, or that other factors are integral in determining the outcome of Hex signaling (the anti-angiogenic effects of homeobox genes will be subsequently detailed in this review). The precise hierarchy and interactions between each of these Hox factors during EC differentiation remains to be established, but suggests a complex network of factors required for EC development.
Hox genes also serve important roles in vascularization post-development in response to injury or other pathological conditions. HOXD3, through binding of cofactor PBX1, directly enhances EC expression of the α5 and β3 integrin subunits in response to FGF2.50-52 Heterodimerization of these subunits with separate subunits form the fibronectin receptors α5β1 and αvβ3, which allow adhesion and migration through fibronectin-rich matrices found in wound healing and tumor microenvironments. HOXD1 was recently shown to provide a synergistic effect to this by directly enhancing the EC expression of the β1 integrin subunit.53 Disruption of HOXD3 expression inhibited the expression of the α5 and β3 integrin subunits, and was sufficient to inhibit angiogenesis, consistent with previous findings that these integrins are essential for angiogenesis.54,55 Additionally, HOXD3 upregulated the expression of the serine protease urokinase plasminogen activator (uPA), which locally activates plasmin during the degradation of the provisional matrix,50 suggesting that EC HOXD3 is associated with the invasive phases of angiogenesis. Subsequent studies revealed that the paralogous Hox members Hoxa3 and Hoxb3 also display pro-angiogenic capabilities, albeit through distinct pathways, consistent with the notion of functional redundancy among Hox paralogs.34 EC expression of Hoxa3 correlated with increased EC migration and expression of the proteases uPA receptor (uPAR) and Mmp14,56 while HOXB3 increased the expression of the angiogenic ligand ephrin A1 and induced capillary morphogenesis in endothelial sprouts.57 Because adult angiogenesis is generally limited to pathological conditions, it is not surprising that these pro-angiogenic HOX3 paralogs are not normally expressed in quiescent vasculature, but are induced in response to cytokines linked to tissue injury or tumors.50,51,56,58
Originally thought to be a potential inhibitor of angiogenesis, HOXA9 was actually found to promote it. Early studies found that post-transcriptional modification in ECs generated a HoxA9 isoform that was unique from that found in fetal tissue.59 This isoform was found to be downregulated in response to the pro-angiogenic TNFα, and therefore, thought to possibly suppress angiogenesis. Subsequent studies however, found that HOXA9, in fact, promotes EC migration and tube formation in culture through direct upregulation of the Ephrin receptor B4 (EPHB4),60 along with direct or indirect upergulation of eNOS and VEGFR2.45
Anti-Angiogenic Homeobox Genes
Angiogenesis is a highly regulated process that consists of many control mechanisms that temporally initiate and terminate the progression through its different phases. In contrast to the pro-angiogenic genes described above, a subset of homeobox genes normally acts to terminate the angiogenic response and/or maintain the vasculature in a quiescent state.
The 5′ Hox gene HOXD10 was shown to be expressed in normal, quiescent vascular endothelium, but not in highly angiogenic, tumor-associated vasculature.20 Overexpression of HOXD10 in ECs mitigated their angiogenic response to FGF2 and VEGFA in two and three-dimensional cultures. Furthermore, HOXD10-expressing ECs showed a marked decrease in the angiogenic factors RHOC, β4 and α3 integrins, MMP14, uPAR and cyclin D1. The expressions of the pro-angiogenic HOXA3, HOXB3 and HOXD3 were not altered, suggesting that HOXD10 functions to mute the downstream effects of those genes, rather than attenuate their expression.
In addition to its direct anti-angiogenic effect on ECs, HOXD10 also attenuates tumor angiogenesis by acting within tumorigenic epithelial cells to reduce expression of angiogenic factors including VEGFA.61 Restoring expression of HOXD10 in mammary tumor cells, from which it is frequently lost, reestablished polarity and the formation of acinar structures in three-dimensional cultures and reduced EC recruitment.61 It is also worth noting that the ability of HOXD10 to reduce VEGFA expression was not due to a direct inhibition of VEGFA transcription, or interference with binding of the transcriptional activator HIF1A, but arose indirectly through the re-establishment of tissue polarity. When HOXD10 was expressed, but tissues were prevented from adopting basolateral polarity, expression of VEGFA and other angiogenic factors remained high. Thus, the ability of Hox genes to modulate the angiogenic capacity of tissues can arise directly through transcriptional modulation of angiogenic effector genes, or indirectly via the Hox mediated changes in tissue architecture.
The findings that HOXD10, whether expressed in epithelial or endothelial cells has a similar inhibitory effect on angiogenesis, also highlights the fact that Hox genes have the capacity to coordinate the responses of different cell populations within a tissue. These coordinated actions represent one means by which these morhpregulatory genes can work to collectively maintain differentiated tissue function.
HOXA5 is another gene expressed in quiescent, but not activated, endothelium.62 Similar to HOXD10, HOXA5-expressing ECs did not respond to the angiogenic stimuli provided by either VEGFA or FGF2. Among the angiogenic genes that were downregulated in ECs were VEGFR2, ephrin A1, HIF1A and COX2. Correspondingly, these cells also upregulated the expression of the anti-angiogenic thrombospondin 2. Further evidence demonstrated that EC HOXA5 stabilized adherens junctions through β-catenin retention.63 Presumably, this prevents endothelium dissociation in the parent vessel—the first step in angiogenesis. Interestingly, paralogous Hoxb5 in ECs was shown to promote both vasculogenesis42 and angiogenesis,64 counterintuitive to the paralog redundancy theory.
The homeobox gene GAX (MEOX2) is also largely confined to quiescent cardiovascular tissue in adults. Originally found to induce G0/G1 cell cycle arrest in VSMCs,65,66 GAX, and its related gene MEOX1, were also found to be expressed in the endothelium and induced similar growth arrest in ECs.67,68 Additionally, GAX-expressing ECs, through NFκB inhibition, exhibited an impaired angiogenic response to stimuli by VEGFA, FGF2 and TNFα.69 Translation of both GAX and HOXA5 is directly regulated by MIR130A (located outside of the Hox clusters).70 EC expression of MIR130A downregulates these homeobox genes and effectively restores EC activation by angiogenic stimuli.
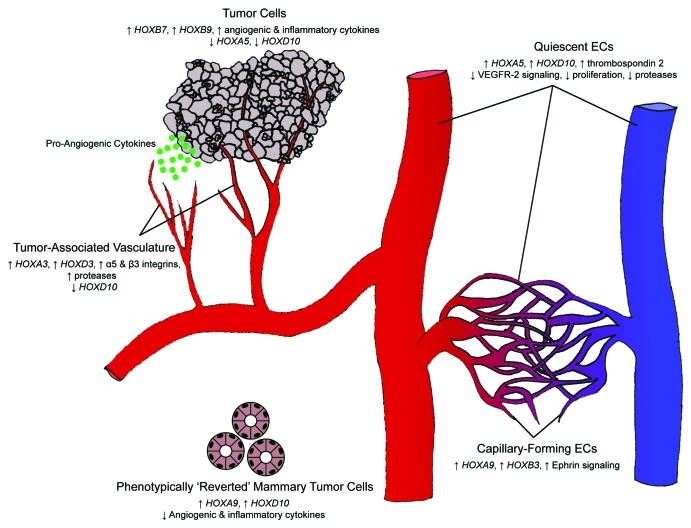
Thus, the ability of tissues to activate the angiogenic switch relies upon upregulation of pro-angiogenc genes and suppression of anti-angiogenic or vascular-stabilizing genes (Fig. 2).
Figure 2. Hox gene control of the angiogenic switch. Differential expression of Hox genes is key in transitioning between homeostasis and angiogenesis. During homeostasis, HOXA5 and HOXD10 upregulate anti-angiogenic cytokines and inhibit pro-angiogenic signals, thereby maintaining a quiescent endothelial phenotype. Tumor cells can initiate an angiogenic response as a result of a change in Hox gene expression. Loss of HOXA5 and HOXD10 expression (along with the upregulation of HOXB7 and HOXB9) leads to the secretion of pro-angiogenic and pro-inflammatory cytokines. Responsive ECs subsequently downregulate the expression of HOXD10 and upregulate the expression of HOXA3 and HOXD3, thereby assuming an invasive phenotype. Tumor-associated endothelium maintains this invasive phenotype, whereas normal angiogenesis is an ordered process that concludes with a closed capillary network. ECs undergoing capillary morphogenesis express HOXA9 and HOXB3 and are phenotypically distinct from their invasive counterparts.
Homeobox Genes in Wound Healing
Wound healing is the coordinated process by which the body repairs damaged tissue and is generally divided into three overlapping phases: inflammation, proliferation and remodeling.71 In the initial inflammatory phase, the body functions to prevent blood loss through vasoconstriction, platelet aggregation and fibrin clot formation—a process known as hemostasis. Platelet secretion of inflammatory cytokines recruits neutrophils and macrophages, which function to remove foreign debris from the wound site. Macrophages also initiate the transition to the proliferative phase by secreting factors that recruit fibroblasts, which in turn secrete collagen and fibronectin at the wound site. Concurrently, ECs undergo angiogenesis within this freshly-laid granulation tissue. Differentiated myofibroblasts contract the wound margins and epithelial cells migrate across the vascularized granulation site to close the wound. Epithelialization of the wound bed does not occur over necrotic tissue;72 therefore, angiogenesis is critical in this phase and poorly healing wounds have been linked to impaired angiogenic responses. The remodeling phase, which can last years, is characterized by myofibroblasts depositing and reorganizing collagen fibers so as to increase tissue tensile strength.
Given the essential role of angiogenesis in wound healing, it is not surprising that several homeobox genes contribute to effective wound repair. Gene delivery of the pro-angiogenic Hoxd3 accelerated wound closure in poorly healing diabetic mice and was accompanied by a robust induction of angiogenesis.58 As transgene uptake is evident in ECs, as well as keratinocytes and fibroblasts, wound closure was accompanied by increased collagen (Col1a1) production mediated by fibroblasts. Similar to Hoxd3, the paralogous Hoxa3 was found to be upregulated in ECs in a wound healing mouse model, whereas diabetic mice were unable to induce its expression;56 Hoxa3 transfer into the wounds of these diabetic mice significantly improved angiogenesis and wound closure. Mechanistically, in addition to the angiogenic response by ECs detailed previously, HOXA3 promoted uPAR-dependent keratinocyte migration and subsequent epithelialization—a feature that was not directly enhanced by HOXD3. Furthermore, HOXA3 expression within wound tissue recruited bone marrow-derived progenitor cells,73 and promoted their differentiation into pro-angiogenic, CD11b+ Gr-1+ myeloid cells.74 The synergistic functions of HOXA3 and HOXD3 in wound healing mimic their complementary roles in angiogenesis, and further support the functional redundancy concept conceived from double Hoxa3/Hoxd3-knockout mice.29
While wound contraction is a normal part of wound healing, excessive or prolonged contraction leads to diminished tissue function and appearance. Enhanced angiogenesis within the wound bed reduces the contractile activity of myofibroblasts and scarring.75 Building from this concept, we have engineered composite skin grafts consisting of a dermal fibroblast layer, which produces a secretable form of HOXA3, overlaid with an epidermal keratinocyte layer. Following wounding and engraftment onto mice, the secretable HOXA3 is taken up by keratinocytes and ECs within the wound tissue, and these grafts displayed increased angiogenesis and reduced contraction compared with controls (unpublished observations, manuscript submitted). Additionally, cells within the graft produced less inflammatory cytokines, consistent with the reduced inflammatory response in wounds treated with the HOXA3 transgene.
In contrast to HOXA3 and HOXD3, the 5′ Hoxb13 was found to impair wound healing.76,77 Hoxb13 transgenic mice displayed increased fibrin clot and immune cell retention at the wound site, leading to a prolonged inflammatory response.77 Additionally, increased expression of VEGFA and TNFα in the wounds of these transgenic mice may be responsible for the formation of irregular, enlarged vasculature. Forced expression of anti-angiogenic HOXA5 or HOXD10 in wounds also impairs angiogenesis and healing (unpublished observations), but whether these genes are normally sustained in poorly healing or chronic wounds that are deficient in angiogenesis, remains to be seen.
Angiogenic Hox Genes and Pathological Angiogenesis
Whereas endogenous HOXA3 and HOXD3 can contribute to wound-induced angiogenesis, diseases characterized by the formation of excessive vasculature, including endotheliomas and tumor vascularization, are often the result of the sustained expression of pro-angiogenic Hox genes.
One of the cornerstones of physiological angiogenesis is the transition from vessel invasion to vessel maturation. ECs in the maturation phase show decreased proliferation relative to their invasive counterparts, and also intimately associate with supporting pericytes, which function to regulate vessel tone and leakage. As mentioned, EC HOXD3 expression correlates with the invasive angiogenic phenotype, and correspondingly, Hoxd3 overexpressing vessels formed in the chick chorioallantoic membrane (CAM) assay did not mature, but formed large endothelima-like masses.50 This result is in agreement with the finding that HOXD3 is expressed in proliferating hemangiomas, but not involuting ones.78 Similarly, tumor-associated vasculature, which is typically comprised of immature, leaky vessels, also highly expressed HOXD3.51 It is also worth noting that HOXA5, which can stabilize vessels and reduce permeability, is absent in both endothelioma tissues and tumor vessels,62,63 suggesting that the unopposed actions of HOXD3 contribute to aberrant vascular morphology. Indeed, ectopic expression of HOXA5 in endotheliomas leads to a reduction in cavernous cyst formation and restores the phenotype of normal, differentiated vessels.79
In addition, to promoting an angiogenic phenotype within the EC compartment, Hox gene expression within tumor cells can further sustain a pro-angiogenic response. An early study found that HOXB7 was expressed in each of 25 melanoma cell lines, as well as in primary melanoma tumors and their metastases.80 Furthermore, HOXB7 was found to increase expression of the angiogenic factor FGF2. This observation was also evident in other glioblastoma, carcinoma and leukemia cell lines; introduction of HOXB7 to the non-HOXB7-expressing SkBr3 breast cancer cell line also lead to increased FGF2 expression.81 A subsequent study by this group found that the HOXB7-transduced SkBr3 cells also upregulated the expression of additional pro-angiogenic factors including VEGFA, IL-8 and CXCL1.82 ANG2 is also elevated in these cells, which, in conjunction with sustained VEGFA expression, produces a robust angiogenic response.83 Correspondingly, tumor explants of SkBr3-HOXB7 xenografts revealed greater levels of vascularization than their control counterparts.82 Another group found that HOXB9 expression was elevated in several breast carcinoma biopsies.84 Culture studies with HOXB9-transduced MCF10A cells revealed increased expression of VEGFA, FGF2, IL8 and ANGPTL2—nearly identical to the effect of the non-paralogous HOXB7. Functionally, these cells, with an additional HRAS mutation, rapidly gave rise to large, vascularized tumors compared with MCF10A cells displaying the HRAS mutation alone. In addition, the HOXB9-expressing cells developed several lung metastases, whereas non-expressing cells developed none—a likely consequence of the enhanced angiogenesis induced by HOXB9.
Homeobox Genes and Regulation of Inflammatory Responses
During the inflammatory phase of wound healing, recruited leukocytes can initiate or augment an angiogenic response through the secretion of several pro-angiogenic factors.85 In addition to directly stimulating ECs to undergo angiogenesis, tumors also frequently induce an indirect angiogenic response via leukocyte recruitment. Macrophages are recruited to the tumor microenvironment via tumor secretion of chemotactic factors such as CCL2 and VEGFA.10 Once within the microenvironment, macrophages increase the local concentration of VEGFA through de novo secretion13 and by catalyzing the release of the matrix-bound form through protease secretion.86 Additionally, a different subset of leukocytes, responding to primary tumor secretion of VEGFA, TNFα and TGFβ, was found to colonize lung tissue and prime it for the seeding of metastases.87-89 As detailed throughout this review, homeobox genes regulate the expression of several key leukocyte chemoattractants.
Tumor expression of HOXB7 or HOXB9 has correlated with an increase in VEGFA, and the neutrophil attractants CXCL1 and IL8.80-82,84 HOXB13-mediated inflammation within wound tissue is characterized by neutrophil infiltration and the development of irregular vasculature.77 Additionally, HOXA9 was found to bind the CCL2 promoter and enhance its transcription in individuals displaying a single nucleotide polymorphism within the promoter region.90 Conversely, we have found the anti-angiogenic HOXD10 reduces tumor cell production of CCL2 and CXCL12, and abates leukocyte recruitment in culture (unpublished data). Interestingly, while HOXA3 also attenuates recruitment of CD45+ cells in wound tissues, it selectively promotes recruitment/differentiation of endothelial precursors and pro-angiogenic myeloid cells.73,74
Given that homeobox genes play a fundamental role in regulating tissue remodeling and organogenesis, it is not surprising that they can profoundly alter the immune microenvironment to facilitate tissue remodeling or maintain normal homeostasis. Interestingly, Hox genes have also been implicated in directly regulating the phenotype of immune cells exposed to activating stimuli,91 thereby suggesting a broader role for Hox genes in regulating the inflammatory milieu in remodeling tissues (for comprehensive reviews on the role of Hox genes on immune cell function, see refs. 92 and 93).
Regulation of Homeobox Genes
Considering the powerful role of Hox genes in normal and pathological tissue responses, it follows that strict regulatory mechanisms have evolved to maintain the correct spatiotemporal expression pattern of Hox genes. Hox genes are commonly regulated epigenetically via histone methylation and acetylation. Chromatin trimethylation by trithorax-group (TrxG) proteins are associated with activating and maintaining Hox gene expression, while trimethylation by polycomb-group (PcG) proteins are known to repress it.94-97 Whereas pluripotent stem cells do not display any significant Hox gene expression, cell lineage commitment is linked to changes in PcG and TrxG genes leading to tissue specific patterns of Hox gene expression.98,99 Several studies have also identified significant roles for TrxG and PcG proteins in regulating expression of Hox proteins in adult cells in the context of angiogenesis and wound healing.
Expression of HOXA9 during EPC differentiation and angiogenesis is reliant on histone deacetylase (HDAC) activity,45 while upregulation of both HOXA9 and HOXD3 has been shown to occur via histone trimethylation by the TrxG protein MLL.100 The expression of the PcG gene MEL18 (PCGF2) is linked to an upregulation in VEGFR2 and ANGPTL2, with a corresponding increase in EC migration and tube formation;101 however, its downstream effectors and global function is still debated, as another group was able to show that MEL18 exhibits anti-angiogenic activity.102
The PcG genes Eed, Ezh2 and Suz12 are downregulated in wound epidermis, corresponding with a decrease in target histone trimethylation.103 Although this study did not explore the effects of this downregulation on Hox gene expression, other reports have demonstrated that protein products of these three PcG genes assemble to form the polycomb repressor complex 2 (PRC2) and attenuate the expression of HOXC8 and HOXA9.104-106 In separate studies, HOXC8 expression has been associated with the promotion of wound epithelialization by keratinocytes,107 while decreased expression of HOXA9 was found in keloid fibroblasts that are associated with aberrant wound healing.108
Deregulation of Hox Genes and Oncogenesis
Although mutations in Hox genes, per se, are relatively rare, the regulatory mechanisms that control Hox gene expression are common targets of mutagenesis, and the subsequent misexpression or deregulation of Hox genes is associated with the formation or progression of several tumors.109-115
An exhaustive study of Hox gene expression in several esophageal carcinomas revealed abnormally high expression of several 5′ Hox genes in a tissue that normally displays low levels of 5′ Hox genes and high levels of 3′ Hox genes.112 Similarly, expression of the 5′ HOXB13 is lost in colorectal tumor cells, leading to increased cell proliferation,116,117 The 3′ HOXB7 was shown to directly upregulate EGFR and confer tamoxifen resistance to estrogen receptor-positive breast cancer cells,118 while loss of the 5′ HOXA9 and HOXD10 are frequently associated with breast cancer progression.109,119 Contrastingly, overexpression of HOXA9 in several leukemias correlates with chemotherapy resistance and poor patient prognosis. Transgenic studies with Hoxa9−/− mice demonstrated an inhibition of leukemogenesis, but also impaired normal hematopoiesis in the process;120 Hoxa9+/− mice developed normally with no leukemogenesis, demonstrating a dose-dependent response of Hox gene expression mediating the shift between organogenesis and oncogenesis.121 Thus, the potential of HoxA9 to act as either a tumor suppressor in breast tissue or an oncogene in leukocytes underscores the tissue-specific and context-dependent function of Hox proteins. Whether tissue-specific repertoires of Hox cofactors and/or differences in tissue architecture (adherent vs. circulating cells) contribute to the differential responses has not been directly addressed. It is worth noting again, that the ability of HOXD10 to reduce VEGFA expression and revert the tumorigenic phenotype of breast epithelial cells is highly context-dependent, as it requires cell adhesion to the basement membranes and acquisition of apical-basal polarity.61
As mentioned in the previous section, the expression of several Hox genes is regulated by histone trimethylation by TrxG or PcG proteins. Changes in chromatin conformation allow CpG motifs in Hox promoter regions to be further methylated or demethylated. Epigenetic deregulation occurs when members of this regulatory process become mutated or misexpressed. The TrxG gene MLL is mutated in some leukemias, causing irregular histone methylation and increased expression of HOXA4, HOXA5, HOXA7 and HOXA9.114,115 The inactivation or loss of the tumor suppressor gene P16 was shown to upregulate the PRC2 proteins and consequently repress HOXA9 in mammary epithelial cells, thereby perpetuating tumorigenesis.106
Additionally, miRNAs that regulate Hox expression may also have altered expression patterns in tumor cells through methylation or other means.122,123 For instance, upregulation of MIR10B in mammary tumor cells inhibits translation of HOXD10 mRNA and produces robust invasion and metastasis.124
Hox gene dysregulation arises through a variety of mechanisms and produces a wide array of aberrant responses in several tumor types; it is still unclear whether Hox dysregulation is a cause or consequence of tumor progression. However, in many instances, acquisition of the tumorigenic phenotype by Hox misexpression results in the upregulation of angiogenic factors that act on adjacent ECs and alter expression of endogenous Hox genes, leading to the formation of new vessels (for a comprehensive review of Hox genes and cancer, see ref. 125).
It is not currently known whether any of the previously mentioned regulatory mechanisms are linked to Hox gene misexpression in non-tumorigenic tissues such as endotheliomas, which exhibit constitutive HOXD3, or in poorly healing wounds, which fail to induce Hoxd3 or Hoxa3.50,56,58
Determinants of Functional Dominance among Hox Genes
During embryogenesis, Hox genes are expressed in a sequentially overlapping pattern with 3′ Hox genes activated prior to 5′ genes, corresponding with anteroposterior axis development. In anatomical regions where the expression of non-paralogous Hox genes overlap, the more 5′ paralogs generally tend to exert dominance over the more 3′ paralogs. This occurrence is thought to be the reason why Hox gene mutations commonly result in homeotic defects found in locations anterior to the normal expression domain of the mutant Hox genes, but not posterior to it.25,126,127 These mechanisms remain active in adult tissue, as observed in ECs expressing HOXA3 and HOXD10. Sustained expression of the 5′ HOXD10 inhibits the expression of angiogenic factors such as uPAR and MMP14,20 both of which are upregulated by the 3′ HOXA3.56
This phenomenon, termed “phenotypic suppression” in invertebrates126,128 and “posterior prevalence” in vertebrates,129 is believed to be part of an ancient, poorly understood, cross-regulatory mechanism, in which posterior Hox transcription factors can inactivate anterior Hox proteins possibly through mechanisms that do not involve transcriptional repression (i.e., even if both genes are expressed, the posterior one is functionally dominant).130 Alternatively, two groups of miRNA sequences, MIR10 and MIR196, which downregulate, several 3′ Hox genes have been identified within the Hox chromosomal clusters, and possibly contribute toward this phenomenon (Fig. 1).124,131,132
However, regardless of the mechanistic basis, the posterior prevalence theory has its share of shortcomings, as several groups have reported instances where the 3′ Hox gene exerts dominance,133 3′ and 5′ Hox genes act cooperatively,134 or the functional response is dose-dependent.135 In all of these models, however, the prevailing theme is that repression of an effector gene product takes precedence over its activation, as is the case for other transcriptional control pathways.136
One plausible model for repression involves the requirement of cofactors for Hox proteins to efficiently bind DNA. Most 3′ Hox proteins contain a hexapeptide motif that allows them to bind members of the PBX family of transcription factors, while most 5′ Hox proteins (members of paralog groups 10–13) lack this motif.137,138 3′ Hox-PBX-DNA complexes exhibited dramatically increased stability compared with 3′ Hox-DNA complexes alone. Many of the Hox genes discussed in this review have been characterized to complex with PBX proteins, including HOXA9,139 HOXB3, HOXB5, HOXB7, HOXB9138 and HOXD3.52 The common theme among these factors is that they are all activators of angiogenesis, wound healing or both (Table 1). In contrast, anti-angiogenic HOXD10, and related paralogs, can bind target DNA with much higher affinity in the absence of PBX proteins. Thus, by requiring additional factors for transcriptional activation by the pro-angiogenic Hox genes, it is evident how repressor Hox proteins could exert functional dominance over activators. Interestingly, ectopic expression of Hoxd10 in Drosophila antennal imaginal discs inhibited nuclear translocation of EXD (Drosophila homolog of PBX1 and PBX2) and rendered it unavailable for cooperative DNA binding with 3′ Hox genes.140 It remains to be seen if this mechanism is recapitulated in mammalian cells, but it does suggest a possible means by which the anti-angiogenic HOXD10 can dominantly counteract Hox3-mediated pathological vascularization.
Table 1. Homeobox activators/repressors of vascularization/wound healing.
| Gene/ protein |
Pro/Anti-vascularization/wound healing |
Observation (expressed in) |
Reference(s) |
|---|---|---|---|
| Hox family genes | |||
| HOXA3 |
Pro-Vasc. |
• Knockout mice developed abnormally sized heart chambers • Expression concomitant with CCL2, MMP14 and β3 integrin expression in ESC → EC differentiation (ESCs) • Expression concomitant with EC migration along with uPAR and MMP14 expression (ECs) |
39, 43 and 56 |
| Pro-WH |
• Natively upregulated in keratinocytes within wound tissue. Enhances uPAR-mediated keratinocyte migration and accelerates wound closure in diabetic mice (dermal wound tissue) • Promotes differentiation of bone marrow progenitor cells into an M2-phenotype myeloid-like cell (dermal wound tissue) |
56 73 and 74 |
|
| HOXA5 |
Anti-Vasc. |
• Expressed in quiescent endothelium. Dowregulates VEGFR2, ephrin A1, HIF1A and COX2. Increases thrombospondin 2 (ECs) • Stabilizes adherens junctions via β-catenin retention (ECs) |
62 and 63 |
| HOXA9 |
Pro-Vasc. |
• Promotes EC migration and tube formation in culture through direct upregulation of EPHB4 (ECs) • Upregulates eNOS, VEGFR2 and VE-cadherin during EPC → EC differentiation (EPCs) |
45 and 60 |
| Pro-WH |
• Decreased expression in keloid fibroblasts associated with abnormal wound healing (dermal fibroblasts) |
108 |
|
| HOXB3 |
Pro-Vasc. |
• Increases expression of ephrin A1 and induces capillary morphogenesis in endothelial sprouts (ECs) |
57 |
| HOXB5 |
Pro-Vasc. |
• Triggers differentiation of embryoid bodies into angioblasts through VEGFR2 transactivation (ESCs) • Upregulates ANG2 and promotes vessel sprouting (ECs) |
42 and 64 |
| HOXB7 |
Pro-Vasc. |
• Directly enhances expression of FGF2, VEGFA, IL8, CXCL1 and ANG2 (many tumor lines) |
80–82 |
| HOXB9 |
Pro-Vasc. |
• Increased expression of VEGFA, FGF2, IL8 and ANGPTL2. Produced highly vascularized tumors which developed metastases (MCF10A breast epithelial cells) |
84 |
| HOXB13 |
Anti-WH |
• Longer retention of fibrin clot, sustained inflammatory response and formation of large, irregular vasculature (dermal wound tissue) • Downregulated following wounding of fetal dermis (fetal wound tissue) |
77 and 144 |
| HOXC8 |
Pro-WH |
• Expression correlates with keratinocyte proliferation and migration, leading to wound epithelialization (keratinocytes) |
107 |
| HOXD1 |
Pro-Vasc. |
• Directly increases β1 integrin expression, allowing formation of pro-angiogenic α5β1 integrin (ECs) |
53 |
| HOXD3 |
Pro-Vasc. |
• Correlates with invasive phase of angiogenesis. Directly enhances expression of the α5 and β3. Indirectly increases uPA. Expressed in immature tumor vasculature (ECs) • Expressed in proliferating hemangiomas (hemangioma tissue) |
50, 51 and 78 |
| Pro-WH |
• Increases angiogenesis and COL1A1 production, leading to accelerated wound closure (ECs, fibroblasts, keratinocytes) • Upregulated in wounded fetal skin (fetal wound tissue) |
58 and 145 |
|
| HOXD8 |
Pro-WH |
• Upregulated in wounded fetal skin (fetal wound tissue) |
145 |
| HOXD10 | Anti-Vasc. | • Expressed in quiescent endothelium. Overexpression inhibits angiogenic respone to FGF2 and VEGFA. Decreases expression of RHOC, β4 and α3 integrins, MMP14, uPAR and cyclin D1 (ECs) • Overexpression in mammary tumor cells decreases VEGFA expression and EC recruitment (mammary tumor cells) |
20 and 61 |
| Non-Hox homeobox genes | |||
|---|---|---|---|
| GAX (MEOX2) |
Anti-Vasc. |
• Induces cell cycle arrest (VSMCs and ECs) • Impairs angiogenic response to VEGFA, FGF2 and TNFα through NFκB inhibition. (ECs) |
66, 67 and 69 |
| HEX |
Pro-Vasc. |
• Required for EC and hematopoietic cell differentiation during vasculogenesis (blood islands) • Knockout mice develop several cardiovascular defects. |
46 and 47 |
| Anti-Vasc. |
• Inhibits EC tube formation in culture and dowregulates VEGFR1, VEGFR2, TIE1, TIE2 and αV integrin (ECs) |
48 and 49 |
|
| MEOX1 |
Anti-Vasc. |
• Induces cell cycle arrest (ECs) |
68 |
| Pro-WH |
• Expressed in fetal epidermis and dermis, but not in adult skin |
146 |
|
| MSX1 |
Pro-WH |
• Expressed in fetal epidermis and dermis, but only in adult epidermis |
146 |
| MSX2 |
Pro-WH |
• Expressed in fetal epidermis and dermis, but only in adult epidermis |
146 |
| PRX1 |
Pro-Vasc. |
• Knockout mice develop malformations of arteries |
40 |
| Pro-WH |
• Increased following wounding of fetal dermis (fetal wound tissue) |
144 |
|
| PRX2 | Pro-Vasc. | • Double knockout with PRX1 results in severe vessel abnormalities | 40 |
| Trithorax and polycomb-group genes | |||
|---|---|---|---|
| EED |
Anti-WH |
• Complexes with EZH2 and SUZ12 to form PRC2. Downregulated in wound epidermis. • Inhibits expression of HOXA9 and HOXC8 (HeLa cells and leukemia cells) |
103–105 |
| EZH2 |
Anti-WH |
• Complexes with EED2 and SUZ12 to form PRC2. Downregulated in wound epidermis. • Inhibits expression of HOXA9 and HOXC8 (HeLa cells and leukemia cells) |
103–105 |
| MEL18 (PCGF2) |
Pro-Vasc. |
• Overexpression causes EC migration and tube formation in culture. Upregulation of VEGFR2 and ANGPTL2 (ECs) |
101 |
| Anti-Vasc. |
• Another study contradicts above. Overexpression inhibits EC tube formation in culture and decreases VEGFA production (ECs) |
102 |
|
| MLL |
Pro-Vasc. |
• Upregulates HOXA9 and HOXD3 via trimethylation of lysine 4 of histone H3 (ECs) |
100 |
| SUZ12 | Anti-WH | • Complexes with EED and EZH2 to form PRC2. Downregulated in wound epidermis. • Inhibits expression of HOXA9 and HOXC8 (HeLa cells and leukemia cells) |
103–105 |
| Hox Cofactors | |||
|---|---|---|---|
| PBX1 | Pro-Vasc. | • Complexes with HOXD3 to more efficiently elicit its angiogenic functions (ECs) | 52 |
Phosphorylation of certain residues may also be required for Hox protein activation or nuclear translocation, and its absence may trigger transcriptional repression. While Hox phosphorylation has not yet been reported in mammals, consensus phosphorylation sites have been identified in Drosophila Hox homologs, and mutation of these sites have led to the repression of limb development.141,142
Future Directions
Many of the studies covered in this review have demonstrated that spatial or temporal misexpression of homeobox genes often results in malformations or oncogenesis; therefore, therapies intent on modulating cellular homeobox gene expression must consider the extent and duration of such a change.
For instance, overexpression of HOXD3 in healthy endothelium may trigger an unwanted inflammatory response or generate large cavernous vessels reminiscent of endotheliomas. To address this issue, our group commonly uses Hox DNA expression plasmids embedded within methylcellulose pellets to limit gene uptake to cells within wound tissue and their duration of expression.56,58
While topical application presents a straightforward approach for cutaneous tissues, targeted delivery strategies must be developed for tumors in internal organs. The mammary tumor suppressing properties of HOXA5 and HOXD10 have been documented,109,143 but a clinically viable treatment for breast cancer would need to target delivery of these Hox genes/proteins to tumors and their metastases in an effective manner. Like most systemic chemotherapies, systemic Hox delivery would likely produce unwanted side effects; therefore, engineering targeted-gene delivery vehicles can reduce off-target effects, but will require identifying appropriate tumor biomarkers for targeting. Moreover, while localized delivery to large tumors may be effective in reducing primary tumor angiogenesis, additional approaches must be considered to limit further progression linked to metastases.
Homeobox gene therapy can also be leveraged in the field of regenerative medicine to induce vascularization within engineered tissue; however, in these cases it will be important to temporally regulate expression levels to effectively transition through the different phases of angiogenesis. In situations such as these, designing inducible homeobox gene expression systems, placing homeobox expression cassettes downstream of phase-specific promoters, or delivering Hox proteins rather than genes may prove beneficial.
In addition, continuing research should also investigate if and how Hox genes regulate the activity of TrxG and PcG genes/proteins and Hox-regulating miRNAs. This will help shed light on Hox cross-regulatory biology and functional dominance and allow researchers to more efficiently manipulate Hox expression for therapeutic purposes.
Acknowledgments
This work was supported by a grant from the NIH/NCI (R01 CA140655) to N.J.B.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/22164
References
- 1.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 2.Radisic M, Deen W, Langer R, Vunjak-Novakovic G. Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers. Am J Physiol Heart Circ Physiol. 2005;288:H1278–89. doi: 10.1152/ajpheart.00787.2004. [DOI] [PubMed] [Google Scholar]
- 3.Patan S. Vasculogenesis and angiogenesis as mechanisms of vascular network formation, growth and remodeling. J Neurooncol. 2000;50:1–15. doi: 10.1023/A:1006493130855. [DOI] [PubMed] [Google Scholar]
- 4.Springer ML, Chen AS, Kraft PE, Bednarski M, Blau HM. VEGF gene delivery to muscle: potential role for vasculogenesis in adults. Mol Cell. 1998;2:549–58. doi: 10.1016/S1097-2765(00)80154-9. [DOI] [PubMed] [Google Scholar]
- 5.Asahara T, Masuda H, Takahashi T, Kalka C, Pastore C, Silver M, et al. Bone marrow origin of endothelial progenitor cells responsible for postnatal vasculogenesis in physiological and pathological neovascularization. Circ Res. 1999;85:221–8. doi: 10.1161/01.RES.85.3.221. [DOI] [PubMed] [Google Scholar]
- 6.Ghajar CM, George SC, Putnam AJ. Matrix metalloproteinase control of capillary morphogenesis. Crit Rev Eukaryot Gene Expr. 2008;18:251–78. doi: 10.1615/CritRevEukarGeneExpr.v18.i3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–64. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 9.Folkman J, Hanahan D. Switch to the angiogenic phenotype during tumorigenesis. Princess Takamatsu Symp. 1991;22:339–47. [PubMed] [Google Scholar]
- 10.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9:259–70. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zumsteg A, Christofori G. Corrupt policemen: inflammatory cells promote tumor angiogenesis. Curr Opin Oncol. 2009;21:60–70. doi: 10.1097/CCO.0b013e32831bed7e. [DOI] [PubMed] [Google Scholar]
- 12.Leek RD, Harris AL. Tumor-associated macrophages in breast cancer. J Mammary Gland Biol Neoplasia. 2002;7:177–89. doi: 10.1023/A:1020304003704. [DOI] [PubMed] [Google Scholar]
- 13.Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66:11238–46. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 14.Giraudo E, Inoue M, Hanahan D. An amino-bisphosphonate targets MMP-9-expressing macrophages and angiogenesis to impair cervical carcinogenesis. J Clin Invest. 2004;114:623–33. doi: 10.1172/JCI22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegué E, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–20. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubota Y, Takubo K, Shimizu T, Ohno H, Kishi K, Shibuya M, et al. M-CSF inhibition selectively targets pathological angiogenesis and lymphangiogenesis. J Exp Med. 2009;206:1089–102. doi: 10.1084/jem.20081605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeisberger SM, Odermatt B, Marty C, Zehnder-Fjällman AH, Ballmer-Hofer K, Schwendener RA. Clodronate-liposome-mediated depletion of tumour-associated macrophages: a new and highly effective antiangiogenic therapy approach. Br J Cancer. 2006;95:272–81. doi: 10.1038/sj.bjc.6603240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good DJ, Polverini PJ, Rastinejad F, Le Beau MM, Lemons RS, Frazier WA, et al. A tumor suppressor-dependent inhibitor of angiogenesis is immunologically and functionally indistinguishable from a fragment of thrombospondin. Proc Natl Acad Sci U S A. 1990;87:6624–8. doi: 10.1073/pnas.87.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dameron KM, Volpert OV, Tainsky MA, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–4. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 20.Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N. Sustained expression of homeobox D10 inhibits angiogenesis. Am J Pathol. 2002;161:2099–109. doi: 10.1016/S0002-9440(10)64488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy JA, Chang SH, Shih SC, Dvorak AM, Dvorak HF. Heterogeneity of the tumor vasculature. Semin Thromb Hemost. 2010;36:321–31. doi: 10.1055/s-0030-1253454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 23.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 24.Wellik DM. Hox genes and vertebrate axial pattern. Curr Top Dev Biol. 2009;88:257–78. doi: 10.1016/S0070-2153(09)88009-5. [DOI] [PubMed] [Google Scholar]
- 25.Lufkin T, Mark M, Hart CP, Dollé P, LeMeur M, Chambon P. Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature. 1992;359:835–41. doi: 10.1038/359835a0. [DOI] [PubMed] [Google Scholar]
- 26.Kessel M, Balling R, Gruss P. Variations of cervical vertebrae after expression of a Hox-1.1 transgene in mice. Cell. 1990;61:301–8. doi: 10.1016/0092-8674(90)90810-2. [DOI] [PubMed] [Google Scholar]
- 27.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-N. [DOI] [PubMed] [Google Scholar]
- 29.Condie BG, Capecchi MR. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature. 1994;370:304–7. doi: 10.1038/370304a0. [DOI] [PubMed] [Google Scholar]
- 30.Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–5. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 31.Horan GS, Ramírez-Solis R, Featherstone MS, Wolgemuth DJ, Bradley A, Behringer RR. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995;9:1667–77. doi: 10.1101/gad.9.13.1667. [DOI] [PubMed] [Google Scholar]
- 32.Horan GS, Kovàcs EN, Behringer RR, Featherstone MS. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function. Dev Biol. 1995;169:359–72. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 33.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, et al. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 1996;122:461–72. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 34.Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–5. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 35.Davis AP, Capecchi MR. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development. 1996;122:1175–85. doi: 10.1242/dev.122.4.1175. [DOI] [PubMed] [Google Scholar]
- 36.de la Cruz CC, Der-Avakian A, Spyropoulos DD, Tieu DD, Carpenter EM. Targeted disruption of Hoxd9 and Hoxd10 alters locomotor behavior, vertebral identity, and peripheral nervous system development. Dev Biol. 1999;216:595–610. doi: 10.1006/dbio.1999.9528. [DOI] [PubMed] [Google Scholar]
- 37.Rancourt DE, Tsuzuki T, Capecchi MR. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 1995;9:108–22. doi: 10.1101/gad.9.1.108. [DOI] [PubMed] [Google Scholar]
- 38.Favier B, Rijli FM, Fromental-Ramain C, Fraulob V, Chambon P, Dollé P. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development. 1996;122:449–60. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- 39.Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991;350:473–9. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 40.Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, et al. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–9. doi: 10.1007/PL00008193. [DOI] [PubMed] [Google Scholar]
- 41.Miano JM, Firulli AB, Olson EN, Hara P, Giachelli CM, Schwartz SM. Restricted expression of homeobox genes distinguishes fetal from adult human smooth muscle cells. Proc Natl Acad Sci U S A. 1996;93:900–5. doi: 10.1073/pnas.93.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Moser M, Bautch VL, Patterson C. HoxB5 is an upstream transcriptional switch for differentiation of the vascular endothelium from precursor cells. Mol Cell Biol. 2003;23:5680–91. doi: 10.1128/MCB.23.16.5680-5691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahrami SB, Veiseh M, Dunn AA, Boudreau NJ. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adh Migr. 2011;5:133–41. doi: 10.4161/cam.5.2.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iacovino M, Chong D, Szatmari I, Hartweck L, Rux D, Caprioli A, et al. HoxA3 is an apical regulator of haemogenic endothelium. Nat Cell Biol. 2011;13:72–8. doi: 10.1038/ncb2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rössig L, Urbich C, Brühl T, Dernbach E, Heeschen C, Chavakis E, et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–35. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crompton MR, Bartlett TJ, MacGregor AD, Manfioletti G, Buratti E, Giancotti V, et al. Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 1992;20:5661–7. doi: 10.1093/nar/20.21.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hallaq H, Pinter E, Enciso J, McGrath J, Zeiss C, Brueckner M, et al. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development. 2004;131:5197–209. doi: 10.1242/dev.01393. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa T, Abe M, Yamazaki T, Miyashita H, Niwa H, Kokubun S, et al. HEX acts as a negative regulator of angiogenesis by modulating the expression of angiogenesis-related gene in endothelial cells in vitro. Arterioscler Thromb Vasc Biol. 2003;23:231–7. doi: 10.1161/01.ATV.0000052670.55321.87. [DOI] [PubMed] [Google Scholar]
- 49.Minami T, Murakami T, Horiuchi K, Miura M, Noguchi T, Miyazaki J, et al. Interaction between hex and GATA transcription factors in vascular endothelial cells inhibits flk-1/KDR-mediated vascular endothelial growth factor signaling. J Biol Chem. 2004;279:20626–35. doi: 10.1074/jbc.M308730200. [DOI] [PubMed] [Google Scholar]
- 50.Boudreau N, Andrews C, Srebrow A, Ravanpay A, Cheresh DA. Induction of the angiogenic phenotype by Hox D3. J Cell Biol. 1997;139:257–64. doi: 10.1083/jcb.139.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boudreau NJ, Varner JA. The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis. J Biol Chem. 2004;279:4862–8. doi: 10.1074/jbc.M305190200. [DOI] [PubMed] [Google Scholar]
- 52.Charboneau A, East L, Mulholland N, Rohde M, Boudreau N. Pbx1 is required for Hox D3-mediated angiogenesis. Angiogenesis. 2005;8:289–96. doi: 10.1007/s10456-005-9016-7. [DOI] [PubMed] [Google Scholar]
- 53.Park H, Choi HJ, Kim J, Kim M, Rho SS, Hwang D, et al. Homeobox D1 regulates angiogenic functions of endothelial cells via integrin β1 expression. Biochem Biophys Res Commun. 2011;408:186–92. doi: 10.1016/j.bbrc.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–71. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 55.Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin alpha5beta1 with the central cell-binding domain of fibronectin. Am J Pathol. 2000;156:1345–62. doi: 10.1016/S0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mace KA, Hansen SL, Myers C, Young DM, Boudreau N. HOXA3 induces cell migration in endothelial and epithelial cells promoting angiogenesis and wound repair. J Cell Sci. 2005;118:2567–77. doi: 10.1242/jcs.02399. [DOI] [PubMed] [Google Scholar]
- 57.Myers C, Charboneau A, Boudreau N. Homeobox B3 promotes capillary morphogenesis and angiogenesis. J Cell Biol. 2000;148:343–51. doi: 10.1083/jcb.148.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen SL, Myers CA, Charboneau A, Young DM, Boudreau N. HoxD3 accelerates wound healing in diabetic mice. Am J Pathol. 2003;163:2421–31. doi: 10.1016/S0002-9440(10)63597-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel CV, Sharangpani R, Bandyopadhyay S, DiCorleto PE. Endothelial cells express a novel, tumor necrosis factor-alpha-regulated variant of HOXA9. J Biol Chem. 1999;274:1415–22. doi: 10.1074/jbc.274.3.1415. [DOI] [PubMed] [Google Scholar]
- 60.Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ Res. 2004;94:743–51. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- 61.Chen A, Cuevas I, Kenny PA, Miyake H, Mace K, Ghajar C, et al. Endothelial cell migration and vascular endothelial growth factor expression are the result of loss of breast tissue polarity. Cancer Res. 2009;69:6721–9. doi: 10.1158/0008-5472.CAN-08-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3:240–52. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
- 63.Arderiu G, Cuevas I, Chen A, Carrio M, East L, Boudreau NJ. HoxA5 stabilizes adherens junctions via increased Akt1. Cell Adh Migr. 2007;1:185–95. doi: 10.4161/cam.1.4.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winnik S, Klinkert M, Kurz H, Zoeller C, Heinke J, Wu Y, et al. HoxB5 induces endothelial sprouting in vitro and modifies intussusceptive angiogenesis in vivo involving angiopoietin-2. Cardiovasc Res. 2009;83:558–65. doi: 10.1093/cvr/cvp133. [DOI] [PubMed] [Google Scholar]
- 65.Gorski DH, LePage DF, Patel CV, Copeland NG, Jenkins NA, Walsh K. Molecular cloning of a diverged homeobox gene that is rapidly down-regulated during the G0/G1 transition in vascular smooth muscle cells. Mol Cell Biol. 1993;13:3722–33. doi: 10.1128/mcb.13.6.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith RC, Branellec D, Gorski DH, Guo K, Perlman H, Dedieu JF, et al. p21CIP1-mediated inhibition of cell proliferation by overexpression of the gax homeodomain gene. Genes Dev. 1997;11:1674–89. doi: 10.1101/gad.11.13.1674. [DOI] [PubMed] [Google Scholar]
- 67.Gorski DH, Leal AJ. Inhibition of endothelial cell activation by the homeobox gene Gax. J Surg Res. 2003;111:91–9. doi: 10.1016/S0022-4804(03)00042-8. [DOI] [PubMed] [Google Scholar]
- 68.Douville JM, Cheung DY, Herbert KL, Moffatt T, Wigle JT. Mechanisms of MEOX1 and MEOX2 regulation of the cyclin dependent kinase inhibitors p21 and p16 in vascular endothelial cells. PLoS One. 2011;6:e29099. doi: 10.1371/journal.pone.0029099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel S, Leal AD, Gorski DH. The homeobox gene Gax inhibits angiogenesis through inhibition of nuclear factor-kappaB-dependent endothelial cell gene expression. Cancer Res. 2005;65:1414–24. doi: 10.1158/0008-5472.CAN-04-3431. [DOI] [PubMed] [Google Scholar]
- 70.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–26. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;341:738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 72.DiPietro LA, Burns AL. Wound Healing: Methods and Protocols. Humana Press, 2003. [Google Scholar]
- 73.Mace KA, Restivo TE, Rinn JL, Paquet AC, Chang HY, Young DM, et al. HOXA3 modulates injury-induced mobilization and recruitment of bone marrow-derived cells. Stem Cells. 2009;27:1654–65. doi: 10.1002/stem.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mahdipour E, Charnock JC, Mace KA. Hoxa3 promotes the differentiation of hematopoietic progenitor cells into proangiogenic Gr-1+CD11b+ myeloid cells. Blood. 2011;117:815–26. doi: 10.1182/blood-2009-12-259549. [DOI] [PubMed] [Google Scholar]
- 75.Maclauchlan S, Skokos EA, Agah A, Zeng J, Tian W, Davidson JM, et al. Enhanced angiogenesis and reduced contraction in thrombospondin-2-null wounds is associated with increased levels of matrix metalloproteinases-2 and -9, and soluble VEGF. J Histochem Cytochem. 2009;57:301–13. doi: 10.1369/jhc.2008.952689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mack JA, Abramson SR, Ben Y, Coffin JC, Rothrock JK, Maytin EV, et al. Hoxb13 knockout adult skin exhibits high levels of hyaluronan and enhanced wound healing. FASEB J. 2003;17:1352–4. doi: 10.1096/fj.02-0959fje. [DOI] [PubMed] [Google Scholar]
- 77.Mack JA, Maytin EV. Persistent inflammation and angiogenesis during wound healing in K14-directed Hoxb13 transgenic mice. J Invest Dermatol. 2010;130:856–65. doi: 10.1038/jid.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hansen SL, Dosanjh A, Young DM, Boudreau N, Hoffman WY. Hemangiomas and homeobox gene expression. J Craniofac Surg. 2006;17:767–71. doi: 10.1097/00001665-200607000-00031. [DOI] [PubMed] [Google Scholar]
- 79.Zhu Y, Cuevas IC, Gabriel RA, Su H, Nishimura S, Gao P, et al. Restoring transcription factor HoxA5 expression inhibits the growth of experimental hemangiomas in the brain. J Neuropathol Exp Neurol. 2009;68:626–32. doi: 10.1097/NEN.0b013e3181a491ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caré A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, et al. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Mol Cell Biol. 1996;16:4842–51. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caré A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16:3285–9. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 82.Carè A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, et al. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61:6532–9. [PubMed] [Google Scholar]
- 83.Lobov IB, Brooks PC, Lang RA. Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proc Natl Acad Sci U S A. 2002;99:11205–10. doi: 10.1073/pnas.172161899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N, et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci U S A. 2010;107:1100–5. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sunderkötter C, Steinbrink K, Goebeler M, Bhardwaj R, Sorg C. Macrophages and angiogenesis. J Leukoc Biol. 1994;55:410–22. doi: 10.1002/jlb.55.3.410. [DOI] [PubMed] [Google Scholar]
- 86.Giraudo E, Inoue M, Hanahan D. An amino-bisphosphonate targets MMP-9-expressing macrophages and angiogenesis to impair cervical carcinogenesis. J Clin Invest. 2004;114:623–33. doi: 10.1172/JCI22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–7. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian B, Deng Y, Im JH, Muschel RJ, Zou Y, Li J, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4:e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–75. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 90.Page SH, Wright EK, Jr., Gama L, Clements JE. Regulation of CCL2 expression by an upstream TALE homeodomain protein-binding site that synergizes with the site created by the A-2578G SNP. PLoS One. 2011;6:e22052. doi: 10.1371/journal.pone.0022052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 92.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 93.Mahdipour E, Mace KA. Hox transcription factor regulation of adult bone-marrow-derived cell behaviour during tissue repair and regeneration. Expert Opin Biol Ther. 2011;11:1079–90. doi: 10.1517/14712598.2011.579096. [DOI] [PubMed] [Google Scholar]
- 94.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci U S A. 1998;95:10632–6. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Déjardin J, Cavalli G. Epigenetic inheritance of chromatin states mediated by Polycomb and trithorax group proteins in Drosophila. Prog Mol Subcell Biol. 2005;38:31–63. doi: 10.1007/3-540-27310-7_2. [DOI] [PubMed] [Google Scholar]
- 96.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 97.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 98.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 100.Diehl F, Rössig L, Zeiher AM, Dimmeler S, Urbich C. The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood. 2007;109:1472–8. doi: 10.1182/blood-2006-08-039651. [DOI] [PubMed] [Google Scholar]
- 101.Jung JH, Choi HJ, Maeng YS, Choi JY, Kim M, Kwon JY, et al. Mel-18, a mammalian Polycomb gene, regulates angiogenic gene expression of endothelial cells. Biochem Biophys Res Commun. 2010;400:523–30. doi: 10.1016/j.bbrc.2010.08.086. [DOI] [PubMed] [Google Scholar]
- 102.Park JH, Lee JY, Shin DH, Jang KS, Kim HJ, Kong G. Loss of Mel-18 induces tumor angiogenesis through enhancing the activity and expression of HIF-1α mediated by the PTEN/PI3K/Akt pathway. Oncogene. 2011;30:4578–89. doi: 10.1038/onc.2011.174. [DOI] [PubMed] [Google Scholar]
- 103.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–6. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 105.Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114:2733–43. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, et al. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281:24790–802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- 107.Huo R, Ma Q, Wu JJ, Chin-Nuke K, Jing Y, Chen J, et al. Noninvasive electromagnetic fields on keratinocyte growth and migration. J Surg Res. 2010;162:299–307. doi: 10.1016/j.jss.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 108.Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, et al. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–96. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005;65:7177–85. doi: 10.1158/0008-5472.CAN-04-1717. [DOI] [PubMed] [Google Scholar]
- 110.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–34. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 111.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–32. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takahashi O, Hamada J, Abe M, Hata S, Asano T, Takahashi Y, et al. Dysregulated expression of HOX and ParaHOX genes in human esophageal squamous cell carcinoma. Oncol Rep. 2007;17:753–60. [PubMed] [Google Scholar]
- 113.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 114.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–8. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 115.Armstrong SA, Golub TR, Korsmeyer SJ. MLL-rearranged leukemias: insights from gene expression profiling. Semin Hematol. 2003;40:268–73. doi: 10.1016/S0037-1963(03)00196-3. [DOI] [PubMed] [Google Scholar]
- 116.Jung C, Kim RS, Lee SJ, Wang C, Jeng MH. HOXB13 homeodomain protein suppresses the growth of prostate cancer cells by the negative regulation of T-cell factor 4. Cancer Res. 2004;64:3046–51. doi: 10.1158/0008-5472.CAN-03-2614. [DOI] [PubMed] [Google Scholar]
- 117.Jung C, Kim RS, Zhang H, Lee SJ, Sheng H, Loehrer PJ, et al. HOXB13 is downregulated in colorectal cancer to confer TCF4-mediated transactivation. Br J Cancer. 2005;92:2233–9. doi: 10.1038/sj.bjc.6602631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci U S A. 2012;109:2736–41. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120:1535–50. doi: 10.1172/JCI39534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–30. [PubMed] [Google Scholar]
- 121.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 122.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–85. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 123.Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–8. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 124.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 125.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–71. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 126.González-Reyes A, Morata G. The developmental effect of overexpressing a Ubx product in Drosophila embryos is dependent on its interactions with other homeotic products. Cell. 1990;61:515–22. doi: 10.1016/0092-8674(90)90533-K. [DOI] [PubMed] [Google Scholar]
- 127.Capovilla M, Botas J. Functional dominance among Hox genes: repression dominates activation in the regulation of Dpp. Development. 1998;125:4949–57. doi: 10.1242/dev.125.24.4949. [DOI] [PubMed] [Google Scholar]
- 128.González-Reyes A, Urquia N, Gehring WJ, Struhl G, Morata G. Are cross-regulatory interactions between homoeotic genes functionally significant? Nature. 1990;344:78–80. doi: 10.1038/344078a0. [DOI] [PubMed] [Google Scholar]
- 129.Duboule D. Patterning in the vertebrate limb. Curr Opin Genet Dev. 1991;1:211–6. doi: 10.1016/S0959-437X(05)80072-3. [DOI] [PubMed] [Google Scholar]
- 130.Schöck F, Reischl J, Wimmer E, Taubert H, Purnell BA, Jäckle H. Phenotypic suppression of empty spiracles is prevented by buttonhead. Nature. 2000;405:351–4. doi: 10.1038/35012620. [DOI] [PubMed] [Google Scholar]
- 131.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 132.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–96. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heuer JG, Li K, Kaufman TC. The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development. 1995;121:3861–76. doi: 10.1242/dev.121.11.3861. [DOI] [PubMed] [Google Scholar]
- 134.Wang BB, Müller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-X. [DOI] [PubMed] [Google Scholar]
- 135.Lamka ML, Boulet AM, Sakonju S. Ectopic expression of UBX and ABD-B proteins during Drosophila embryogenesis: competition, not a functional hierarchy, explains phenotypic suppression. Development. 1992;116:841–54. doi: 10.1242/dev.116.4.841. [DOI] [PubMed] [Google Scholar]
- 136.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–81. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 137.Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–74. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 138.Shen WF, Chang CP, Rozenfeld S, Sauvageau G, Humphries RK, Lu M, et al. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen WF, Rozenfeld S, Kwong A, Köm ves LG, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–61. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Azpiazu N, Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 1998;12:261–73. doi: 10.1101/gad.12.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jaffe L, Ryoo HD, Mann RS. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev. 1997;11:1327–40. doi: 10.1101/gad.11.10.1327. [DOI] [PubMed] [Google Scholar]
- 142.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–7. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 143.Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24:924–35. doi: 10.1128/MCB.24.2.924-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stelnicki EJ, Arbeit J, Cass DL, Saner C, Harrison M, Largman C. Modulation of the human homeobox genes PRX-2 and HOXB13 in scarless fetal wounds. J Invest Dermatol. 1998;111:57–63. doi: 10.1046/j.1523-1747.1998.00238.x. [DOI] [PubMed] [Google Scholar]
- 145.Jain K, Sykes V, Kordula T, Lanning D. Homeobox genes Hoxd3 and Hoxd8 are differentially expressed in fetal mouse excisional wounds. J Surg Res. 2008;148:45–8. doi: 10.1016/j.jss.2008.02.053. [DOI] [PubMed] [Google Scholar]
- 146.Stelnicki EJ, Kömüves LG, Holmes D, Clavin W, Harrison MR, Adzick NS, et al. The human homeobox genes MSX-1, MSX-2, and MOX-1 are differentially expressed in the dermis and epidermis in fetal and adult skin. Differentiation. 1997;62:33–41. doi: 10.1046/j.1432-0436.1997.6210033.x. [DOI] [PubMed] [Google Scholar]