Abstract
Cuticular wax composition greatly impacts plant responses to dehydration. Two parallel pathways exist in Triticeae for manipulating wax composition: the acyl elongation, reduction, and decarbonylation pathway that is active at the vegetative stage and yields primary alcohols and alkanes, and the β-diketone pathway that predominates at the reproductive stage and synthesizes β-diketones. Variation in glaucousness during the reproductive stage of wheat is mainly controlled by the wax production genes, W1 and W2, and wax inhibitor genes, Iw1 and Iw2. Little is known about the metabolic and physiological effects of the genetic interactions among these genes and their roles in shifting wax composition during plant development. We characterized the effect of W1, W2, Iw1, and Iw2 and analyzed their interaction using a set of six near-isogenic lines (NILs) by metabolic, molecular and physiological approaches. Loss of functional alleles of both W genes or the presence of either Iw gene depletes β-diketones and results in the nonglaucous phenotype. Elimination of β-diketones is compensated for by an increase in aldehydes and primary alcohols in the Iw NILs. Accordingly, transcription of CER4-6, which encodes an alcohol-forming fatty acyl-CoA reductase, was elevated 120-fold in iw1Iw2. CER4-6 was transcribed at much higher levels in seedlings than in adult plants, and showed little difference between the glaucous and nonglaucous NILs, suggesting that Iw2 counteracts the developmental repression of CER4-6 at the reproductive stage. While W1 and W2 redundantly function in β-diketone biosynthesis, a combination of both functional alleles led to the β-diketone hydroxylation. Consistent with this, transcription of MAH1-9, which encodes a mid-chain alkane hydroxylase, increased seven-fold only in W1W2. In parallel with the hydroxyl-β-diketone production patterns, glaucousness was intensified and cuticle permeability was reduced significantly in W1W2 compared to the other NILs. This suggests that both W1 and W2 are required for enhancing drought tolerance.
Introduction
The aerial organs of terrestrial plants are coated with an extracellular layer of hydrophobic lipids, termed cuticle. Produced by epidermal cells, the cuticle plays important roles in plant growth and development and, as the interface between sessile plants and the environments they live in, in the interaction with abiotic and biotic elements [1], [2]. Based on solubility in organic solvents, the cuticle is composed of insoluble cutin and soluble cuticular wax. Cutin, a cell wall-bound ester polymer of modified fatty acids and glycerol, serves as the backbone of the cuticle [3], [4], [5]. Intracuticular wax is embedded in the underlying cutin framework and epicuticular wax is overlaid on the cutin matrix and intracuticular wax [2], [6]. Wax composition varies with developmental stage, between organs, and with genetic and environmental conditions [1], [4], [6], causing the plant to be bluish-white (glaucous) or nonglaucous. Glaucousness is the visible form of densely distributed epicuticular wax crystallites.
Our current knowledge of wax biosynthesis (Figure S1), which is mainly derived from the model plant Arabidopsis, indicates that this process begins with the release of C16 and C18 fatty acids from the acyl carrier protein (ACP) by fatty acyl–ACP thioesterase B (FATB) [7] and their subsequent activation to CoA thioesters by a long-chain acyl-CoA synthetase (LACS) [8]. The activated forms of these fatty acids are transferred from plastids to the endoplasmic reticulum (ER), where they are made available for fatty acid elongase (FAE), which extends them to very-long-chain fatty acids (VLCFAs). The FAE complex consists of four types of enzymes: β-keto acyl-CoA synthases (KCS), β-keto acyl-CoA reductase (KCR), 3-hydroxy-acyl-CoA dehydratase (HCD), and enoyl-CoA reductase (ECR). In Arabidopsis, ECERIFERUM 6 (CER6) [9], [10], [11] and KCS1 [12] encode KCS and KCR1 [13], PAS2 [14], and CER10 [15] encode KCR, HCD, and ECR, respectively.
The resulting VLCFAs can be released from the FAE complex as free fatty acids, or converted either to primary alcohols by acyl reduction [16] or to alkanes by acyl decarbonylation through an aldehyde intermediate [6], [17] and further to secondary alcohols and ketones by hydroxylation [18]. In the acyl reduction branch, fatty acyl-CoA reductase CER4 exclusively reduces VLCFAs to the corresponding primary alcohols [19], and wax ester synthase/acyl-CoA:diacylglycerol acyltransferase 1 (WSD1) uses the long-chain and very-long-chain primary alcohols and C16 fatty acid for wax ester production [20]. In the decarbonylation branch, CER1 physically interacts with the wax-associated protein CER3 and ER-localized cytochrome b5 isomers to catalyze the redox-dependent biosynthesis of alkanes [21]. Subsequently, midchain alkane hydroxylase 1 (MAH1), a cytochrome P450 enzyme, oxidizes the alkanes to generate secondary alcohols and ketones [18].
All of the cuticular lipid species synthesized in the ER need to be deposited extracellularly. The ABC transporters ABCG12/CER5 [22] and ABCG11/WBC11 [23], [24] are involved in exporting wax through the plasma membrane to the apoplastic space. A glycosylphosphatidylinositol-anchored lipid transfer protein, LTPG, functions as part of the machinery, either as a regulatory component or by creating the appropriate conditions for cuticular lipid exportation [25]. Recent efforts have started to shed light on the regulatory network underlying wax production by identifying transcription factors involved in this process. Several transcription factors of the AP2/EREBP [26], [27], [28], [29] and MYB family [30], [31], [32], [33], [34] regulate genes involved in cuticle biosynthesis. CER7, a 3′-to-5′ exoribonuclease, conditions wax synthesis by degrading a specific mRNA species of a CER3 repressor [35], [36].
In grasses, transposon tagging identified several maize GLOSSY (GL) loci. GL1 is homologous to CER3/WAX2 [37], [38], GL2 is homologous to CER2 [39], GL4 is homologous to CER6/CUT1 [40], and GL8 is homologous to KCR [41], [42], [43]. More recently, screening of a rice T-DNA tagging population isolated a CER6/CUT1 homolog, WSL1 [44]. Furthermore, GL15, which encodes a transcription factor of the AP2 family and is involved in the transition from juvenile to adult leaves [45], and OCL1, which encodes a HD-ZIP IV family transcription factor that activates a fatty acyl-CoA reductase and a putative wax transporter [46], [47], were found to regulate cuticular wax biosynthesis.
In the tribe Triticeae, including wheat and barley, the acyl elongation, reduction, and decarbonylation pathway for the synthesis of alcohol-rich waxes is active in the vegetative stages. A parallel pathway, which predominates in the reproductive stages, is responsible for biosynthesis of β-diketones, i.e., hentriacontane-14,16-dione and its hydroxyl derivatives [48], [49], [50]. The β-diketone pathway differs from the acyl elongation, reduction, and decarbonylation pathway in substrate and inhibitor specificity [49] and in pathway-specific genes [51]. Because β-diketones are not detected in wax extracts from the model plants Arabidopsis and rice, the β-diketone biosynthetic pathway draws much less attention.
Common wheat or bread wheat (Triticum aestivum L) is an important, widely adapted crop that is cultivated in many arid or semiarid areas. The inheritance of wheat glaucousness is mainly governed by two sets of dominant genes, W1 and W2, which promote glaucous formation, and Iw1 and Iw2, which inhibit it. W1 and Iw1 are located on the short arm of chromosome 2B (2BS), and W2 and Iw2 on 2DS [52]. Genetic mapping using molecular markers has localized the Iw1 [53] and Iw2 locus [54], [55] to the distal regions of the chromosome arms. While the non-glaucous phenotype prevails in the wild diploid ancestors [52], [56], the glaucous type predominates in cultivated polyploid wheat, suggesting that glaucousness was favored during wheat domestication. Field physiology studies showed that glaucousness is positively related to wheat yield, especially under drought-stressed conditions [57], [58], [59]. Despite its adaptive importance, the molecular mechanisms underlying variation in glaucousness and its association with drought tolerance remain largely unknown. We characterized a set of six near isogenic lines (NILs) by scanning electron microscopy (SEM), metabolite profiling of wax extracts, measuring the rate of water loss and chlorophyll efflux, and transcriptional profiling of 72 cuticle genes. Here, we report the results of this study and their implications in our understanding of cuticular wax pathways and drought tolerance.
Results
Genetic Background Analysis of Wax NILs
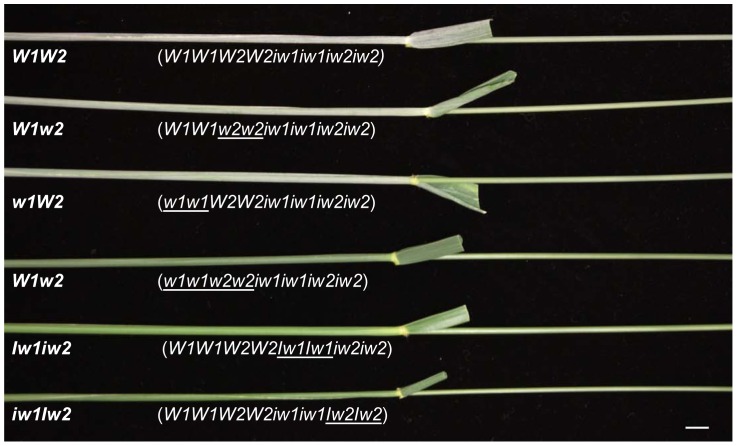
Six wax NILs were developed by ten backcrosses to wheat cultivar S-615 [52] and their genotypes and glaucousness on flag leaf sheaths and peduncles are shown in Figure 1. The NILs W1W2, W1w2, and w1W2 are glaucous, whereas w1w2, Iw1iw2 and iw1Iw2 are not, indicating that W1 and W2 are functionally redundant in forming glaucousness, but that a single Iw gene is sufficient to suppress glaucousness formation. Before physiological, metabolic, and expressional characterization, we inspected the genetic background by single nucleotide polymorphism (SNP) and simple sequence repeat (SSR) genotyping. Of the 9,000 SNP probes assayed, over 5,200 produced positive signals in each line and 18 probes (0.35% of 5,200) or fewer detected polymorphisms among the NILs, suggesting that the NILs are identical over 99.5% of the genome. The lowest level of polymorphism was found between NILs W1W2 and iw1Iw2, where only one probe detected a polymorphism. We also genotyped the six NILs using SSR markers previously mapped at the tips of 2BS and 2DS arms, where the W and Iw genes are located. All of the NILs were positive for all SSR markers, indicating that the mutations were not caused by chromosome deletions. These results indicate that the NILs differ from each other only at a very small fraction of the genome and suggest that there are probably no polymorphisms in wax genes other than at the W and Iw loci.
Figure 1. Flag leaf sheaths and peduncles of the NILs examined in this study.
NIL designations are indicated beneath each peduncle and genotypes are specified in parentheses. The introduced alleles in the genotypes are underlined. The bar indicates 1 cm.
Wax Morphology
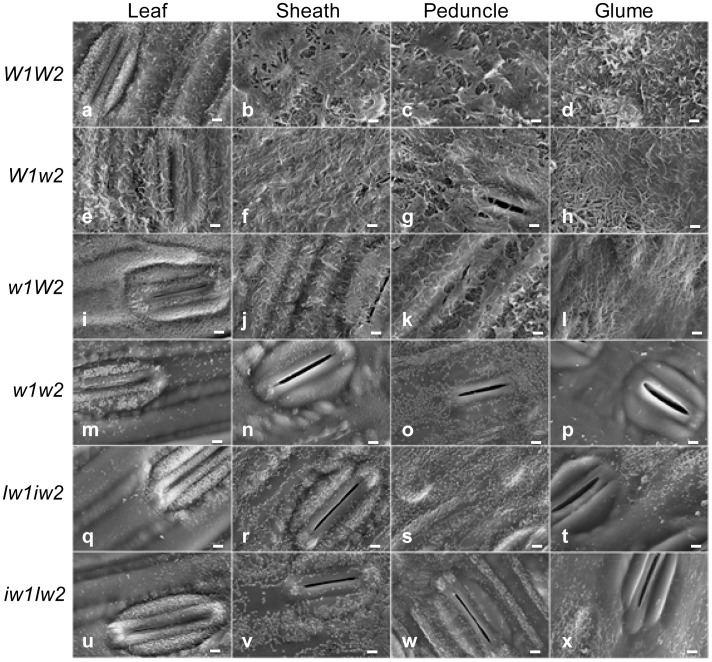
When flowering begins at Fakes’ stage 10.5.1 (F10.5.1), glaucousness is fully developed in the upper part of the wheat plant. To gain insight into the organization of epicuticular wax, we examined the wax crystallites deposited on the abaxial surface of the flag leaf blades and sheaths, the peduncles, and the glumes of the NILs at this stage under SEM. The morphology of wax crystallites in glaucous NILs was clearly different from that of nonglaucous ones (Figure 2). In all tissue types, the cuticle surfaces of glaucous NILs were covered with a meshwork of wax tubes, and the density was higher in W1W2 (Figure 2a, 2b, 2c, and 2d) and W1w2 (Figure 2e, 2f, 2g, and 2h) than in w1W2 (Figure 2i, 2j, 2k, and 2l). Besides wax tubes, large wax sheets were also observed above the wax meshwork in the sheath and peduncle of the W1W2 plants (Figure 2b and 2c). On the leaf cuticles of the w1W2 plants, sparse wax tubes were seen over the ground cells, whereas fine wax granules densely covered the guard cells (Figure 2i).
Figure 2. Electron micrographs of the cuticle surfaces of flag leaf blades, sheaths, peduncles, and glumes of the NILS.
The tissues are indicated on the top and the NIL designations at the left. The bars indicate 2.5 µm.
In contrast to the glaucous NILs, the cuticles of the nonglaucous NILs were either smooth (Figure 2p) or carried fine wax particles. On the leaf cuticles, wax crystallites were mainly found over the guard cells (Figure 2m, 2q, and 2u). However, the guard cells of glumes of the Iw NILs were devoid of wax crystallites (Figure 2t and 2x). Wax crystallites were distributed on both ground and guard cells in the sheath (Figure 2n, 2r and 2v) and peduncle cuticles (Figure 2o, 2s and 2w).
Wax morphology is determined by wax composition. The dramatic difference in wax morphology suggests possible differences in wax content, composition, and cuticle permeability between the glaucous and nonglaucous NILs. Considering that sheath wax morphology was a good indicator of the differences among the NILs, we concentrated our research on the sheath wax.
Wax Composition
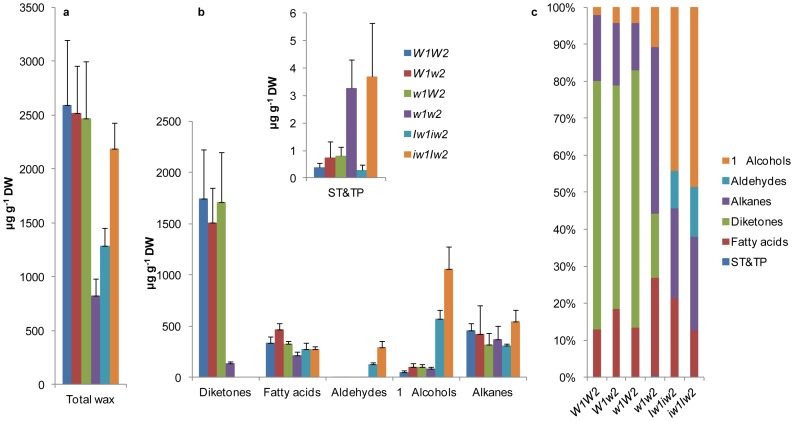
We extracted cuticular wax from the flag leaf sheaths of the wax NILs at stage F10.5.1 and profiled it using gas chromatography-mass spectrometry (GC-MS). The results showed that ∼90% of the wax extract consisted of known wax species and the remaining ∼10% consisted of unknown species. As expected, the NILs varied greatly in total wax load and composition. All glaucous NILs carried the same wax load (P>0.82980; Figure 3a) and had a similar wax composition, with β-diketones accounting for >60%, fatty acids and alkanes for ∼15% each, primary alcohols for 2 to 4%, and cyclic lipids for trace amounts of total wax (Figures 3b and 3c). Wax load varied by ∼60% among the nonglaucous NILs. Compared to W1W2, no significant reduction was found in iw1Iw2 (P = 0.42065); however, a 50% and 68% reduction was detected in Iw1iw2 (P = 0.0412) and w1w2 (P = 0.01577), respectively (Figure 3a). This indicates that wax composition, instead of wax load, determines glaucousness. In contrast to the glaucous NILs, β-diketones were depleted in the nonglaucous NILs, being reduced to ∼8% of total wax species in w1w2 (P = 0.00925) and completely eliminated in Iw1iw2 and iw1Iw2 (Figure 3c). There was little net increase in w1w2 for the remaining components (P>0.12834), which elevated the alkane proportion to ∼45% of the total wax species (Figure 3c), indicating that W genes are specific for β-diketone synthesis. In contrast to the situation in w1w2, loss of β-diketones in Iw1iw2 and iw1Iw2 was compensated by an increase of aldehydes and primary alcohols. Compared to W1W2, aldehydes and primary alcohols were increased ∼250-fold and ∼10-fold in Iw1iw2, and ∼600-fold and 20-fold in iw1Iw2, respectively (P<0.00254; Figure 3b). This indicates that the Iw genes inhibit the synthesis of β-diketones and shunt their substrates to the fatty acyl reduction pathway. In this respect, the action of Iw2 is much stronger than that of Iw1. As a result, three different types of waxes are recognized in this NIL set: β-diketone-rich wax in the glaucous NILs, alkane-rich wax in w1w2, and primary alcohol-rich wax in Iw1iw2 and iw1Iw2 (Figure 3c). In contrast to the aliphatic wax species, cyclic wax species, such as sterols and triterpenes (ST&TP) were detected at much lower levels. Compared to W1W2, ST&TP, which mainly consisted of β-sitosterol and β-amyrin, were increased nine-fold in w1w2 (P = 0.0171; Figure 3b).
Figure 3. Wax composition of the six NILs.
(a) Total wax load of the flag leaf sheaths was measured by GC-MS. (b) β-diketone, fatty acid, aldehyde, primary (1°) alcohol, alkane, and sterol and triterpene (ST&TP) content. The numbers on the y-axes indicate average content expressed as µg per g dried tissue (dry weight, DW). The bars indicate standard deviation of the mean estimated from six biological replicates. (c) The percentage of wax species in each genotype was calculated from the means.
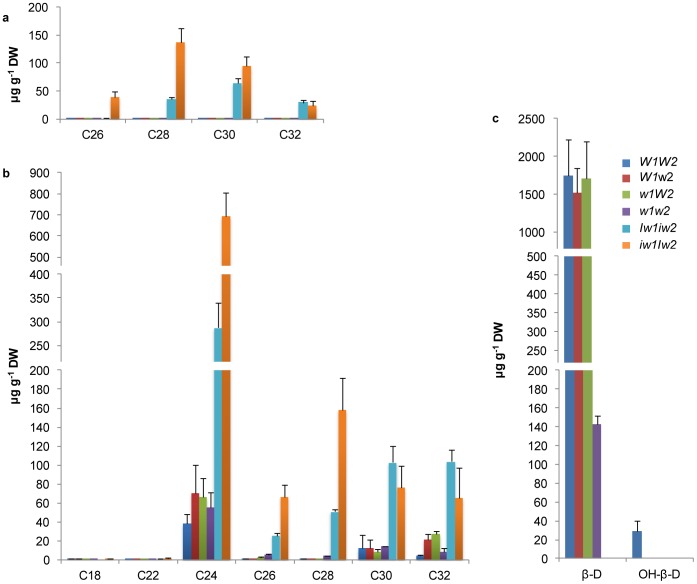
The carbon chain length ranges from C12 to C32 in fatty acids, C25 to C31 in alkanes (Figure S2), C26 to C32 in aldehydes, and C18 to C32 in primary alcohols (Figure 4a and 4b). Compared to W1W2, aldehyde homologs were increased by ∼14- to 485-fold in Iw1iw2 and by ∼170- to 1110-fold in iw1Iw2 (P<0.00928; Figure 4a). VLCFA-derived primary alcohol homologs were increased by 7- to 75-fold in Iw1iw2 and by 6- to 112-fold in iw1Iw2 (P<0.03231; Figure 4b). Although tetracosan-1-ol (C24) was the most abundant homolog, the maximal increase was found in octocosan-1-ol (C28) in Iw1iw2 and in hexacosan-1-ol (C26) in iw1Iw2 (Figure 4b).
Figure 4. Homolog variation of major wax species.
Carbon atom numbers of aldehydes (a) and primary (1°) alcohols (b), and β-diketones (c) are indicated on the x-axes. Their contents are indicated on y-axes as µg per g dried tissue. The bars indicate standard deviation of the mean calculated from six biological replicates. β-D, β-diketone; and OH-β, hydroxyl-β-diketones.
A single carbon chain length, C31, was detected for β-diketones; however, three substituted derivatives were identified: 8- and 9-hydroxyl isomers, and enolic isomer. Considering keto-enol tautomerism, we combined enolic β-diketone with β-diketone. No difference in the unsubstituted β-diketone was found among the glaucous NILs; however, that amount was reduced >10-fold in w1w2 compared with in the glaucous NILs (P<0.00985; Figure 4c). Hydroxyl-β-diketones were only detected in W1W2, and not in W1w2 and w1W2 (Figure 4c), indicating that an interaction between W1 and W2 is required for β-diketone hydroxylation at C8 or C9. Depending on the presence of hydroxyl β-diketones, the β-diketone-rich wax can be further divided into two types, OH-β wax in W1W2 and β-D wax in W1w2 and w1W2.
Transcriptional Profiling of Cuticle Genes
When flag leaves are fully emerged from the whorl (stage F9.0), flag leaf sheaths elongate rapidly. Because cuticle genes are highly expressed in the elongating epidermal cells [60], [61], we selected flag leaf sheaths at this stage for transcription profiling. We chose nine candidate reference genes (Table S1) from previous publications [62], [63], [64] and validated their expression stability in 18 cDNA samples from the NIL set using programs qBasePlus (Biogazelle, Belgium) and NormFinder [65]. Both programs demonstrated that TaRPII36 was the best reference gene for quantifying cuticle gene expression (Figure S3).
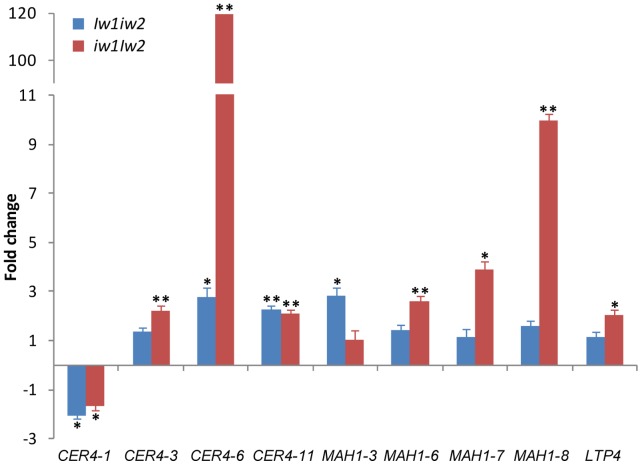
The increase of aldehydes and primary alcohols in Iw1iw2 and iw1Iw2 suggests that the Iw genes activate CER4 homologs; the abundance of β-diketones in the glaucous NILs suggests that the W genes up-regulate special KCS homologs and the decarbonylation components and the Iw genes suppress them; and the presence of hydroxyl-β-diketones only in W1W2 suggests that the interaction between W1 and W2 activates MAH1 homologs. To test these hypotheses, we profiled the transcription of wheat wax candidate genes with an emphasis on the CER1, CER3, CER4, KCS, and MAH1 gene families. To test if cutin is also involved in the phenotype variation, we selected five cutin biosynthetic genes. We designed primers for 64 unigenes and adopted primers for eight additional genes from a previous publication [66]. Thus, we evaluated the transcription of 72 genes involved in cutin and wax biosynthesis, transport, and transcription regulation (Table S1). Compared with W1W2, the expression of 11 genes in Iw1iw2 and 29 genes in iw1Iw2 was significantly altered, of which the expression patterns of seven genes were shared by Iw1iw2 and iw1Iw2 (Table S2). Of these 72 genes examined, nine, including four CER4 members, four MAH1 members, and LTP4, showed over a two-fold difference in expression. The most dramatic change was observed in CER4-6, which was up-regulated ∼120-fold in iw1Iw2 and 2.8-fold in Iw1iw2 (Figure 5). Searching the D-genome physical mapping database (http://probes.pw.usda.gov/WheatDMarker) revealed that CER4-6 is located on chromosome 5D. This indicates that CER4-6 is not Iw2 itself, which is located on chromosome 2D, but an Iw2 target that possibly plays a role in increasing aldehyde and primary alcohol content. CER4-11 was also up-regulated two-fold in both Iw1iw2 and iw1Iw2. In addition, four MAH1 members, MAH1-4, MAH1-7, MAH1-8, and MAH1-9, were differentially expressed in Iw1iw2 and iw1Iw2 (Figure 5 and Table S2). Together, these results indicate that Iw1 differs from Iw2 in terms of the molecular mechanisms for increasing aldehyde and primary alcohol content.
Figure 5. Expression of cuticular wax genes in the wheat flag leaf sheath of the Iw NILs compared to W1W2.
Genes with two- or higher-fold changes are depicted and expression data for all genes analyzed are listed in Table S2. The bars represent standard deviation of the mean fold-change of mRNA levels. Asterisks indicate that the difference is significant at P<0.05 (*) or at P<0.01 (**).
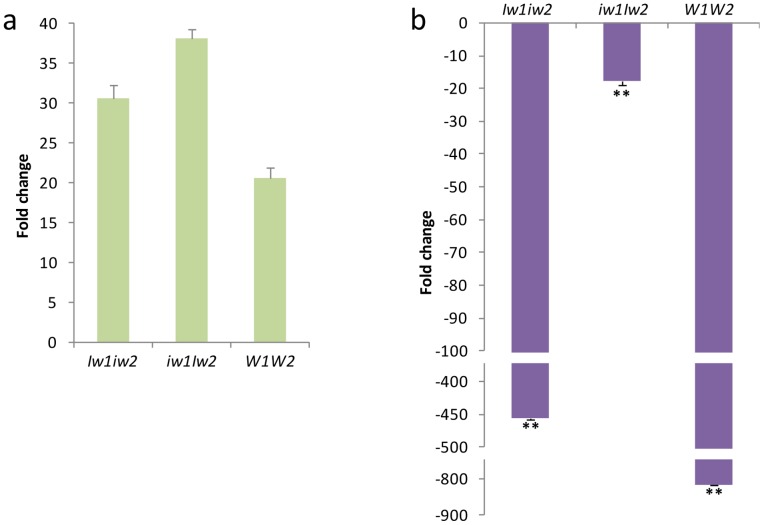
We further asked if the up-regulation of CER4-6 was the result of activation or de-repression by Iw2. To test this, we examined the transcription of CER4-6 in sheaths of W1W2, Iw1iw2, and iw1Iw2 plants at the seedling stage (F4.0). Wax composition changed dramatically during wheat development, from alcohol-rich wax in the vegetative stage to β-diketone-rich wax in the reproductive stage, particularly in leaf sheaths [48]. We found that CER4-6 transcription was increased by less than two-fold in Iw1iw2 and iw1Iw2 compared to W1W2 at stage F4.0 (P<0.0002; Figure 6a), which was much smaller than the difference detected at stage F9.0 (Figure 5). Compared to stage F4.0, CER4-6 was down-regulated 815-fold in W1W2 (P = 1×10−7) and 450-fold in Iw1iw2 (P = 2×10−7), but only 17-fold in iw1Iw2 at stage F9.0 (P = 4×10−7; Figure 6b). This indicates that expression of CER4-6 is under developmental regulation and is suppressed at the reproductive stages, and that Iw2 counteracts this developmental suppression.
Figure 6. CER4-6 expression in sheaths of Iw1iw2, iw1Iw2, and W1W2 at different developmental stages.
(a) Transcription levels of CER4-6 at stage F4.0 compared to that of the reference gene TaRPII36. (b) Fold changes of CER4-6 transcription at stage F9.0 compared to that at stage F4.0. The bars represent standard deviation of the mean fold-change of mRNA levels. Asterisks indicate that the difference is significant at P<0.01 (**).
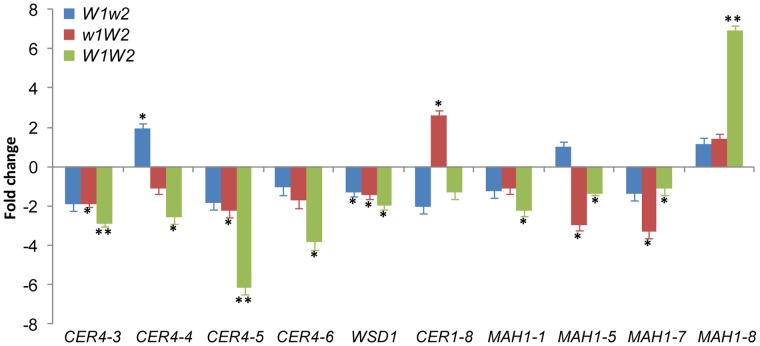
Compared to the situation in w1w2, expression of four genes in W1w2, 19 genes in w1W2, and 26 genes in W1W2 was significantly altered (Table S3). The expression patterns of three genes overlapped between W1w2 and W1W2, 15 between w1W2 and W1W2, and two among all three glaucous NILs. While transcriptional changes of five genes relative to w1w2 were detected in W1w2 or w1W2 but not in W1W2, changes of another 10 genes were detected in W1W2 but not in W1w2 or w1W2. This indicates that W1 and W2 had a non-additive effect on the expression of these cuticular wax-related genes. The expression of four genes in w1W2 and six genes in W1W2 was altered by two-fold or more, including four CER4 members, five MAH1 members, and CER1-8 (Table S3; Figure 7). The expression of MAH1-8 matches the production pattern of hydroxyl-β-diketones: no change in W1w2 and w1W2 were observed, but the expression increased seven-fold in W1W2. MAH1 is responsible for alkane hydroxylation in Arabidopsis [18], suggesting a role for this gene in generating hydroxyl-β-diketones in wheat.
Figure 7. The expression of cuticular wax- and cutin-related genes in the wheat flag leaf sheath of W1w2, w1W2, and W1W2 plants compared to w1w2.
Genes with two- or higher-fold changes are depicted and expression data for all genes analyzed are listed in Table S3. The bars represent standard deviation of the mean fold-change of mRNA levels. Asterisks indicate that the difference is significant at P<0.05 (*) or at P<0.01 (**).
Cuticle Permeability
Nonstomal transpiration is directly correlated with cuticle permeability. We evaluated the cuticle permeability of this NIL set by measuring the rates of water loss and chlorophyll efflux of the flag leaf sheaths at stage F10.5.1. Simultaneously, we inspected stomatal density and aperture size under a light microscope. All of the NILs had a similar stomal density, i.e., ∼60 stomata in a field of view at a magnification of 10×20, and most stomata were closed 1 h after detachment at room temperature and under lab conditions. Therefore, the differences observed in water-loss rate were presumably attributed to cuticle permeability.
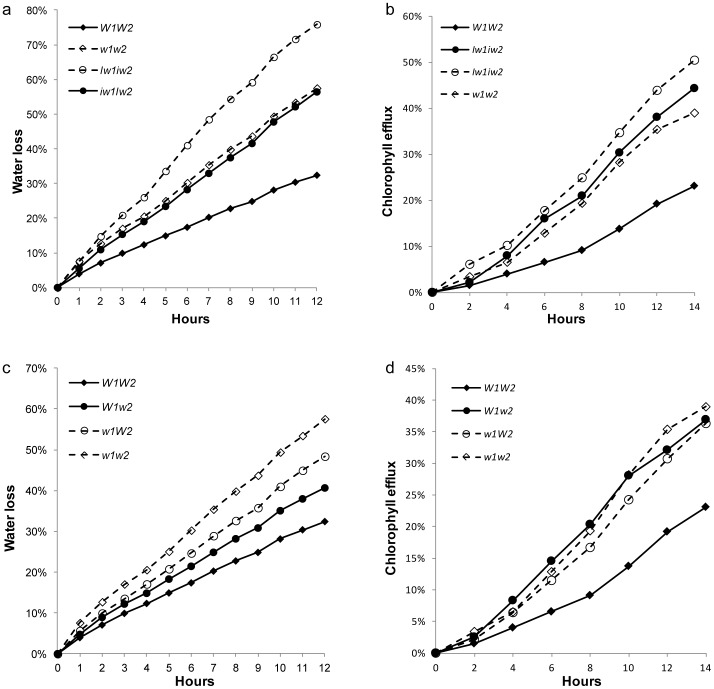
Compared to W1W2, Iw1iw2 and iw1Iw2 started showing higher rates of water loss 1 h after detachment (P<0.03694) and the differences remained and even increased thereafter (Figure 8a). The Iw NILs also showed significantly higher chlorophyll leaching rates than W1W2 after 4 h of treatment (P<0.04285; Figure 8b). Although Iw1iw2 and iw1Iw2 exhibited similar chlorophyll efflux rates (P>0.05503; Figure 8b), Iw1iw2 showed a higher water-loss rate after 4 h of treatment (P<0.03272; Figure 8a). These findings echo the wax data and suggest that the increased amount of primary alcohols and aldehydes in iw1Iw2 have some effect on blocking nonstomatal transpiration, but not on preventing chlorophyll efflux.
Figure 8. Cuticle permeability of the NILs.
Cuticle permeability was evaluated by air drying at room temperature (a and c) and chlorophyll leaching in 80% ethanol (b and d). The numbers on the x-axes represent hours of treatment. Water loss or chlorophyll leaching at each time point is represented on the y-axes as percentages of the total water content or total chlorophyll content in the tissue. Measurements taken from four individuals were averaged.
Similar to iw1Iw2, w1w2 showed a significantly higher rate of both water loss and chlorophyll leaching compared to W1W2 (Figure 8a and 8b). The difference in water loss between w1w2 and W1W2 was detectable as early as 1 h after detachment (P = 0.03607; Figure 8c) and that in chlorophyll leaching at 6 h of treatment (P = 0.04527; Figure 8d). In both experiments, the performance of w1W2 was similar to that of w1w2 (P>0.13101) and significantly different from W1W2 after 3 h of desiccation and 6 h of chlorophyll-leaching (P<0.0494). The findings for W1w2 were not consistent in the two experiments. The water-loss rate of this NIL was intermediate, being higher than that of W1W2 (P<0.04174), but lower than that of w1w2 (P<0.0356 (Figure 8c). In the chlorophyll-leaching experiment, W1w2 showed high similarity to w1w2 (P>0.11128) and significant difference from W1W2 after 6 h of treatment (P<0.02202; Figure 8d). Collectively, W1W2 showed significantly lower rates of water loss and chlorophyll efflux than the nonglaucous and other glaucous NILs, suggesting that cuticle permeability is not inversely proportional to wax load, but rather that it is closely related to wax composition, and that W1 and W2 function epistatically to reduce cuticular transpiration and chlorophyll leaching.
Discussion
Glaucousness is a classic genetic marker trait of wheat. Recent drought threat renewed interest in this trait. Despite several reports on the molecular mapping of Iw genes [53], [54], [55] and quantitative trait loci that affect glaucousness [67], this is the first systematic characterization of wheat glaucousness. In this study, we combined SEM, metabolite profiling, gene expression profiling, and physiologic approaches in a near isogenic background to gain insight into the genetic regulation of cuticular wax biosynthesis and its perturbation during drought tolerance.
Wax Composition and Glaucousness
Numerous cuticular wax genes were identified based on the loss- or reduction-of-glaucousness phenotypes, such as the cer mutants of Arabidopsis. The CER genes participate in almost all steps of cuticle biosynthesis, indicating that glaucousness formation requires multiple wax components. Comparative analyses of the cuticular wax profiles of glaucous and nonglaucous durum lines and of wheat aneuploids found that β-diketone was critical for glaucousness formation [68], [69]. Our research showed that wax composition is more important than wax load in glaucousness determination and that the nonglaucous NILs differ from the glaucous ones in terms of β-diketone content. Iw1iw2 and iw1Iw2 completely lacked β-diketones and w1w2 exhibited a ∼90% reduction in β-diketones. These findings confirm that β-diketones are essential for glaucousness formation. SEM observation also confirmed that wax composition was directly related to wax morphology: β-diketones form wax tubes in the glaucous NILs and primary alcohols form wax chips in the Iw NILs. The glaucous NILs and w1w2 carried trace aldehydes and low levels of primary alcohols, suggesting that these wax species play a limited role in glaucousness formation.
Compared to W1w2 and w1W2, W1W2 showed slightly higher levels of glaucousness (Figure 1), but no difference in total β-diketone load. The major difference among the glaucous NILs is the presence of 8- and 9-hydroxyl β-diketones in W1W2 and the absence of these hydroxy isomers in W1w2 and w1W2. It is believed that hydroxyl-β-diketone is derived from β-diketone through mid-chain hydroxylation. In barley, this hydroxylation is defined by the cer-u mutants [70]. In the genus Triticum, 25-isomer is the typical hydroxyl-β-diketone in durum wax, but is substituted by 8- and 9-isomers in hexaploid wheat [71], suggesting that the generation of these substituted β-diketones involves the interaction of several genes. The absence of hydroxyl β-diketones in W1w2 and w1W2 suggests that the W1-W2 interaction is required for β-diketone hydroxylation and that these substituted β-diketones are important for intensifying glaucousness and forming wax crystallite sheets in W1W2 (Figure 2b and 2c).
Wax Pathways
During plant development, multiple wax pathways operate in parallel to produce different wax types [49], [51], [72], and wax composition changes as plants develop and grow. In the Triticeae, primary alcohols and alkanes are the predominant wax species at the seedling stage [61], [66] and β-diketones are abundant at the reproductive stage, particularly in the sheaths and spike [48], [61], [70]. Genetic and biochemical studies in barley supported the existence of two parallel pathways, the acyl elongation, reduction, and decarbonylation pathway and the β-diketone pathway [50]. Mutations in CER-cqu of barley [51] and in the W genes of wheat (Figure 3) specifically affect β-diketone synthesis. However, we know little about the cross-talk between these two pathways. In the present research, we identified three major types of waxes in a set of six NILs: alkane-rich wax in w1w2, alcohol-rich wax in the Iw NILs, and β-diketone-rich wax in the glaucous NILs (Figure 3c). A survey of Triticeae species found that many wild species produce alcohol-rich wax at the reproductive stage [73], implying that the Iw alleles prevail in nature and that the iw mutations led to the production of β-diketone-rich wax. The similar wax loads in W1W2 and iw1Iw2 (Figure 3a) suggest that precursors are shunted between the acyl reduction pathway and the β-diketone pathway by Iw2. Consistent with this, we found that expression of CER4-6 was elevated 120-fold in iw1Iw2. The wax composition in the Iw NILs at the adult stage (F10.5.1) resembles that at the seedling stage [48], being rich in primary alcohols and lacking β-diketones. Transcription quantification further demonstrated that CER4-6 is active in the seedling stage (F4.0) in W1W2, Iw1iw2, and iw1Iw2 (Figure 6a). Expression of CER4-6 was reduced in all three NILs at stage F9.0; however, the reduction was most pronounced in W1W2 (Figure 6b). Similarly, expression of four FAE genes showed no difference between iw1Iw2 and W1W2 in the seedling stage, but was reduced at the adult plant stage to a much greater extent in W1W2 than in iw1Iw2 (Figure S4). This suggests that the genes involved in the acyl elongation and reduction pathway are repressed by reproductive development and de-repressed by the Iw genes, which act as cross-talkers that regulate the acyl reduction and β-diketone pathway. Iw2 maintains an active CER4-6 at the reproductive stage and suppresses β-diketone production. It would be fascinating to determine how these two opposite activities are coupled by one gene. This is probably achieved by an interaction between Iw2 and different sets of genes. Molecular cloning of Iw2 and an in-depth analysis of CER4-6 expression will provide insight into the developmental regulation of the wax composition shift.
Much work has focused on the biosynthesis of β-diketones in barley by genetic and metabolic analyses of the cer mutants. Wax profiling found that cer-q, cer-c, and cer-u mutants define chain elongation, decarbonylation, and hydroxylation reactions in β-diketone synthesis, respectively [50], [70]. However, genetic analyses of these mutants in barley support the hypothesis that β-ketoacyl elongation, decarbonylation, and hydroxylation are carried out by one gene, cer-cqu [50], [51]. It is difficult to imagine how one gene might govern three different reactions. cer-cqu has been redesignated as gsh1 and was mapped to the subtelomeric region of the barley chromosome arm 2HS (http://wheat.pw.usda.gov/ggpages/bgn/26/BGS351), which is colinear to the W1 locus in wheat [52]. In the present research, we found that the W1 and W2 genes in wheat are each sufficient for the deposition of β-diketone and complement each other, but that both are required for the synthesis of hydroxyl-β-diketones. One scenario would be that the W genes encode transcription factors belonging to the same family of proteins that activate the transcription of the β-diketone biosynthetic genes. We profiled 18 FAE genes; however, none of them were upregulated in the glaucous NILs. This is probably because β-diketones are synthesized by a separate enzyme system [50], but that the genes tested were chosen based on their function in model plants, which do not synthesize β-diketones. The proposed β-diketone biosynthetic pathway includes a decarbonylation reaction [50]. We examined the expression of eight CER1 and five CER3 members of the decarbonylation pathway, and found that only the expression of CER1-8 was increased in w1W2 (2.6-fold). To explore the possibility that MAH1 participates in β-diketone hydroxylation, we evaluated the expression of eight MAH1 members and found that MAH1-8 matched the production pattern of hydroxyl-β-diketones (Figures 4c and 7). Compared to W1W2, transcription of MAH1-8 was further elevated (∼10-fold) in iw1Iw2, but hydroxyl-β-diketone was not detected, because Iw2 inhibits the biosynthesis of β-diketone, the substrate of hydroxyl-β-diketones.
The wheat W1 gene resembles the barley cer-cqu locus in terms of chromosomal location and regulation of β-diketone synthesis and hydroxylation. Considering the close phylogenetic relationship between barley and wheat, cer-cqu is probably orthologous to W1 and may also need to interact with other genes for β-diketone hydroxylation.
β-Diketones and Drought Tolerance
Previous physiological studies in wheat found that glaucousness significantly increased grain and biomass yield in irrigated and rainfed field experiments [74], and increased the photosynthesis to transpiration ratio and reduced the photosynthetic surface temperature in greenhouse experiments [58]. Glaucousness significantly reduced transpiration at night, which caused a relatively greater reduction in transpiration than photosynthesis and the increase of water-use efficiency [58]. In the present research, we measured cuticle permeability in terms of water loss and chlorophyll leaching in six NILs of four wax genes, and profiled their wax composition and inspected their wax morphologies. This allowed us to compare the effect of the individual genes and to analyze their interaction. We found that cuticle traits are closely associated with wax composition, mainly with respect to the β-diketones. The nonglaucous NILs had little or no β-diketones and showed significantly higher water-loss and chlorophyll-leaching rates (Figures 8a and 8b). A small but significant effect was also seen for other wax species. Although Iw1iw2 and iw1Iw2 had similar chlorophyll-leaching rates (fig. 8b), the former had a higher water-loss rate (Figure 8a), probably because iw1Iw2 wax had a significantly higher content of primary alcohols, aldehydes, and alkanes (Figure 3). Cuticle trait differences were also observed among the glaucous NILs. W1w2 and w1W2 had higher chlorophyll-leaching and water-loss rates than W1W2 (Figures 8c and 8d). The most significant difference in the glaucous NILs in terms of wax composition is the presence of 8- and 9-hydroxyl-β-diketones in W1W2, suggesting a role of these hydroxyl isomers in reducing cuticle permeability and improving drought tolerance. Compared to β-diketone, the hydroxyl-β-diketones were much less abundant in the sheath wax. It is hard to imagine how hydroxyl-β-diketones reduce cuticle permeability. One explanation is that the addition of hydroxyl-β-diketones may underlie the changes in wax crystal organization of W1W2 (e.g., the formation of wax crystal sheets), which result in reduced water loss and chlorophyll efflux. We hypothesize that the hydroxyl-β-diketones function as the “glue” that cross-links the β-diketone tubes via the formation of hydrogen bonds between the hydroxyl and keto groups. Application of matrix-assisted laser desorption/ionization–MS imaging to the NILs and re-crystallization analysis of combinations of β-diketone and substituted β-diketone at different ratios may shed light on this possibility.
Glaucousness is an adaptive trait to dry cultivation conditions and will play an important role in developing cuticle-based strategies to improve drought tolerance. The Iw genes have a negative impact on drought tolerance and need to be eliminated from wheat breeding programs. Due to the dominant mode of inheritance, Iw-mediated nonglaucousness can be eradicated by one selection in early generations. The manipulation of the W genes is more challenging. Our results indicate that one W gene is sufficient to restore glaucousness, but not to prevent nonstomal transpiration. Therefore, both W genes are required for enhancing drought tolerance. In this respect, marker-assisted selection will help improve breeding efficiency. To this end, user-friendly molecular markers tightly linked to the W genes need to be developed. Molecular mapping and cloning of these W genes will open novel routes to manipulate cuticle permeability for drought tolerance.
Conclusions
In summary, characterization of a set of six NILs demonstrated that β-diketones contribute to glaucousness formation in the reproductive stage. A single W gene is sufficient for β-diketone synthesis, but both W1 and W2 are required for β-diketone hydroxylation. The Iw genes suppress β-diketone synthesis, but promote the production of aldehydes and primary alcohols. Consistent with the wax profiles, CER4-6 was de-repressed by Iw2, and MAH1-8 was activated by the interaction between W1 and W2. W1W2 showed the lowest cuticle permeability, suggesting that hydroxyl-β-diketones play a role in drought tolerance.
Materials and Methods
Plant Materials and Growth Conditions
One set of six NILs in the S-615 background was developed by Tsunewaki and Ebana [52] and the seeds were obtained from the corresponding author. The NILs were planted in 4×4 inch pots containing Sunshine® Container Potting Mix 3 (Sun Gro Horticulture) supplemented with Multicote® 8 Controlled-Release Fertilizer (Haifa) and grown in a greenhouse at a temperature of 20°C in the day and 18°C at night and with a day length of 16 h. Total genomic DNA was isolated from the NIL set using the Plant DNeasy Kit (Qiagen), following the manufacturer’s instructions, and used for SNP and SSR genotyping. SNP genotyping was conducted commercially by Infinium Assay (Illumina, CA).
Microscopic Observation
Stoma counting and aperture observations were performed using an imprinted slide. Briefly, both sides of flag leaf sheaths were coated with 10% cellulose acetate dissolved in acetone using a paint brush. When dried, the cellulose acetate film (∼2×1 cm) was carefully peeled, mounted on a slide, covered with a cover glass, and observed using a light microscope at a magnification of 10×20. Two imprints were taken from each side of the sheath and three independent inspections were carried out on each of the imprints.
For SEM imaging of cuticle surfaces, a 0.5-cm tissue fragment was harvested from the base of the flag leaf, the uppermost part of the flag leaf sheath, and the peduncle, and glumes were collected from the spikelets in the middle of spikes at anthesis (F10.5.1). The tissue samples went through 10 glycerol gradients, from 10% to 100%, with 2 h in each glycerol solution, to replace the cellular water. The pretreated samples were sputtered with gold powder using the CrC-150 Sputtering System, and inspected with a Hitachi S-3400N SEM (Hitachi). Images were captured with the voltage set at 5 kV.
GC-MS Profiling of Wax Composition
Cuticular wax was extracted from two flag leaf sheaths of similar age from the same plant by submerging tissues in a glass tube containing 10 ml of HPLC grade chloroform (Fisher Scientific) and 4 µg of tetracosane (Sigma-Aldrich) as an internal standard and agitating manually for 1 min. The tissue was rinsed with an additional 5 ml of chloroform, and the two extracts were combined. The wax extract was filtered through a polytetrafluoroethylene filter with a 10-mL SGE syringe (Supelco) into a new glass tube and dried under a nitrogen stream. The dried wax was resuspended in 500 µl of acetonitrile and silylation was performed in 6% bis-trimethylsilyl-trifluoroacetamide and 10% trimethyl-chlorosilane at 80°C for 30 minutes to transform the hydroxyl and carboxylic groups into trimethylsilyl derivatives. The suspension was concentrated to 200 µl, and 1 µl was used for GC-MS analysis. Six biological replicates were included for each genotype. Wax silylation, GC-MS profiling, and substance identification were performed at the W.M. Keck Metabolomics Research Laboratory of Iowa State University (Ames, IA) on a fee-for-service basis.
Quantification of Cuticle Traits
To evaluate the effect of cuticular wax on water loss rate, flag leaf sheaths were excised from wheat plants at stage F10.5.1, dehydrated for 12 h at room temperature with a relative humidity of 44%, and weighed every hour using an analytical balance with a readability of 0.001 mg. The dry weight of tissues was measured after incubation at 37°C for 72 h. For the chlorophyll efflux assay, a flag leaf sheath was placed in a 50-ml tube containing 30 ml of 80% ethanol and the tube was gently agitated on a rotator at 50 rpm. A 150-µl aliquot of chlorophyll extract was transferred to a well in a microplate for quantification and then returned to the same tube. Absorbance was measured at 647 and 664 nm using a Synergy 2 Multi-Mode Microplate Reader (Biotek) and total chlorophyll micromoles were calculated as described [75]. Two measurements were adopted as technical replicates and four biological replicates were included for both experiments.
Transcription Quantification
Wheat cuticle gene homologs were identified by BLASTn searches of the wheat gene index database (http://compbio.dfci.harvard.edu/tgi) using known wax gene sequences as queries. The maximal cut-off value was set at E-20. qPCR primers were designed using the Primer3 program (http://frodo.wi.mit.edu/), with the PCR product size ranging from 80 to 200 bp (Table S1). Total RNA was isolated from flag leaf sheaths using Trizol® reagent (Invitrogen), following the manufacturer’s instructions. After RNA integrity evaluation by agarose gel electrophoresis and quantification using Nanodrop ND-1000 (Thermo Scientific), 1 µg of total RNA was used for cDNA synthesis in a 20-µl reaction using the QuantiTect Rev Transcription Kit (Qiagen). After dilution, ∼5 ng of cDNA was used as template for qPCR in a volume of 20 µl. qPCR was performed in 96-well plates with an ABI 7900HT High-Throughput Real-Time Thermocycler (Life Tech) using the iTaq™ Fast SYBR® Green Supermix with ROX (Bio-Rad). Two technical and four biological replicates were included for each NIL. The comparative ΔΔCT method was used to evaluate the relative quantities of each amplified product using TaRPII36 as an internal reference in the same run. PCR specificity was determined by melt curve analysis of the amplified products.
Data Analysis and Statistics
Measured values from replicates were averaged and their standard deviations (SD) were estimated using Microsoft® Excel functions. Student’s t-test was performed using pooled SDs to evaluate the statistical significance of the differences among isogenic lines. The cut-off for statistical significance was set to a P-value of 0.05 or less.
Supporting Information
A diagram showing our current understanding of cuticular wax deposition.
(DOCX)
Homolog variation of major wax species among the NILs.
(DOCX)
Validation of reference genes.
(DOCX)
Transcriptional changes of fatty acyl elongation genes in iw1Iw2 compared to W1W2 at the seedling (F4.0) and adult plant (F9.0) stages.
(DOCX)
qPCR primers designed from wheat ESTs homologous to the wax genes characterized in Arabidopsis, maize, and rice.
(DOCX)
Transcription fold changes of cutin- and cuticular wax-related genes in Iw1iw2 and iw1Iw2 in comparison to W1W2.
(DOCX)
Transcription fold changes of cutin- and cuticular wax-related genes in the glaucous NILs compared to w1w2.
(DOCX)
Acknowledgments
We are grateful to Prof. Koichiro Tsunewaki for providing the seeds of the NIL set and Huilan Zhu for technical assistance.
Funding Statement
Funding for this research was provided by South Dakota Agricultural Experiment Station and South Dakota Wheat Commission. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shepherd T, Wynne Griffiths D (2006) The effects of stress on plant cuticular waxes. New Phytol 171: 469–499. [DOI] [PubMed] [Google Scholar]
- 2.Jenks MA, Ashworth EN (2010) Plant Epicuticular Waxes: Function, Production, and Genetics, in Horticultural Reviews, Volume 23 (ed J. Janick), John Wiley & Sons, Inc., Oxford, UK. doi: 10.1002/9780470650752.ch1.
- 3. Kolattukudy PE (1980) Biopolyester membranes of plants: cutin and suberin. Science 208: 990–1000. [DOI] [PubMed] [Google Scholar]
- 4. Nawrath C (2002) The biopolymers cutin and suberin. The Arabidopsis Book 1: e0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pollard M, Beisson F, Li Y, Ohlrogge JB (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246. [DOI] [PubMed] [Google Scholar]
- 6. Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707. [DOI] [PubMed] [Google Scholar]
- 8. Schnurr JA, Shockey J, deBoer G-J, Browse J (2002) Fatty acid export from the chloroplast: Molecular characterization of a major plastidial acyl-coenzymeA synthetase from Arabidopsis. Plant Physiol 129: 1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, et al. (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hooker TS, Millar AA, Kunst L (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, et al. (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Todd J, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17: 119–130. [DOI] [PubMed] [Google Scholar]
- 13. Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, et al. (2009) Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol 150: 1174–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, et al. (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105: 14727–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng H, Rowland O, Kunst L (2005) Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vioque J, Kolattukudy PE (1997) Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch Biochem Biophys 340: 64–72. [DOI] [PubMed] [Google Scholar]
- 17. Cheesbrough TM, Kolattukudy PE (1984) Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum . Proc Natl Acad Sci USA 81: 6613–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greer S, Wen M, Bird D, Wu X, Samuels L, et al. (2007) The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol 145: 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, et al. (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li F, Wu X, Lam P, Bird D, Zheng H, et al. (2008) Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol 148: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernard A, Domergue F, Pascal S, Jetter R, Renne C, et al.. (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis. Plant Cell doi/10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, et al. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704. [DOI] [PubMed] [Google Scholar]
- 23. Bird D, Beisson F, Brigham A, Shin J, Greer S, et al. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498. [DOI] [PubMed] [Google Scholar]
- 24. Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, et al. (2007) The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol 145: 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeBono A, Yeats TH, Rose JK, Bird D, Jetter R, et al. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, et al. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–248064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, et al. (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42: 689–707. [DOI] [PubMed] [Google Scholar]
- 29. Zhang JY, Broeckling CD, Sumner LW, Wang ZY (2007) Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol 64: 265–278. [DOI] [PubMed] [Google Scholar]
- 30. Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53: 53–64. [DOI] [PubMed] [Google Scholar]
- 31. Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, et al. (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, et al. (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, et al. (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, et al. (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hooker TS, Lam P, Zheng H, Kunst L (2007) A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lam P, Zhao L, McFarlane HE, Aiga M, Lam V, et al. (2012) RDR1 and SGS3, components of rna-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol 159: 1385–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hansen JD, Pyee J, Xia Y, Wen TJ, Robertson DS, et al. (1997) The GLOSSY1 locus of maize and an epidermis-specific cDNA from Kleinia odora define a class of receptor-like proteins required for the normal accumulation of cuticular waxes. Plant Physiol 113: 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sturaro M, Hartings H, Schmelzer E, Velasco R, Salamini F, et al. (2005) Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol 138: 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tacke E, Korfhage C, Michel D, Maddaloni M, Motto M, et al. (1995) Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J 8: 907–917. [DOI] [PubMed] [Google Scholar]
- 40. Liu S, Dietrich CR, Schnable PS (2009) DLA-based strategies for cloning insertion mutants: cloning the gl4 locus of maize using Mu transposon tagged alleles. Genetics 183: 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu X, Dietrich CR, Delledonne M, Xia Y, Wen TJ, et al. (1997) Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a beta-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol 115: 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu X, Dietrich CR, Lessire R, Nikolau BJ, Schnable PS (2002) The Endoplasmic reticulum-associated maize GL8 protein is a component of the acyl-coenzyme A elongase involved in the production of cuticular waxes. Plant Physiol 128: 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dietrich CR, Perera MA, D Yandeau-Nelson M, Meeley RB, Nikolau BJ, et al. (2005) Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 42: 844–861. [DOI] [PubMed] [Google Scholar]
- 44. Yu D, Ranathunge K, Huang H, Pei Z, Franke R, et al. (2008) Wax Crystal-Sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228: 675–685. [DOI] [PubMed] [Google Scholar]
- 45. Moose SP, Sisco PH (1996) Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10: 3018–3027. [DOI] [PubMed] [Google Scholar]
- 46. Javelle M, Vernoud V, Depège-Fargeix N, Arnould C, Oursel D, et al. (2010) Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol 154: 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Javelle M, Klein-Cosson C, Vernoud V, Boltz V, Maher C, et al. (2011) Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol 157: 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tulloch AP (1973) Composition of leaf surface waxes of Triticum species: Variation with age and tissue. Phytochemistry 12: 2225–2232. [Google Scholar]
- 49. Mikkelsen JD (1978) The effects of inhibitors on the biosynthesis of the long chain lipids with even carbon numbers in barley spike epicuticular wax. Carlsberg Res Commun 43: 15–35. [Google Scholar]
- 50.von Wettstein-Knowles P (1995) Biosynthesis and genetics of waxes. In: Hamilton RJ, ed. Waxes: chemistry, molecular biology and functions. Dundee, UK: The Oily Press 91–129. [Google Scholar]
- 51. von Wettstein-Knowles P, Søgaard B (1980) The cer-cqu region in barley: gene cluster or multifunctional gene. Carlsberg Res Commun 45: 125–141. [Google Scholar]
- 52. Tsunewaki K, Ebana K (1999) Production of near-isogenic lines of common wheat for glaucousness and genetics basis of the trait clarified by their use. Genes Genet Syst 74: 33–41. [Google Scholar]
- 53. Simmonds JR, Fish LJ, Leverington-Waite MA, Wang Y, Howell P, et al. (2008) Mapping of a gene (Vir) for a non-glaucous, viridescent phenotype in bread wheat derived from Triticum dicoccoides, and its association with yield variation. Euphytica 159: 333–341. [Google Scholar]
- 54. Nelson JC, van Deynze AE, Autrique E, Sorrells ME, Lu YH, et al. (1995) Molecular mapping of wheat: homoeologous group 2. Genome 38: 516–524. [DOI] [PubMed] [Google Scholar]
- 55. Liu Q, Ni Z, Peng H, Song W, Liu Z, et al. (2007) Molecular mapping of a dominant non-glaucousness gene from synthetic hexaploid wheat (Triticum aestivum L.). Eupytica 155: 71–78. [Google Scholar]
- 56. Dudnikov AJ (2012) Geographic patterns of histone H1 encoding genes allelic variation in Aegilops tauschii Coss. (Poaceae). Mol Biol Rep 39: 2355–2363. [DOI] [PubMed] [Google Scholar]
- 57.Richards RA (1984) Glaucousness in wheat, its effect on yield and related characteristics in dryland environments, and its control by minor genes. In: Sakanoto S, ed. Proceedings of the 6th International Wheat Genetics Symposium Kyoto, Japan, 447–451. [Google Scholar]
- 58. Richards RA, Rawson HM, Johnson DA (1986) Glaucousness in wheat: its development and effect on water use efficiency, gas exchange and photosynthetic tissue temperatures. Aust. J. Plant Physiol 13: 465–473. [Google Scholar]
- 59. Monneveux P, Reynolds MP, Gonzalez-Santoyo H, Pena RJ, Mayr L, et al. (2004) Relationships between grain yield, flag leaf morphology, carbon isotope discrimination and ash content in irrigated wheat. J Agron Crop Sci 190: 395–401. [Google Scholar]
- 60. Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, et al. (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139: 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Richardson A, Boscari A, Schreiber L, Kerstiens G, Jarvis M, et al. (2007) Cloning and expression analysis of candidate genes involved in wax deposition along the growing barley (Hordeum vulgare) leaf. Planta 226: 1459–73. [DOI] [PubMed] [Google Scholar]
- 62. Xue GP, McIntyre CL, Chapman S, Bower NI, Way H, et al. (2006) Differential gene expression of wheat progeny with contrasting levels of transpiration efficiency. Plant Mol Biol 61: 863–881. [DOI] [PubMed] [Google Scholar]
- 63. Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Long XY, Wang JR, Ouellet T, Rocheleau H, Wei YM, et al. (2010) Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol Biol 74: 307–311. [DOI] [PubMed] [Google Scholar]
- 65. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 66. Kosma DK, Nemacheck JA, Jenks MA, Williams CE (2010) Changes in properties of wheat leaf cuticle during interactions with Hessian fly. Plant J 63: 31–43. [DOI] [PubMed] [Google Scholar]
- 67. Bennett D, Izanloo A, Edwards J, Kuchel H, Chalmers K, et al. (2012) Identification of novel quantitative trait loci for days to ear emergence and flag leaf glaucousness in a bread wheat (Triticum aestivum L.) population adapted to southern Australian conditions. Theor Appl Genet 124: 697–711. [DOI] [PubMed] [Google Scholar]
- 68. Barber HN, Netting AG (1968) Chemical genetics of β-diketone formation in wheat. Phytochem 7: 2089–2093. [Google Scholar]
- 69. Bianchi G, Figini ML (1986) Epicuticular waxes of glaucous and nonglaucous durum wheat lines. J. Agr Food Chem 34: 429–433. [Google Scholar]
- 70. von Wettstein-Knowles P (1972) Genetic control of β-diketone and hydroxyl-β-diketone synthesis in epicuticular waxes of barley. Planta 106: 113–130. [DOI] [PubMed] [Google Scholar]
- 71.Bianchi G (1995) Plant waxes. In: Hamilton RJ, ed. Waxes: chemistry, molecular biology and functions. Dundee, UK: The Oily Press, 175–222. [Google Scholar]
- 72. Bianchi G, Avato P, Salamini F (1984) Surface Waxes from Grain, Leaves, and Husks of Maize (Zea mays L.). Cereal Chem 61: 45–47. [Google Scholar]
- 73. Tulloch AP, Baum BR, Hoffman LL (1980) A survey ofepicuticular waxes among generaof Triticeae. 2. Chemistry. Can J Bot 58: 2602–2615. [Google Scholar]
- 74. Johnson DA, Richards RA, Turner NC (1983) Yield, water relations, gas exchange, and surface reflectances of near-isogenic wheat lines differing in glaucousness. Crop Sci 23: 318–325. [Google Scholar]
- 75. Lolle SJ, Berlyn GP, Engstrom EM, Krolikowski KA, Reiter WD, et al. (1997) Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: a role for the epidermal cell wall and cuticle. Dev Biol 189: 311–321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A diagram showing our current understanding of cuticular wax deposition.
(DOCX)
Homolog variation of major wax species among the NILs.
(DOCX)
Validation of reference genes.
(DOCX)
Transcriptional changes of fatty acyl elongation genes in iw1Iw2 compared to W1W2 at the seedling (F4.0) and adult plant (F9.0) stages.
(DOCX)
qPCR primers designed from wheat ESTs homologous to the wax genes characterized in Arabidopsis, maize, and rice.
(DOCX)
Transcription fold changes of cutin- and cuticular wax-related genes in Iw1iw2 and iw1Iw2 in comparison to W1W2.
(DOCX)
Transcription fold changes of cutin- and cuticular wax-related genes in the glaucous NILs compared to w1w2.
(DOCX)