Abstract
An adenosine diphosphate sugar pyrophosphatase (ASPPase, EC 3.6.1.21) has been characterized by using Escherichia coli. This enzyme, whose activities in the cell are inversely correlated with the intracellular glycogen content and the glucose concentration in the culture medium, hydrolyzes ADP-glucose, the precursor molecule of glycogen biosynthesis. ASPPase was purified to apparent homogeneity (over 3,000-fold), and sequence analyses revealed that it is a member of the ubiquitously distributed group of nucleotide pyrophosphatases designated as “nudix” hydrolases. Insertional mutagenesis experiments leading to the inactivation of the ASPPase encoding gene, aspP, produced cells with marginally low enzymatic activities and higher glycogen content than wild-type bacteria. aspP was cloned into an expression vector and introduced into E. coli. Transformed cells were shown to contain a dramatically reduced amount of glycogen, as compared with the untransformed bacteria. No pleiotropic changes in the bacterial growth occurred in both the aspP-overexpressing and aspP-deficient strains. The overall results pinpoint the reaction catalyzed by ASPPase as a potential step of regulating glycogen biosynthesis in E. coli.
Glycogen is a branched homopolysaccharide of α-1,4-linked glucose subunits with α-1,6-linked glucose at the branching points. Synthesized by glycogen synthase (EC 2.4.1.21) using ADP-glucose (ADPG) as the sugar donor nucleotide, the accumulation of this polysaccharide in Escherichia coli occurs as a result of limited growth conditions in the presence of an excess of a carbon source (1).
Regulation of bacterial glycogen synthesis involves a complex assemblage of factors that adjust the rate of synthesis according to the physiological status of the cell. It is generally argued that these factors affect the gluconeogenic process at the level of allosteric regulation of the committed step of the pathway, catalyzed by ADPG pyrophosphorylase (AGPase) (EC 2.7.7.27), and at the level of regulation of the expression of the genes encoding gluconeogenic enzymes such as AGPase, phosphoglucomutase, glycogen synthase, and the glycogen branching enzyme (1, 2).
Recent investigations have demonstrated that glycogen can be simultaneously synthesized and degraded during bacterial growth, thus making up a futile cycle wherein AGPase has a dual role in catalyzing the de novo synthesis of ADPG and in recycling the glucose units derived from the glycogen breakdown (3–5). Simultaneous synthesis and degradation of glycogen and starch have been reported to occur also in animals and plants, respectively (6–10), thus indicating that the operation of futile cycling may entail advantages such as sensitive regulation and channeling of excess gluconeogenic intermediates toward various metabolic pathways in response to physiological and biochemical needs. In this context, it should be emphasized that nucleotide-sugar pyrophosphatases from both plants and animals have been suggested to play a crucial role in diverting carbon flow from gluconeogenesis to other metabolic pathways (11–13). According to this implicated hypothetical mechanism, the extent to which both nucleotide-sugars as well as typical gluconeogenic end products such as starch or glycogen accumulate in the cell will be inversely correlated to the activities of nucleotide-sugar hydrolases.
Nucleotide-sugar pyrophosphatases catalyzing the hydrolytic breakdown of ADPG were first reported to occur in E. coli (14). The possible involvement of such an activity in the control of carbon flow toward glycogen has prompted us to identify a molecular entity, adenosine diphosphate sugar pyrophosphatase (designated as ASPPase), that hydrolyzes ADPG. Results presented in this work relating to changes of glycogen content in E. coli as a result of altered expression of the ASPPase encoding gene, aspP, strongly indicate that enzymatic breakdown of ADPG may play a pivotal role in controlling the gluconeogenic process and in interconnecting gluconeogenesis with other metabolic pathways.
Materials and Methods
Bacterial Strains and Culture Media.
Purification of ASPPase was performed by using E. coli BL21 grown in M9 minimal medium (96 mM Na2HPO4/44 mM KH2PO4/15 mM NaCl/35 mM NH4Cl/0.1 mM CaCl2/2 mM MgSO4) supplemented with 5 mM glucose. All plasmid constructs were electroporated and propagated in E. coli DH5α grown in LB medium with the appropriate selection. aspP disruption and overexpression experiments were performed by using BL21 and BL21(DE3) cells, respectively, that were grown in M9 minimal medium supplemented with the indicated amount of glucose. In every case, the bacteria were grown with rapid gyratory shaking at 37°C after inoculation with 1 volume of an overnight culture per 100 volumes of fresh medium.
Enzyme Assays.
Measurements of ADPG-dependent ASPPase activities were performed by using the two-step spectrophotometric determination of glucose-1-phosphate described by Rodríguez-López et al. (12) to measure plant ADPG pyrophosphatase, but with the following modifications. In step one, the reaction mixture contained 50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/2 mM ADPG and protein extract. ADP-ribose- and ADP-mannose-dependent ASPPase activities were measured by the chromatographic determination of AMP as described (12).
The unit is defined as the amount of enzyme that catalyzes the production of 1 μmol of product per minute. Kinetic parameters such as Km and Vmax were evaluated by Lineweaver–Burk plots.
ASPPase Purification.
For purification of ASPPase, 30 liters of an overnight grown bacterial culture were centrifuged at 10,000 × g for 10 min. The pelleted bacteria were then resuspended in 200 ml of extraction buffer (50 mM Tris⋅HCl, pH 7.5), and the supernatant obtained after sonication and ultracentrifugation at 100,000 × g for 30 min was heated in a water bath at 58°C for 10 min, cooled on ice, and centrifuged at 30,000 × g for 30 min. Proteins from the supernatant (166 ml) were precipitated with 30–50% (NH4)2SO4 and resuspended in 2.7 ml of extraction buffer. The sample was then subjected to gel filtration on a Superdex 200 column (Amersham Pharmacia) preequilibrated with 50 mM Tris⋅HCl, pH 7.5/150 mM NaCl. The partially purified enzyme was then applied to a Mono-Q column (HR 5 × 5, Amersham Pharmacia) equilibrated with extraction buffer and eluted with a 45-ml gradient of 0–1 M KCl in 30 mM Tris⋅HCl, pH 8.0, at a flow rate of 0.75 ml/min. Enzymatically active fractions were then subjected to isoelectrofocusing separation by using a Rotofor Cell apparatus (Bio-Rad).
The molecular mass of the native ASPPase was determined from a plot of Kav (partition coefficient) versus log molecular mass of protein standards.
Protein Sequence Analyses.
Purified ASPPase was electrophoretically separated in a 10% SDS/PAGE and blotted onto polyvinylidene difluoride membrane (Immobilon P; Millipore). The protein band was then excised and used for N-terminal sequencing analyses with an Applied Biosystems model 473A amino acid sequencer.
Overexpression and Insertional Disruption of aspP.
aspP was amplified by PCR using the primers 5′-GGACATTAACAATGCTTAAGCC-3′ (forward) and 5′-GGTGTGTAACGCTTCATTTATGCCCACTC-3′ (reverse) and the chromosomal DNA of E. coli BL21. The 640-bp PCR product obtained was blunt ended and cloned into the SmaI–EcoRV sites of pBluescript SK− (Stratagene) to create pASPP.
For aspP overexpression experiments, the HindIII–NotI fragment of pASPP was ligated with the corresponding restriction sites of the pET-28b(+) expression vector (Novagen) to create pET-ASPP. E. coli BL21(DE3) transformed with this plasmid were grown to an absorbance at 600 nm of about 0.6, and then 1 mM isopropyl-β-D-thiogalactopyranoside was added to the culture medium.
Insertional mutagenesis was carried out by using the chromosomal mutagenesis procedure developed by Link et al. (15). Toward this end, the SmaI cassette of pKT1 (16) containing the thermostable kanamycin adenyl transferase kat gene was cloned into the internal SmaI site of pASPP to create pASPPkat. The NotI–SalI fragment of pASPPkat containing the mutant allele of aspP was then cloned into the pKO3 gene replacement vector to create pKO3-ASPPkat, whose cloned fragment was integrated into the E. coli BL21 chromosome by homologous recombination with aspP.
Analytical Procedures.
Bacterial growth was spectrophotometrically determined by absorbance at 600 nm. Quantitative assay of biofilm formation was performed as described by O'Toole and Kolter (17). ASPPase was analyzed for potential cellular localization by using PSORT algorithm (18). Glycogen was determined by using a glucose oxidase-based test kit (Boehringer Mannheim). Protein content was measured by the Bradford method using a Bio-Rad prepared reagent.
Results and Discussion
ASPPase Purification and Characterization.
Consistent with the results of previous investigations reported by Melo and Glaser (14), preliminary analyses using crude extracts of E. coli revealed the existence of an enzymatic activity catalyzing the hydrolytic breakdown of ADPG. We then attempted to purify the enzyme molecule responsible for this activity. As presented in Table 1, it was purified over 3,000-fold. SDS/PAGE and subsequent staining of the purified protein revealed a single band of about 26 kDa (data not shown). The apparent molecular mass of the native enzyme measured by gel filtration was estimated to be 40–50 kDa, indicating that the enzyme responsible for the ADPG breakdown is a homodimer.
Table 1.
Purification of ASPPase from E. coli
| Total volume, ml | Total protein, mg | Total activity, milliunits | Specific activity, milliunits/mg protein | Purification, -fold | Yield, % | |
|---|---|---|---|---|---|---|
| Crude extract | 198 | 4,206 | 12,905 | 3 | — | 100 |
| Sup. 100,000 × g (58°C) | 166 | 462 | 54,499 | 118 | 38 | 422* |
| Ammonium sulfate | 2.7 | 101 | 29,719 | 293 | 95 | 155 |
| Superdex 200 | 15 | 7.4 | 19,125 | 2,584 | 842 | 148 |
| Mono-Q | 2.7 | 1.9 | 6,023 | 3,170 | 1,032 | 47 |
| Isoelectrofocusing | 0.6 | 0.2 | 1,807 | 9,511 | 3,098 | 15 |
Consistent with the results of Melo and Glaser (14), ASPPase in crude extracts of E. coli was shown to be elicidated by mild heating.
Substrate specificity was tested by using a wide range of compounds at a concentration of 5 mM. These preliminary analyses showed that the enzyme, designated as ASPPase, is a pyrophosphatase that specifically hydrolyzes adenosine diphosphate sugars to produce equimolar amounts of the corresponding sugar monophosphate and AMP (Table 2). ASPPase does not hydrolyze UDP-glucose, CDP-glucose, GDP-mannose, adenosine 5′-phosphosulfate, PPi, synthetic phosphodiester bond containing compounds such as bis-p-nitrophenyl-phosphate, and phosphomonoester bond containing compounds such as p-nitrophenyl-phosphate, sugar phosphates, and nucleoside mono-, di-, and triphosphates.
Table 2.
Substrate specificity of E. coli ASPPase
| Substrates | Km, μM | Vmax/Km, % with respect to ADPG |
|---|---|---|
| ADPG | 167 | 100 |
| ADP-mannose | 183 | 80 |
| ADP-ribose | 125 | 200 |
Kitetic parameters (Km and Vmax) are the mean values from five independent experiments. ASPPase does not hydrolyze UDP-glucose, GDP-mannose, adenosine 5′-phosphosulfate, and nucleoside mono-, di-, and triphosphates.
Essentially similar to the case of other ADPG-cleaving pyrophosphatases of plant origin reported (12), ASPPase is a “regulatable” enzyme that is inhibited by phosphorylated compounds such as AMP, ADP, ATP, 3-phosphoglyceric acid, and PPi, whereas orthophosphate, a strong inhibitor of the reaction catalyzed by AGPase, does not exert a substantial inhibitory effect on ASPPase (data not shown).
Reciprocal Pattern Between ASPPase Activities and Glycogen Content in E. coli.
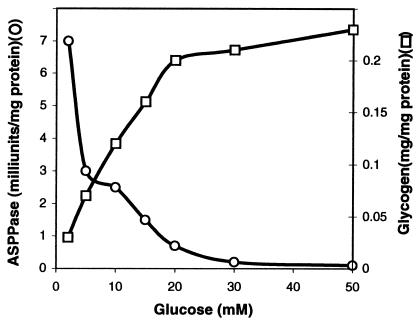
Glycogen accumulation in E. coli occurs as a result of limited growth conditions in the presence of an excess of a carbon source, and it is enhanced as bacterial cultures enter the stationary phase (1). We have thus examined ASPPase activities and compared them with the glycogen content in E. coli BL21 grown with different glucose concentrations. As presented in Fig. 1, the intracellular glycogen contents during the stationary growth phase correlate well with the amount of glucose in the culture medium. In sharp contrast, ASPPase activities are high in cells grown in low glucose concentration in the culture medium, whereas they decrease dramatically as the glucose content increases. Despite the limitations inherent in basing conclusions on in vitro activities, these results showing a glucose-dependent reciprocal pattern between glycogen content and ASPPase indicate that, especially under conditions of low carbon source, the reaction catalyzed by this enzyme may act as a decisively important limiting step of the gluconeogenic process.
Figure 1.
ASPPase activities and glycogen content in E. coli BL21. Bacteria were grown in 100 ml of M9 minimal medium supplemented with different glucose concentrations. Values presented correspond to bacteria entering the stationary phase of growth.
ASPPase Is a Member of the “Nudix” Hydrolases.
Purified ASPPase was subjected to peptide sequencing. Comparison of the 15 amino acids from its N-terminal against sequences existing in the EMBL data bank showed that ASPPase corresponds to the product of orf209 (accession number P36651), a member of a previously characterized group of proteins designated as “nudix” (nucleoside diphosphate linked to some other moiety, X) hydrolases that exist in organisms ranging from viruses to mammalians, whose suggested role is to cleanse the cell from potentially deleterious endogenous metabolites (19).
Although the product of orf209 remained to be identified in E. coli, analyses consisting of the systematic expression of nudix hydrolase-encoding ORFs have revealed that the product of orf209 is able to hydrolyze ADP-ribose (20). Our results demonstrate that orf209 is indeed a gene, which will be designated as aspP in this work, encoding an ASPPase.
Insertional Mutagenesis of aspP Induces a Dramatic Reduction of ADPG Hydrolytic Activities and Increase of Glycogen Content.
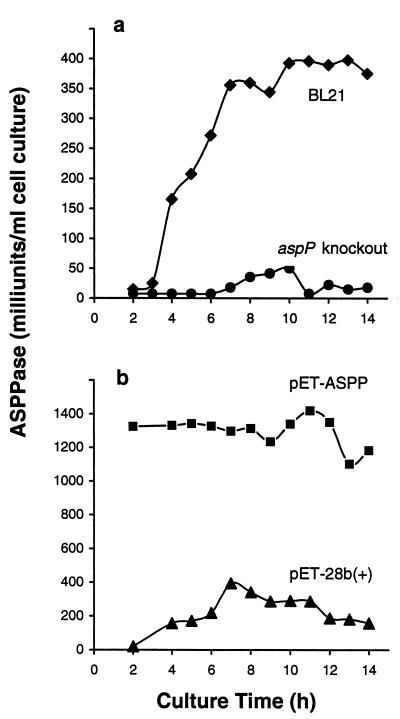
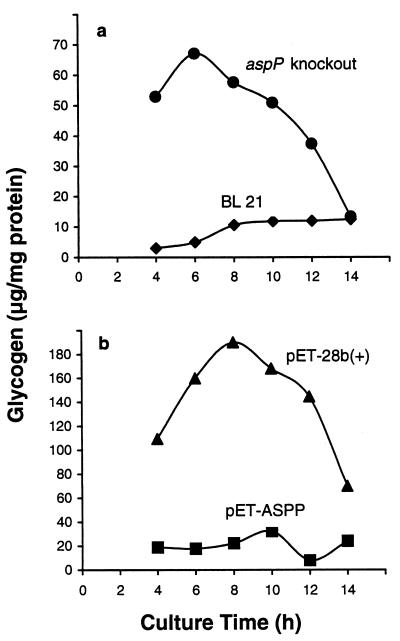
aspP was disrupted by insertional mutagenesis, and a comparison of the ADPG hydrolytic activities and glycogen content between the mutant and wild-type cells was carried out. As presented in Fig. 2a, whereas the untransformed cells showed readily detectable activities, they were almost undetectable in the aspP knockout mutants in any of the stages of the cell growth. These results thus demonstrate that most, if not all, ADPG hydrolytic activity in E. coli is catalyzed by the product of aspP, ASPPase. Although ASPPase can hydrolyze ADP-ribose (see Table 2), aspP insertional mutants showed no significant reduction of ADP-ribose hydrolytic activities (data not shown), which is ascribable to various enzymes existing in E. coli that catalyze the hydrolytic breakdown of this nucleotide-sugar (21, 22). Remarkably, aspP knockout mutants grown in low glucose concentrations exhibited a significantly higher glycogen content than the wild-type cells at any stage of the cell growth (Fig. 3a), the overall results thus strongly indicating that the reaction catalyzed by ASPPase is a limiting step of the gluconeogenic process.
Figure 2.
Maximum ASPPase catalytic activities in aspP-overexpressing and aspP-deficient bacteria. Cells were grown in batch cultures containing 100 ml of M9 supplemented with 15 mM glucose. At the indicated incubation periods, the cells were harvested for measurement of ADPG hydrolytic activities. (a) ASPPase activities in E. coli BL21 and in the aspP knockout mutant. (b) ASPPase activities in BL21(DE3) bacteria transformed with pET-28b(+) or with pET-ASPP. Zero time of the incubation time starts when the overnight grown culture is inoculated to fresh medium.
Figure 3.
Glycogen content in aspP insertional mutants (a) and in aspP-overexpressing bacteria (b). The cells were grown in the presence of 2 mM and 50 mM glucose, respectively. At the indicated incubation periods, the cells were harvested for measurement of the glycogen content.
Enhanced Expression of aspP Results in a Dramatic Reduction of Intracellular Levels of Glycogen.
We then attempted to explore the effect of increasing ASPPase activities on the glycogen content. Toward this end, aspP was cloned into the pET-28b(+) expression vector, and the resulting plasmid (pET-ASPP) was used to transform E. coli BL21(DE3). These cells were found to accumulate an approximate 30-kDa protein that showed kinetic properties identical to those presented in Table 2 (data not shown). Furthermore, bacteria transformed with pET-ASPP exhibited significantly higher ASPPase activities than the control cells transformed with pET-28b(+) at any stage of the growth (Fig. 2b).
Cells transformed with pET-28b(+) were shown to accumulate detectable amounts of glycogen at any time of their culture. Following the pattern of glycogen accumulation described in several bacterial species (4, 5, 23), these cells exhibited a maximum of their glycogen content when entering into the stationary phase of growth (Fig. 3b). Most importantly, pET-ASPP transformants showed a dramatic reduction of glycogen content at any stage of the cell growth even under conditions of high glucose concentration in the culture medium (Fig. 3b), which further strengthens the view that the reaction catalyzed by ASPPase is a limiting step of the gluconeogenic process.
Changes in aspP Expression Are Not Accompanied by Pleiotropic Changes in Bacterial Growth, Biofilm Formation, and Activities of Enzymes Involved in Glycogen Biosynthesis.
Mutations affecting the glycogen biosynthetic process in E. coli have been reported to occur that are accompanied by changes in cell size, growth, morphology, surface adherence, and synthesis of exopolysaccharides necessary for biofilm production (23, 24). Therefore, to explore whether or not aspP mutations may result in such pleiotropic changes, comparison of the cell growth patterns and biofilm formation capacities were performed by using both transformed and untransformed bacteria.
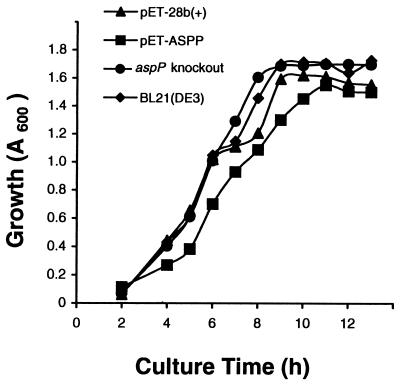
As shown in Fig. 4, both aspP-overexpressing and aspP-deficient cells exhibited normal growth behavior as compared with untransformed bacteria, confirming that, as observed in other mutants with altered glycogen content, this polysaccharide does not appear to be necessary for bacterial growth (25). Furthermore, both aspP-overexpressing and aspP-deficient cells exhibited normal capacities to produce biofilms as compared with the wild-type bacteria (data not shown), the overall results thus indicating that the reaction catalyzed by ASPPase is not a limiting step in the gluconeogenic pathways leading to the production of exopolysaccharides.
Figure 4.
Nonpleiotropic effect of altered aspP expression on bacterial growth. Cells were grown in batch cultures containing 100 ml of M9 supplemented with 50 mM glucose. At the indicated incubation periods, the cells were harvested for determination of growth (A600) in the BL21 wild-type cells, aspP knockout mutant, and BL21(DE3) transformed with pET-ASPP or with pET-28b(+).
The use of mutants has been proved to be a powerful tool to establish the currently prevailing schematic pathway of glycogen biosynthesis, wherein AGPase occupies a decisively important role (26–28). It must be pointed out, however, that all of these studies have not been substantially backed up by determining possible pleiotropic changes in enzymes involved in glycogen biosynthesis. However, our current ongoing investigations have clearly demonstrated that aspP mutations described herein do not affect the activities of enzymes typically considered to be involved in glycogen biosynthesis (unpublished results).
Additional Remarks.
Evidence has been presented that, basically analogous to the role attributable to ADPG pyrophosphatase in plants (12), ASPPase may play a role in preventing carbon flow toward glycogen biosynthesis.
It has been well recognized that futile cycles provide the cell with a useful regulatory mechanism in the control of the flux through a metabolic pathway (29). In this context, turnover of bacterial glycogen has been interpreted by the idea that glycogen acts as a carbon capacitator that regulates the carbon flow through glycolysis (4, 5). Essentially in agreement with this conceptual idea, results presented in this work strongly indicate that ASPPase may play an important role in connecting the cyclic gluconeogenic process with other metabolic pathways such as glycolysis.
Our computer-assisted analyses using the PSORT algorithm (18) have revealed that ASPPase has characteristics of a typical cytosolic protein of Gram-negative bacteria (data not shown). It is therefore highly conceivable that, in contrast to other bacterial nucleotide-sugar pyrophosphatases that act as “ectoenzymes” involved in the scavenging of extracellular nucleotide-sugars (30, 31), ASPPase is in the same compartment of the cell as are the established processes of ADPG synthesis and utilization. Because it exhibits high affinity toward ADPG (Table 2), thus competing with glycogen synthase for the same substrate, it can be readily predicted that, unless ASPPase is highly regulated, ADPG will be instantaneously hydrolyzed to produce glucose-1-phosphate and AMP, thus preventing glycogen biosynthesis. However ASPPase can be inhibited by nonsubstrate molecules. Moreover, results presented in Fig. 1 showing a glucose-dependent reciprocal relationship between ASPPase activities and glycogen content in E. coli strongly indicate that aspP expression can be adjusted to the physiological status of the cell. Needless to say, further research testing possible mechanisms that regulate the expression of aspP is essential to clarify the contribution of ASPPase in the control of glycogen biosynthesis in bacteria.
Acknowledgments
We thank Maria José Villafranca and Amaya Azqueta (Instituto de Agrobiotecnologia y Recursos Naturales, Spain) for expertise technical support. We are indebted to Pepa González-Nicolás (Universidad Autónoma de Madrid, Spain) for helpful and valuable contributions in microsequencing of ASPPase. We thank David Ndlovu for critical examination of the text. T.A. sincerely appreciates the support from the Spanish Ministry of Culture and Education.
Abbreviations
- ADPG
ADP-glucose
- AGPase
ADPG pyrophosphorylase
- ASPPase
ADP-sugar pyrophosphatase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the EMBL database (accession no. AJ298136) and in the SWISS-PROT protein database (accession no. P82969).
References
- 1.Preiss J, Romeo T. Prog Nucleic Acid Res Mol Biol. 1994;47:301–327. doi: 10.1016/s0079-6603(08)60255-x. [DOI] [PubMed] [Google Scholar]
- 2.Liu M Y, Yang H, Romeo T. J Bacteriol. 1995;177:2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudet G, Forano E, Dauphin G, Delort A M. Eur J Biochem. 1992;207:155–162. doi: 10.1111/j.1432-1033.1992.tb17032.x. [DOI] [PubMed] [Google Scholar]
- 4.Belanger A E, Hatfull G F. J Bacteriol. 1999;181:6670–6678. doi: 10.1128/jb.181.21.6670-6678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guedon E, Desvaux M, Petitdemange H. J Bacteriol. 2000;182:2010–2017. doi: 10.1128/jb.182.7.2010-2017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David M, Petit W A, Laughlin M R, Shulman R G, King J E, Barrett E J. J Clin Invest. 1990;86:612–617. doi: 10.1172/JCI114752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozueta-Romero J, Akazawa T. J Exp Bot. 1993;44,Suppl.:297–306. [Google Scholar]
- 8.Massillon D, Bollen M, De Wulf H, Overloop K, Vanstapel F, Van Hecke P, Stalmans W. J Biol Chem. 1995;270:19351–19356. doi: 10.1074/jbc.270.33.19351. [DOI] [PubMed] [Google Scholar]
- 9.Sweetlove L J, Burrell M, ap Rees T. Biochem J. 1996;320:493–498. doi: 10.1042/bj3200493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pozueta-Romero J, Perata P, Akazawa T. Crit Rev Plant Sci. 1999;18:489–525. [Google Scholar]
- 11.Hickman S, Wong-Yip Y P, Rebbe N F, Greco J M. J Biol Chem. 1985;260:6098–6106. [PubMed] [Google Scholar]
- 12.Rodríguez-López M, Baroja-Fernández E, Zandueta-Criado A, Pozueta-Romero J. Proc Natl Acad Sci USA. 2000;97:8705–8710. doi: 10.1073/pnas.120168097. . (First Published July 11, 2000; 10.1073/pnas.120168097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-López M, Baroja-Fernández E, Zandueta-Criado A, Moreno-Bruna B, Muñoz F J, Akazawa T, Pozueta-Romero J. FEBS Lett. 2001;490:44–48. doi: 10.1016/s0014-5793(01)02135-4. [DOI] [PubMed] [Google Scholar]
- 14.Melo A, Glaser L. Biochem Biophys Res Commun. 1966;22:524–531. doi: 10.1016/0006-291x(66)90306-8. [DOI] [PubMed] [Google Scholar]
- 15.Link A J, Phillips D, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasa I, Castón J R, Fernández-Herrero L A, de Pedro M A, Berenguer J. Mol Microbiol. 1992;6:1555–1564. doi: 10.1111/j.1365-2958.1992.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 17.O'Toole G A, Kolter R. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 18.Nakai K, Kanehisa M. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 19.Bessman M J, Frick D N, O'Handley S F. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 20.Dunn C A, O'Handley S F, Frick D N, Bessman M J. J Biol Chem. 1999;274:32318–32324. doi: 10.1074/jbc.274.45.32318. [DOI] [PubMed] [Google Scholar]
- 21.Frick D N, Bessamn M J. J Biol Chem. 1995;270:1529–1534. doi: 10.1074/jbc.270.4.1529. [DOI] [PubMed] [Google Scholar]
- 22.O'Handley S F, Frick D N, Dunn C A, Bessman M J. J Biol Chem. 1998;273:3192–3197. doi: 10.1074/jbc.273.6.3192. [DOI] [PubMed] [Google Scholar]
- 23.Romeo T, Gong M, Liu M Y, Brun-Zinkernagel A-M. J Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Toole G, Kaplan H B, Kolter R. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 25.Preiss J. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh P, Meyer C, Remy E, Peterson D, Preiss J. Arch Biochem Biophys. 1992;296:122–128. doi: 10.1016/0003-9861(92)90553-9. [DOI] [PubMed] [Google Scholar]
- 27.Hill M A, Kaufmann K, Otero J, Preiss J. J Biol Chem. 1991;266:12455–12460. [PubMed] [Google Scholar]
- 28.Meyer C R, Ghosh P, Remy E, Preiss J. J Bacteriol. 1992;174:4509–4512. doi: 10.1128/jb.174.13.4509-4512.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neijssel O M, Buurman E T, Teixeira de Mattos M J. Biochim Biophys Acta. 1990;1018:252–255. doi: 10.1016/0005-2728(90)90260-b. [DOI] [PubMed] [Google Scholar]
- 30.Mauck J, Glaser L. Biochemistry. 1970;9:1140–1147. doi: 10.1021/bi00807a014. [DOI] [PubMed] [Google Scholar]
- 31.Burns D M, Beachman I R. Nucleic Acids Res. 1986;14:4325–4342. doi: 10.1093/nar/14.10.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]