Abstract
Asymmetrical mRNA localization and subsequent local translation provide efficient mechanisms for protein sorting in polarized cells. Defects in mRNA localization have been linked to developmental abnormalities and neurological diseases. Thus, it is critical to understand the machineries mediating and mechanisms underlying the asymmetrical distribution of mRNA and its regulation. The goal of this review is to summarize recent advances in the understanding of mRNA transport and localization, including the assembly and sorting of transport mRNP granules, molecular mechanisms of active mRNP transport, cytoskeletal interactions and regulation of these events by extracellular signals.
Keywords: RNA binding protein, mRNA localization, mRNA transport, RNA granule, molecular motor, dynein, kinesin, myosin, cytoskeleton
Asymmetrical mRNA distribution is a common phenomenon in polarized cells
Asymmetrical mRNA localization is a conserved cellular process observed in diverse types of cells and organisms. Development of refined methods of in situ hybridization led to the now seminal discovery of mRNA localization in somatic cells, wherein β-actin mRNA was localized to the leading edge of primary fibroblasts, yet other mRNAs e.g vimentin assumed a perinuclear localization (1). More recently, mRNAs encoding members of the Arp2/3 complex have been shown localized to the leading edge of fibroblast protrusions (2). Specific mRNAs can show different localization patterns in different cells. For example, vimentin mRNA is initially enriched in perinuclear areas in developing myoblasts, then eventually localized to costameres in colocalization with vimentin proteins in mature myotubes (23). In budding yeast Saccharomyces cerevisiae, Ash1 mRNA is localized to the bud cortex of daughter cells during late anaphase (3). The local translation of Ash1 mRNA restricts the expression of Ash1p in the daughter cell, which controls mating type switching. Asymmetrical mRNA localization during oogenesis and embryonic development is essential for the establishment of body axes, cell fate specification and patterning (4, 5). In the Drosophila oocyte, bicoid mRNA is localized to the anterior pole, whereas oskar and nanos mRNAs are localized to the posterior pole (6). A similar phenomenon is also observed in Xenopus oocytes, in which Vg1 mRNA is localized to the vegetal pole (7, 8). Specific targeting of mRNAs is also demonstrated in neurons. In situ hybridization studies revealed that certain mRNAs are selectively localized to axons and/or dendrites of primary neurons. For example, tau mRNA is specifically localized to developing axons (9), and Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα), microtubule-associated protein 2 (MAP2) mRNAs were shown to be localized to dendrites (10, 11). β-actin mRNA can be localized to both developing axons and dendrites, where it concentrates in growth cones and filopodial protrusions (12, 13); whereas its homolog γ-actin mRNA is largely restricted to the cell body. In oligodendrocytes, myelin basic protein (MBP) mRNA is localized to processes and the myelin-rich peripheral compartments (14). In many of the above examples, mRNAs are localized in the form of particles or granules, which are mRNP complexes that contain one or more mRNAs, RNA-binding proteins, translational factors, and in some cases also ribosomes (15).
Genome-wide analyses reveled that asymmetrical mRNA distribution is a common phenomenon observed for multiple mRNAs in the same type of cells. An examination of 3370 transcripts in Drosophila embryos by high throughput in situ hybridization revealed that 71% of them exhibited distinct non-uniform distribution (16). Fractionation of subcellular compartments coupled with systematic RNA analysis has provided a comprehensive view of mRNA localization in fibroblast protrusions and neuronal axons. Mili et al. identified more than 50 transcripts significantly enriched in purified fibroblast protrusions in response to treatment with lysophosphatidic acid (17). A serial analyses of axonal mRNAs revealed that hundreds of transcripts are localized to axons of primary cortical and sensory neurons (18–22). Interestingly, the pools of mRNAs present in normal and injured axons (18, 22), or axons from different developmental stages (20), exhibit differential composition, revealing that mRNA localization in neurons is dynamically regulated by activities and extracellular stimuli. Of interest, the profile of mRNAs within the axon growth cone differs from the axon process, and is also under developmental control (21). Recently, a stringent analysis of mRNAs isolated from the hippocampal CA1 neuropil, a synaptic region containing axons and dendrites, revealed 2550 mRNA species localized in this brain region (24), a much larger pool of mRNA than pervious estimations of less than a few hundred (25–27). In both fibroblast protrusions and neuronal processes, proteins encoded by localized mRNAs play diverse functions, including RNA and/or translational regulation, active transport, cytoskeleton dynamics, signal transduction, and/or synaptic transmission and regulation in neurons, suggesting that mRNA localization and subsequent local translation are common regulatory mechanisms widely involved in diverse cellular functions.
Local translation of mRNA provides a mechanism for efficient protein sorting shaping cell function
Translation of localized mRNAs enables protein production to be concentrated to restricted locations and allows cells or organisms to generate timely responses to external stimuli or intrinsic developmental programs. During Drosophila oogenesis, for example, translation of the anterior pole-localized bicoid mRNA and posterior localized oskar and nanos mRNAs generate increased levels of encoded proteins in the anterior and posterior poles, respectively (6). Diffusion of these proteins results in protein gradients, which are essential for the establishment of the body axis of Drosophila embryos. Since translation of these mRNAs will not be activated until they are properly localized, the deleterious effect of ectopic expression of these proteins is avoided.
In polarized cells, such as neurons and oligodendrocytes, mRNAs can be targeted to the location where their protein products are enriched. For example, mRNAs encoding axonally enriched proteins e.g. tau can be enriched in axons and mRNAs encoding the dendritically enriched proteins, MAP2, CaMKIIα and PSD-95 can be targeted to dendrites and subsynaptic sites (10, 11, 28). Numerous mRNAs can be localized to growth cones (21), including β-actin and GAP-43 (12, 29), where the protein products are enriched. In oligodendrocytes, MBP mRNA is localized to myelinating compartments where MBP is enriched (14). Although these proteins can be delivered to their target compartments through protein trafficking, the localization and local translation of these mRNAs provides a molecular basis for efficient protein production, which could be temporally controlled, particularly for cells to rapidly respond to extracellular signals.
The regulation and function of mRNA localization and local translation has been well established in neurons. In cultured primary neurons, the localization of β-actin mRNA and local synthesis of β-actin protein mediate chemo-attractive growth cone turning induced by brain-derived neurotrophic factor (BDNF) or Netrin-1 (30–33). In the developing nervous system, axon guidance is an essential targeting process for the formation of functional neuronal circuits that is mediated by growth cone steering responses to both chemo-attractant and chemo-repulsive guidance cues. Recent studies showed that the disruption of β-actin mRNA transport and localization with an antisense oligonucleotides against its localization elements abolished BDNF-induce axonal turning (30) and neutrophin-3-induced axon growth (34). In DRG sensory neurons, the local translation of Par6 mRNA plays a role in nerve growth factor- induced axon elongation (35). Here, the Jaffrey lab has used microfluidic chambers to isolate axons and directly perturb axonal translation, providing definitive proof for the function of local translation in axons (35). Translation of growth cone-localized mRNAs can also be activated by chemo-repulsive guidance cues, such as Slit and Sema3A. Piper et al. showed that Slit incubation triggers the protein synthesis dependent enrichment of cofilin protein prior to the collapse of growth cones of retinal ganglia neurons (36), suggesting a possible local translation of cofilin or associated proteins. Wu et al. demonstrated that Sema3A treatment of dorsal root ganglia neurons induces the local translation of RhoA in axons; this event appears necessary for Sema3A-induced growth cone collapse (37). Both cofilin and RhoA mRNAs are localized to growth cones of developing axons (36, 37). With regard to dendritic protein synthesis, the seminal discovery of polyribosomes in dendritic spines (Steward and Levy, 1982), stimulated decades of research. These numerous studies have revealed that mechanisms of local translation in dendrites and their postsynaptic compartments plays essential roles in synaptic plasticity, which further demonstrates the importance of mRNA localization to modify protein composition in a spatially restricted manner in polarized cells (38, 39).
Direct interaction of localization/zipcode elements and RNA-binding proteins
The active transport and asymmetrical distribution of mRNAs rely on the intrinsic mRNA targeting elements, also known as zipcode sequences, first described for β-actin mRNA (40), which are frequently located within the 3′UTR of a localized transcript and occasionally the 5′UTR and coding regions. Multiple cis-acting elements are often observed in a localized mRNA (41). In many cases, the presence of multiple localization elements provides stronger localization capability. As demonstrated in budding yeast, four cis-acting elements, E1, E2A, E2B and E3 within the coding region and the 3′UTR of Ash1 mRNA, were identified by their ability to localize Ash1 reporter transcripts (42). Although each element alone is sufficient to localize Ash1 reporter transcripts, the presence of four elements dramatically increases the efficiency of localization. In Drosophila oocytes, four cis-acting elements were identified for the posterior pole localization of nanos mRNA. Similar to yeast Ash1 mRNA, each individual nanos localization element possesses only weak mRNA localization ability and the existence of four localization elements provides enhanced localization strength (43). The anterior pole localization of bicoid mRNA is determined by several bicoid localization elements (BLEs) within a 625nt long region of the 3′UTR, which forms 5 stem-loop structures (44, 45). The presence of multiple localization elements allow bicoid mRNA to be properly localized through a few developmental stages of the whole process of oogenesis.
The redundancy of mRNA localization elements has also observed in vertebrate cells. Two conserved zipcode sequences were identified within the 3′UTR of β-actin mRNA: a 54nt zipcode sequence and a 43nt zipcode sequence (40). Both elements are sufficient to localize chimeric mRNA reporters to the leading lamellipodia of primary fibroblasts, although the 43nt zipcode exhibited weaker capability to localize a reporter mRNA in comparison to the 54nt zipcode (40). In mature neurons, CamKIIα mRNA is targeted to dendrites of mature neurons and translated locally (11) and genetic deletion of the CamKIIα 3′UTR in vivo abolished dendritic localization of CamKIIα mRNA and resulted in reduced levels of CamKIIα protein at postsynaptic sites (46). At least four cis-acting targeting elements have been identified up to now, including a 94nt element after the stop codon, a 1200nt long stretch at the 3′end, a cytoplasmic polyadenylation element (CPE), and G-quartet structures (47–50). All four targeting elements are sufficient to direct mRNAs to dendrites. However, how these localization elements may cooperate during the localization and regulation of CaMKIIα mRNA needs to be further investigated.
The physiological importance for the presence of multiple localization elements needs further study. Although the presence of multiple localization elements in some mRNAs provide stronger localization strength, it is also likely that each cis-acting element may encode different information within a multistep localization pathway to enable fine modulation of mRNA localization. In addition, multiple localization elements might be important for cells to differentially respond to different stimuli. However, these models remain to be further tested in biological systems.
The information encoded by mRNA localization elements can be in the form of primary sequence and/or secondary structures. For examples, the cytoplasmic polyadenylation element bears the conserved minimal nucleotide sequence UUUAUU, which is sufficient for the dendritic localization of CaMKIIα mRNA in mature neurons (49); and the conserved RNA sequence GGCAAGGAGCC serves as hnRNP-A2 response element (A2RE) for the binding of hnRNP-A2 to localize MBP mRNA in oligodendrocytes (51). In contrast, the localization of Ash1 mRNA in budding yeast depends on secondary stem-loop structures formed by localization elements (52). Mutations in the stem region disrupting the stem loop structure abolish the localization of Ash1 mRNA; whereas rebuilding of the stem loop structure by mutating the compensatory bases successfully restores Ash1 mRNA localization. Similarly, Drosophila bicoid mRNAs carrying mutations of the primary sequence of BLEs which do not affect stem-loop structures are properly localized (53).
A common feature of cis-acting mRNA localization elements is that each of them provides a unique platform to recruit specific trans-acting protein factors for the assembly of transport mRNP granules, despite the diverse sequence and structural information they carry. Current studies suggest that the specific interaction of trans-acting RNA binding proteins with mRNA localization elements is essential for proper mRNA localization. One early and well characterized example is that the 54nt zipcode sequence of β-actin mRNA, which plays a major role in β-actin mRNA localization, specifically interacts with the RNA binding protein, zipcode binding protein 1 (Zbp1) (54). A recent crystallographic study revealed that the KH3 and KH4 domains of Zbp1 form an anti-parallel pseudodimer binding to the bipartite element (5′-GGACU-3′ and 5′-ACA-3′) within the 54nt zipcode (55). Treatment of cells with anti-sense oligonucleotide against zipcodes abolished β-actin mRNA localization in both primary fibroblasts and primary neurons (34, 40). The stem loop structures of bicoid mRNA recruit the double-strand RNA binding protein, Staufen, which has been demonstrated to be important for the posterior localization of bicoid mRNA (53). She2p directly interacts with Ash1 mRNA localization elements and mediates its localization (56, 57). Fragile X mental retardation protein (FMRP) can bind to its target mRNAs through G-quartet structures formed by G-rich sequences (58, 59). The G-quartet structure was recently demonstrated as a localization element to target mRNAs to dendritic compartment (50). FMRP plays an essential role in dendritic localization of its associated mRNAs in response to activation of group 1 metabotropic glutamate receptors (mGluRs); in neurons cultured from Fmr1 knockout mice, a mouse model of Fragile X syndrome, mGluR-induced dendritic mRNA localization is diminished (60). The cytoplasmic polyadenylation element binding protein (CPEB) was also shown to facilitate dendritic mRNA localization through the interaction with CPEs (49).
mRNA-binding proteins as adaptors for molecular motors for active mRNA transport
Numerous studies have demonstrated that mRNA localization is a transport activity mediated by molecular motors in a cytoskeleton-dependent manner (61, 62). Three classes of molecular motors have been shown to be involved in mRNA transport, namely microtubule-dependent motors, kinesin and dynein, and the microfilament-dependent motors, myosins. In fact, analyses of single mRNA molecules revealed that a larger portion of localized mRNA granules exhibit directional movement versus a non-localized control. Fusco et al. showed that about 20% of LacZ-β-actin chimeric mRNA bearing β-actin 3′UTR exhibit directional movement versus 2–4% for non-localized mRNA (LacZ without β-actin mRNA 3′UTR) (63). This unique feature of localized mRNAs suggests that the presence of a localization element in an mRNP complex with its trans-acting factors may potently engage machineries for active transport. RNA-binding proteins may serve as adaptors to directly, or indirectly through additional factors, recruit motor proteins to form mRNP transport granules.
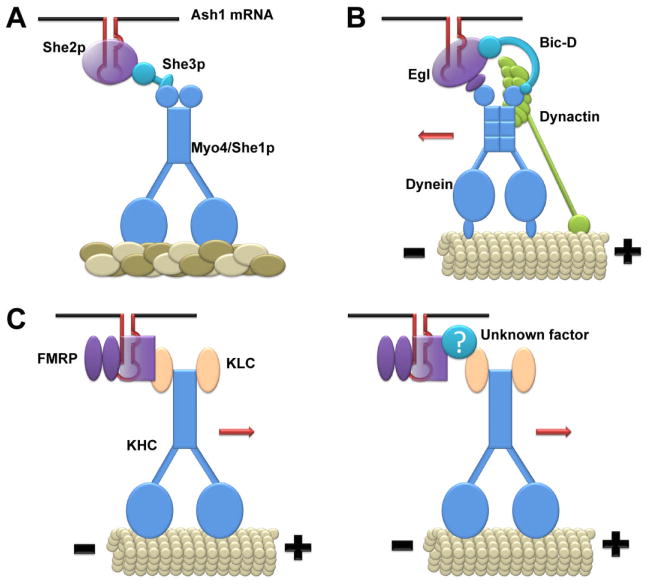
Such a model was well demonstrated in budding yeast for the localization of Ash1 mRNA. A protein complex named “locasome” was identified, including She1p/Myo4p, She2p and She3p (56, 64, 65). She2p is an RNA binding protein that shuttles between the nucleus and cytoplasm and forms mRNP granules with Ash1 mRNA by direct interaction with its cis-acting elements; She1p/Myo4p is a type V myosin (myosin V) which exerts actin filament-mediated transport; She3p acts as an adaptor mediating the association of She2p/Ash1 mRNA complexes with She1p/Myo4p (Figure 1A). The tethering of Ash1 mRNA to She3p is abolished by the loss of She2p, and both She2p and She3p are required for the recruitment of She1p/Myo4p to Ash1 mRNA and Ash1 mRNA localization (56).
Figure 1. mRNA-binding proteins couple mRNAs to molecular motors for active transport.
A, Yeast mRNA-binding protein She2p in a complex with She3p recruits myosin (Myo4/She1p) onto Ash1 mRNA. B, Drosophila non-canonical RNA-binding protein, Egl1, and Bic-D interact with dynein and dynactin complexes, respectively, to couple TLS-containing mRNAs to dynein for microtubule minus-end directed mRNA transport. C, FMRP facilitates the localization of its target mRNAs to dendritic compartments in mammalian neurons in response gp1 mGluR activation. FMRP may directly (left) or indirectly (right) interact with kinesin light chain (KLC) to couple mRNAs to conventional kinesin motors for mRNA localization.
The apical localization of mRNAs in Drosophila syncytical blastoderm embryo is mediated by the microtubule minus end-directed transport process. Factors involved in the apical mRNA localization include Bic-D, Egl, dynein and dynactin. Dienstbier et al. reported that protein complexes composed of Egl and Bic-D couple the mRNA Transport and Localization Signal (TLS) to dynein motors in Drosophila embryos (66) (Figure 1B). Previously, both Egl and Bic-D were shown to associate with dynein light chain and Bic-D associates with dynactin as well (67, 68). Dienstbier et al. demonstrated that Egl is a non-canonical RNA-binding protein, which contains no known RNA binding domain, but can recognize TLS. The authors further showed that the Egl and Bic-D are the only two proteins exhibiting specific affinity to several different TLSs and suggest that Egl and Bic-D in a complex link TLS to dynein motors for microtubule minus end-directed transport.
Early observations of mRNA localization and active transport in neurons demonstrated a microtubule-dependent mechanism for neuronal mRNA localization (69, 70), suggesting the involvement of microtubule-dependent motors. The involvement of kinesin in neuronal mRNA transport was further tested by an attempt to identify proteins associated with the KIF5 cargo binding domain. Results from this studies revealed that mRNP granules are cargos of conventional kinesin (71). RNA binding proteins identified include several well-characterized examples involved in mRNA transport, such as hnRNP-A2, Staufen and FMRP. Overexpression of wild-type or a dominant-negative form of KIF5 promoted or suppressed the transport ability of mRNP granules, respectively, and further demonstrate the direct involvement of kinesin in mRNA transport (71).
Although both mRNAs and RNA binding proteins are co-purified with the cargo-binding domain of conventional kinesin, it remains unclear how mRNAs are directly coupled to kinesin motors in mammalian cells. A recent study showed that the loss of FMRP reduces the coupling efficiency of FMRP target mRNAs to the conventional kinesin heavy chain and subsequently leads to reduced dendritic localization of FMRP target mRNAs upon mGluR receptor activation (60) (Figure 1C). The mRNA localization defects caused by the loss of FMRP may partly underlie dendritic developmental defects associated with Fragile X Syndrome, an inherited form of intellectual disability caused by the silencing of FMRP protein expression (72). The function of RNA-binding proteins in coupling mRNAs to molecular motors may not be unique to FMRP in mammalian cells. As one additional example, CPEB, which regulates dendritic localization and translation of cytoplasmic polyadenylation element-containing mRNAs, was shown to interact with kinesin and dynein as well (49). However, whether CPEB is involved in the coupling of its target mRNAs to molecular motors needs to be further investigated.
Nuclear initiation of mRNA transport
Several lines of evidence suggest that mRNA transport initiates in the nucleus where it is coupled to transcription. This model was well demonstrated for the localization of Ash1 mRNA in budding yeast. Shen et al. showed that the nuclear localization of She2p and the subsequent interaction with Ash1 mRNA in the nucleus are essential for the proper localization of Ash1 mRNA (73). The nuclear interaction between She2p and Ash1 mRNA is mediated by type II RNA polymerase elongation factors, Spt4–Spt5/DSIF, and coupled to Ash1 mRNA transcription (74). Although the nuclear localization of She2p is not required for its interaction with Ash1 mRNA localization elements, the abolishment of its nuclear localization by mutating the She2p NLS completely eliminates its function for Ash1 mRNA localization as demonstrated in a rescue experiment in a She2p null background (73). The initial binding of She2p to Ash1 mRNA during transcription may trigger the stepwise recruitment of other factors and eventually lead to the assembly of Ash1 mRNP granules with localization capability. Indeed, She2p recruits Loc1p and Puf6p to Ash1 mRNA in the nucleus, both Loc1p and Puf6p were known to be required for Ash1 mRNA localization and the binding of these factors results in Ash1 mRNA translational repression (75, 76).
Similar to yeast She2p, many, if not all, mRNA-binding proteins are nuclear-cytoplasmic shuttling proteins containing both nuclear localization signals and nuclear export signals. Zbp1 is one of the well-characterized examples. Although it is not clear whether the binding of Zbp1 to β-actin mRNA is coupled to transcription, studies have shown that Zbp1 is localized to β-actin transcription loci in the nucleus and interacts with β-actin mRNA at the β-actin transcription sites (77). This interaction is facilitated by the initial binding of a predominantly nuclear RNA binding protein, Zbp2, to the nascent β-actin mRNA during transcription (78). The depletion of Zbp2 by RNAi results in reduced Zbp1 and β-actin mRNA association (78).
The involvement of exon-junction complexes in mRNA localization provides another piece of evidence in support of the nuclear initiation of mRNA transport. The EJC, containing Barentsz, Mago, Y14 and eIF4AIII, is deposited onto mRNA about 20nt upstream of the exon junction during splicing (79). In Drosophila oocytes, the splicing of the first intron and subsequent deposition of an EJC are essential for the posterior pole localization of oskar mRNA (80, 81). Thirdly, nuclear mRNA cap binding proteins CBP20/80 are in complexes with mRNAs localized to dendritic compartments and anchored to locations close to dendritic spines in response to stimulation (82). CBP20/80 complexes associate with newly transcribed mRNA and are replaced by a cytoplasmic cap-binding protein, eIF4E, during the pioneer round of translation (83, 84), suggesting these localized mRNAs are translationally quiescent. In addition, biochemical analysis revealed that CBP20/80 are only associated with mRNAs loaded with exon-junction complexes (84), further suggesting that these CBP20/80 bound mRNAs are translational repressed. However, whether the binding of nuclear CBPs and association with EJC is a common requirement for general mRNA transport and localization remain to be tested. At least in mature neurons, eIF4AIII is colocalized with dendritic mRNP granules and associates with dendritic localized Arc mRNA, although its function in Arc mRNA localization is unclear (85).
Cooperation of mRNA-binding proteins in mRNA localization
The specific localization of an mRNA is unlikely achieved by a single trans-acting factor, given that the complex process of mRNA transport involves multiple steps, which may require different combinations of trans-acting factors. In fact, the existence of multiple RNA localization elements may allow the binding of multiple RNA-binding proteins. These targeting elements, by recruiting trans-acting factors, thus provide a foundation for the assembly and maintenance of transport mRNP granules, the efficient recruitment of transport machineries to regulate local translation. These trans-acting RNA binding proteins may play different roles during during the multistep transport process of an mRNA.
Some well understood examples are trans-acting factors involved in oskar mRNA localization to the posterior pole of the Drosophila oocyte (6). In an elegant study from St. Johnson and colleagues, in vivo imaging combined with mutational analysis of oskar mRNA transport demonstrated that mutations of each factor leads to different phenotypes on oskar mRNA distribution or transport capability. These data suggest that oskar mRNA-binding proteins play different functions for oskar mRNA localization, although all binding factors are required for proper oskar mRNA localization (86). For example, the mutation of HRP48, a Drosophila hnRNP-A/B homolog, leads to diffuse distribution of oskar mRNA and the disappearance of GFP-Staufen granules, suggesting a possible role for HRP48 in the assembly of oskar mRNP granules (86). In contrast, loss of exon-junction proteins, Mago and Btz, had no effect on the formation of visible oskar mRNA and GFP-staufen granules, but led to reduced number of actively transporting granules. In addition, loss of Mago and Btz led to reversed directional net movement of oskar mRNA toward the anterior pole; and in these mutants, oskar mRNA distributes as a gradient extended away from the anterior pole (86). Such alterations in the dynamics and distribution of oskar mRNA suggest that EJC may play a role in the coupling of oskar mRNA with kinesin, the molecular motor mediating oskar mRNA localization in the cytoplasm of oocytes. Alternatively, EJC proteins may play a role in inhibiting the activity of dynein complexes, which mediate the export of mRNAs from nurse cells. Staufen seems to play a major role in coupling oskar mRNP with kinesin, as in Staufen mutants, oskar mRNA is mainly restricted to the anterior pole with a small portion properly localized to the posterior pole (86). Live cell imaging analysis revealed that oskar mRNA granules in Staufen mutants exhibit reduced frequency of fast directional movement, suggesting that Staufen plays a role in coupling oskar mRNA to kinesin or regulates kinesin activity. For the small proportion of oskar mRNA exhibiting fast movement, Staufen mutation does not affect posterior bias for proper oskar mRNA localization, indicating additional factors may also mediate the association of oskar mRNA with kinesin for proper localization. However, the above-mentioned hypothetical functions for these trans-acting factors remain to be further tested. Nevertheless, these results indicate that, to ensure proper mRNA localization, multiple trans-acting factors and associated motors are involved, either playing roles cooperatively or in different transport steps to bias the localization of oskar mRNA.
Cooperative interactions between mRNA binding proteins involved in mRNA localization have been observed in mammalian cells, including fibroblasts and neurons. Taking β-actin mRNA as an example, besides Zbp1 and Zbp2, which were first shown to be involved in β-actin mRNA localization by directly interacting with the 54nt zipcode (54, 87), several other RNA-binding proteins have been shown to play roles in the localization of β-actin mRNA in neurons, including hnRNP-R (88), Staufen2 (89), and polypyrimidine tract-binding protein (90). Although the detailed working mechanisms for each RNA binding protein in β-actin mRNA localization has not been revealed in detail, it does suggest a more complex composition of factors that are involved to regulate the assembly and sorting of localized β-actin mRNA granules.
As one example of a disease connection, the axonal and growth cone localization of β-actin mRNA is impaired in motor neurons cultured from a mouse model of spinal muscular atrophy (SMA) (91). Spinal Muscular Atrophy (SMA) is an autosomal recessive neurodegenerative disease specifically affecting motor neurons in the lower spinal cord. SMA is caused by low level of the survival of motor neuron protein (SMN) (91), suggesting a possible link between SMN and the RNA binding proteins involved in mRNA localization. Accumulated evidence suggests that the function of SMN in mRNA localization can be beyond β-actin mRNA, and SMN is likely a general factor involved in the localization of other mRNAs in motor neuron axons (92). Firstly, recombinant fluorescent protein tagged SMN exhibits bidirectional movement in motor neuron axons (92); secondly, axonal SMN interacts with a number RNA binding proteins in granules containing mRNAs (92–96); thirdly, the acute loss of SMN induced by RNAi leads to reduced axonal localization of poly(A) mRNAs, suggesting a broad range of mRNAs that are regulated by SMN (92). Although the precise function of SMN in mRNA localization remains unclear, a plausible model may be to facilitate mRNP assembly. SMN, in complexes containing Gemin proteins and additional factors, facilitates the assembly of transport mRNP granules (97). While it has been well-established that SMN complexes function as a platform for the assembly of small nuclear RNP spliceosomes, increasing support suggests a non-canonical function in mRNP assembly and/or trafficking (98, 99).
The dendritic localization of CaMKIIα mRNA in neurons also likely involves multiple RNA-binding proteins. Besides CPEB, which interacts with the CPE of CaMKIIα mRNA, the possibility for involvement of other factors is an open question. One immediate candidate is FMRP, which binds to the G-quartet structure, a known binding element for FMRP (58). In line with this prediction, Dictenberg et al. showed that the localization of CamKIIα mRNA in response to group I mGluR activation was abolished in neurons lacking FMRP (60). CamKIIα mRNA is a direct target of FMRP (100), and both are colocalized in dendrites, where FMRP is required to regulate mGluR-induced translation of CaMKIIα protein (28).
Co-transport versus single mRNA transport
One important topic in mRNA transport research is to decipher the molecular composition of transport mRNP granules. A few proteomic attempts have been made to identify the protein composition of mRNP granules (71, 101, 102), yet components of a single mRNP granule have not been well understood, particularly the mRNA composition of a single transport mRNP granule. Several studies have described large mRNP granules in neuronal dendrites (71, 103). These large mRNP granules contain mRNAs, RNA binding proteins, translational factors, and ribosomes, with an S value over 1000 (71). These type of mRNP granules can be directly visualized by electron microcopy by negative staining (103). Given the large size of these granules, presumably one granule contains multiple species or copies of mRNAs and these mRNAs may be co-transported into neuronal processes in the same transport mRNP granules. Gao et al. provided supportive evidence for this model by investigating the colocalization and co-transport of CaMKIIα, neurogranin and Arc mRNA using microinjection of labeled RNAs (104). The data suggest that these mRNAs were packed into the same transporting mRNA granules by the A2RE localization elements through the interaction with hnRNP-A2 (104). These mRNA granules do not contain β-actin mRNA which has no A2RE, further suggesting the functional sorting of mRNAs into different types of granules. Yet, whether hnRNP-A2 containing dendritic mRNPs, observed by microscopic visualization, represents the large mRNP granules that have been biochemically characterized is still unclear. An apparent advantage for mRNA co-packaging and co-transport in large mRNA granules is to allow the co-regulation of the transport and translation of these mRNAs in polarized cells. In principle, the RNA granules could represent translational units that make specific proteins on demand within a microenvironment, such as a dendritic spine.
Recent quantitative studies using single molecule imaging has modified the concept of co-transport mechanism that involves obligatory packaging of multiple functionally related mRNAs into single granules, and provide evidence for transport as individual mRNAs or in small copy numbers. A study by Mikl et al. provided evidence in fixed and live cells that MAP2, CaMKII α and β -actin mRNAs are sorted in different mRNA granules containing only a low copy number of each mRNA (105). Detection of endogenous mRNAs by in situ hybridization using antisense probes with labeled with different fluorophores retrieved was consistent with the presence of few mRNA molecules (105). Using single molecule multiplex detection of endogenous mRNAs in fixed cells, Batish et al. revealed no colocalization among the eight mRNAs examined (106). These studies suggest that many mRNAs are likely transported in neuronal dendrites in mRNP granules containing only a single mRNA molecule. More work still needs to be done, and it remains an open question if mRNAs might be sorted in higher copy numbers under certain conditions, such as in response to activity or neuronal stimulation. Although high copy numbers of mRNAs may not be transported in the same granules, the assembly and sorting of mRNP granules seems likely to be a highly selective process. Tubing et al. showed that a much larger fraction of Septin7 mRNA is colocalized with CaMKIIα mRNA (36%) over MAP2 mRNA (14%) in dendrites (107). Interestingly, apparent colocalization was frequently observed for the same transcripts, either for Septin7 and MAP2 mRNAs, when each co-injected mRNAs was labeled in vitro with different fluorophores. It remains unclear how specific sorting of mRNAs to distinct granules is achieved to enable differential mRNA composition e.g. multiple mRNA copies. One speculated mechanism could be through RNA-binding protein dimerization, a process which can bring mRNAs with similar or the same localization element into the same granules. Recent, a cell free system has been established to artificially nucleate the assembly of multiple mRNAs into high order RNA granules, which involves the RNA binding protein, FUS, and its low complexity (LC) RNA binding domain; of interest, the phosphorylation of the LC seemed to inhibit granule assembly (108). Further work is needed to validate this proposed model for RNA granule assembly in cells. It will be important to assess more precisely the circumstances and mechanisms by which mRNAs are sorted and/or transported in low versus high copy number RNA granules. Stress granules provide an example of a higher order structure whereby mRNAs appear to be packaged into larger structures, which are distinct from RNA transport granules (15). It is likely that RNA granules exist along a continuum, which are heterogeneous in size and molecular composition, and serve multiple functions to regulate mRNA metabolism, sorting and trafficking.
The impact of polarized cytoskeletal networks and coordinated role for multiple motors on mRNA localization
The localization of mRNA requires proper assembly of mRNP granules and their coupling with molecular motors. There has been much interest to define the structural basis for asymmetrical mRNA distribution. Recent studies suggest that the polarized organization of cytoskeletal networks is an important contributing factor. Live cell imaging analysis of endogenous fluorescently labeled oskar mRNA by GFP-MS2 or GFP-Staufen, a protein marker for oskar mRNA granules, revealed that the posterior localization of oskar mRNA in Drosophila oocyte is determined by a slightly biased directional transport of oskar mRNA granules on a weakly polarized microtubule network with more plus ends pointing toward the posterior pole (86). Oskar mRNPs exhibit bidirectional fast movement along the microtubule network, with a larger population localized toward the posterior pole of the oocyte. In Xenopus embryos, Vg1 mRNA is localized to the vegetal pole of embryos via kinesins through a population of microtubules with plus ends mostly pointing toward the vegetal pole, and interactions between kinesin-1 and kinesin-2 (130). Although studies imply that mRNA localization is an active and directed process, more recent studies suggest that asymmetrical mRNA distribution may also be a passive effect of asymmetrical orientation of cytoskeletal structures as a means to bias motors. During the process of mRNA localization, the intrinsic targeting elements of localized mRNAs may provide for the assembly of transport competent mRNP granules that can couple to molecular motors. The asymmetric organization of microtubules would then influence whether there is net accumulation of RNA in one part of the cell versus another. Yet, whether this is a general mechanism for mRNA in other types of cells still remains unclear.
Asymmetrical localization of mRNA is a common phenomenon of most polarized cells, which possess highly oriented cytoskeletal filaments. For example, neurons extend highly differentiated processes, axons and dendrites. In axons, microtubules are oriented with plus-end uniformly pointing toward growth cones; whereas in the mid-region of dendrites, equal proportions of microtubule plus-ends point toward and away from the cell body (109, 110). The differential microtubule cytoskeleton in axons and dendrites suggests that the transport machineries underlying mRNA localization in these functionally distinct neuronal processes may be different. A common feature of both axonal and dendritic mRNP transport is that mRNP granules exhibit bidirectional movement as examined by fluorescently-labeled mRNA or RNA binding proteins. However, the detailed mechanism for motor-mediated mRNA transport in neurons largely remains elusive. Given that axonal microtubules are uniformly oriented, the bidirectional transport of mRNP granules in axons suggests that both dynein and kinesin associate with mRNP granules. As an example, GFP-tagged ZBP1, the RNA-binding protein mediating β-actin mRNA transport in developing axons, exhibits bidirectional fast movement in axons (33, 34). One assumption derived from such studies is that the specific enrichment of β-actin/Zbp1 mRNPs in the growth cone may be a consequence of unbalanced anterograde and retrograde forces generated by kinesin and dynein, respectively. Yet, how the activity of dynein and kinesin in transporting mRNP granules is coordinated remains to be studied. There is also the interesting possibility that myosin motors may trap or anchor RNA granules on cortical actin along the neuronal process, as an additional means to compete or influence long distance transport of RNA on microtubules. One likely role for myosins is to promote docking or traffic of mRNAs in peripheral actin-rich compartments, such as growth cones and spines. Myosin II was shown to mediate β-actin mRNA localization in fibroblasts in response to serum stimulation, and Myosin V was shown to be involved in the anchoring of mRNP granules in dendritic spines (111, 112). Thus, a microtubule-dependent motor to microfilament-dependent motor relay may occur locally that allows the enrichment of β-actin mRNA in growth cones or spines.
A similar mechanism may underlie the localization of Translocated in Liposarcoma (TLS) protein containing mRNPs. TLS is an RNA binding protein localized to distal dendrites and dendritic spines of mature hippocampal neurons, and the accumulation of TLS in dendritic spine is mediated by myosin V (112, 113). Perturbation of myosin V function disrupts this specific enrichment of TLS in spines. In addition to myosin V, TLS is also found in neuronal transport mRNP granules associated with kinesin (KIF5 (71)). In addition, the disruption of either microtubules or actin networks abolished the localization of TLS to dendrites (112). For these observations, at least two possible mechanisms underlying TLS localization can be proposed. First, microtubule- and microfilament-mediated TLS transport are two parallel processes requiring kinesin and myosin V, respectively. Second, microtubule-mediated and microfilament-mediated TLS transport and localization may be two sequential steps for proper TLS localization in dendrites, in which, TLS is firstly transported to distal dendrites by kinesin through microtubules and then translocated or anchored to actin filaments to maintain the localization of TLS by myosin V.
Both microtubule- and microfilament-based transport may also be involved in oskar mRNA localization to the posterior pole of Drosophila oocytes. While this specific localization of oskar mRNA and its associating RNA binding protein, staufen, is largely mediated by kinesin through microtubules (86), Krauss et al. observed that myosin V only associates with staufen at the posterior cortex of the Drosophila oocyte (114). Mutations of myosin V and disruption of myosin V function by expressing dominant negative myosin V proteins result in compromised localization of staufen protein, a marker for oskar mRNA, with a portion mislocalized to the center of oocyte (114). This study implies that myosin V regulates the localization of oskar mRNA in the later transport stages, such as local rearrangement or anchorage of oskar mRNPs and further suggest that an intact cytoskeleton composed of microtubule and microfilaments network is necessary for the efficient mRNA transport and localization.
Signaling pathways regulating mRNA localization and mRNA binding proteins
In polarized somatic cells, mRNA localization is actively regulated by extracellular stimuli. In fibroblasts, serum stimulation induces the redistribution of β-actin mRNA to the leading edge of starved cells (115). Mechanistic studies revealed that serum-induced β-actin mRNA localization is mediated through RhoA signaling, which induces both actin filament rearrangement and activation of myosin II (111). The localization of β-actin mRNA to the leading edge can regulate directional cell motility (116, 117). In developing neurons, BDNF and netrin-1 stimulation augments growth cone accumulation of β-actin mRNA as well as the asymmetrical distribution of β-actin mRNPs to the side of the growth cone near the source of BDNF and netrin-1 (30, 31). The asymmetrical distribution and subsequent translation of β-actin mRNA within a single growth cone are required for BDNF and netrin-1 induced growth turning. Zbp1 is necessary for local translation of β-actin and growth cone steering (33). Neurons cultured from Zbp1 KO mice have impaired recruitment of β-actin mRNA into growth cones following neuronal stimulation, absence of netrin-mediated local protein synthesis, and loss of growth cone steering toward a netrin gradient (33). The molecular interaction between Zbp1 and the zipcode is required for β-actin mRNA localization in fibroblasts and neurons (34, 54). Translation of β-actin mRNA is believed to be repressed during transport, which can be deprepressed in the growth cone by Src mediated phosphorylation of Zbp1 (32). Since phosphorylation of Zbp1 by Src releases Zbp1 from β-actin mRNA (118), the regulated phosphorylation in response to receptor signaling is believed to provide a switch to activate local protein synthesis from translationally quiescent RNA granules. Beyond the role of local translation in axon guidance during development of the nervous system, mRNA localization also plays a role in adult axons in response to injuries, which represent another type of external stimuli having complex effects on mRNA localization (18, 119). Most recently, Willis et al. provided evidence showing that both peripheral axotomy and spinal cord injuries can induce the localization of GFP-β-actin mRNA reporters into regenerating axons in vivo (120). The ability to deliver β-actin mRNA and other mRNAs into mature axons and augment nerve regeneration was impaired in Zbp1 heteroygous mice or transgenic mice expressing a dominant interfering β-actin 3′UTR reporter (121). Taken together, these studies underscore the requirement of one important mRNA binding protein and its regulation in response to physiological signals.
Summary and perspective
Exciting research progress has identified factors involved in and mechanisms underlying mRNA transport and localization, and has allowed the formation of multistep models of mRNA localization (Figure 2). In general, mRNA localization may initiate from the assembly of transport mRNP granules within the nucleus, likely coupled to transcription and splicing. However, the requirement of a nuclear step is by no means obligatory, as shown in several model systems and experimental paradigms. The assembly and sorting of transport mRNP granules rely on the internal localization elements residing within mRNA sequences and their specific interaction with trans-acting factors. These trans-acting protein factors provide the molecular bases for the translational status of transporting mRNAs and their tethering to molecular motors, which mediate directional active transport and local anchoring through interactions with the cytoskeleton. In addition, mRNA localization is a highly regulated process, which can be affected by both intracellular microenvironment, such as the asymmetrical organization of the cytoskeleton, and extracellular signaling events which impinge on the trans-acting factors to modulate RNA granule assembly, sorting, transport, anchoring and local translation. It should be noted that although this model summarized above has been well tested in multiple organisms, it is certainly not the solo mechanism by which mRNAs are asymmetrically localized. For example, mRNAs encoding cytosolic proteins may also be directed to their target locations through a cis-acting RNA element-independent mechanism. In budding yeast S. cerevisiae, ABP140 mRNA is localized to the distal pole of the mother cells (122) in contrast to Ash1 mRNA which is targeted to the budding cortex of the daughter cell. The localization of ABP140 mRNA relies on the translation of N-terminal peptide of ABP140 protein rather than specific RNA zipcode sequences as observed in Ash1 mRNA (122). The N-terminal motif of partially translated ABP140 tethers ABP140 mRNA and its associated ribosomes to actin filaments for distal pole localization (122). Similarly in fibroblast, Dia1 mRNA is localized to the endoplasmic reticulum (ER), this localization process is also mediated by the nascent Dia1 peptide undergoing translation through its direct interaction with GTP-bound RhoA (123). These emerging novel mechanisms for mRNA localization underscore our still limited understanding on the integration of mRNA localization mechanisms and encourage further studies.
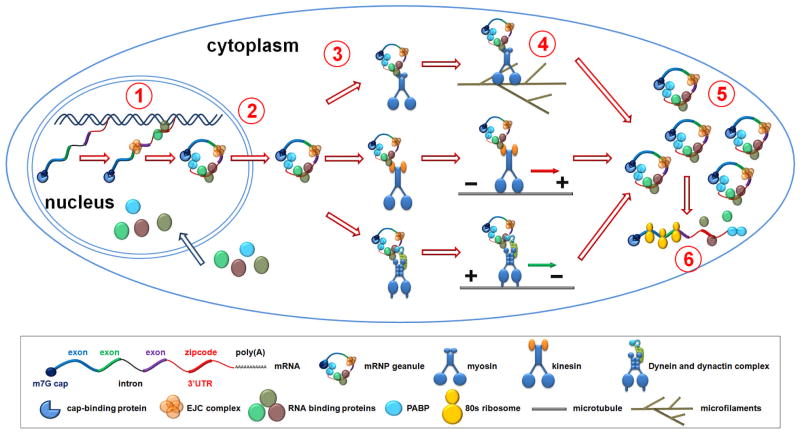
Figure 2. A schematic view of the multistep process for mRNA localization.
Mechanistic studies in different model systems reveal that mRNA localization is a multi-step process involving the assembly of mRNP granules, engagement of transport molecular motors and subsequent active transport and anchorage of mRNPs at the destination. (1) Assembly of transport mRNP granules can initiate from the nucleus, coupled to transcription and splicing. The binding of mRNA-binding proteins to nascent pre-mRNAs may help to maintain mature mRNAs in a translational quiescent state. EJCs are loaded onto mature mRNAs following the incision of introns. (2) mRNPs are exported into the cytoplasm. (3) mRNA-binding proteins interacting with zipcode elements recruit transport machineries by direct or indirect interaction with molecular motors including myosin, kinesin and dynein/dynactin complexes for microfilament or microtubule-mediated cargo transport. (4) Molecular motors couple mRNPs to microfilaments or microtubules for active transport. Molecular motor-mediated mRNA transport and localization requires intact cytoskeletal structures. (5) mRNPs are anchored to their destination. (6) Properly targeted and repressed mRNAs are released for local translation in response to local signaling events.
Although tremendous progress has been made in recent years, many interesting issues remain to be further addressed. It is important to further determine the molecular composition of transport mRNP granules, define the mechanisms that regulate their heterogeneity and elucidate the functional significance of diverse RNA sorting mechanisms for the polarized cell. In light of the complex composition of mRNP granules, it seems likely that there are machineries involved to ensure the proper assembly of granules with transport capability, but these machineries remain unclear. The continued development and applications of endogenous mRNA labeling systems, such as MS2 and λ-N22 systems, will continue to elucidate and further characterize mechanisms of mRNA sorting and trafficking (124, 125). Several MS2 transgenic animal models have been successfully generated to enable the visualization of endogenous mRNA (86, 126). With the MS2-mRNA labeling system, the endogenous mRNA can be purified to characterize the protein and mRNA compositions in detail (127). By analyzing the composition of mRNA granules of a given mRNA, mechanisms underlying development regulation of mRNA localization may also be better understood. In addition, such studies may help to identify the molecular motors mediating the transport of a specific mRNA and, possibly, mechanisms and conditions whereby mRNPs are directly and/or indirectly coupled to motors.
It will be important to continue to advance technology for visualization of endogenous mRNAs without the need for expression of transgenic reporters. The use of multiply labeled tetravalent RNA imaging probes (MTRIPs) has allowed visualization of native mRNA in live cells, including β-actin mRNA (128). Recently, this method was used to visualize microtubule dependent transport of single β-actin mRNA molecules in live cells (129). The opportunity for multiplex applications should shed further light on the sorting, copy number and heterogeneity of native RNA granules.
Further work is needed to understand the cytoskeletal basis for directed mRNA transport, and the role of asymmetric cytoskeletal organization. One important observation made recently is that the posterior pole localization of oskar mRNA in the Drosophila oocyte is mediated through slightly biased microtubules (86). Similarly, the vegetal localization of Vg1 mRNA in Xenopus oocyte is mediated by distinct kinesins through a group of microtubules with plus ends oriented toward the vegetal pole (130). Beyond oskar and Vg1 mRNAs, whether the polarized cytoskeleton constitutes a general mechanism for mRNA asymmetrical distribution remains as a further question to be addressed. Particularly in neurons, how the organization of microtubules contributes to the differential mRNA transport in axons and dendrites is unclear.
As an exciting topic, recent studies demonstrate that miRNAs and RNA-induced silencing complexes are selectively localized to dendritic compartments and distal axons and presumably regulate local protein synthesis (131–134). In neuronal dendrites, miR-134 colocalizes with its target Limk1 mRNA in discrete dendritic granules (135) and miR-125a is in association with PSD-95 mRNA in dendritic compartments (136), suggesting the presence of localized mRNA/miRNA complexes. These observations raise an interesting hypothesis that miRNAs are components of localized mRNA granules and may regulate mRNA localization by repressing mRNA translation. It remains unclear whether miRNAs are co-transported bound with their target mRNAs, or could traffic independently (133). One possible mode could be for microRNAs to sort with P-bodies, which have been described in dendrites to dynamically interact with RNA transport granules (137). The interrelationships between mRNA transport granules and microRNAs that regulate local translation remain to be further investigated.
Further work is needed to understand how extracellular signaling actively regulates the localization of mRNAs. Especially in neurons, stimulation with neurotransmitters and neurotrophic factors can selectively facilitate the localization of certain mRNAs. In mature axons that have limiting supplies of mRNA, injury can surely reactivate this biological process, and promote the localization of mRNAs to regenerating axons. The basis underlying extracellular signaling induced mRNA localization remains an area of great interest. Activity-dependent mRNA localization plays an important role in neuronal differentiation, survival and neural circuit formation (39). The defects in mRNA localization have been linked to a number of neurological disorders, such as Fragile X syndrome (72), amyotrophic lateral sclerosis and frontotemporal dementia (138, 139) and Spinal Muscular Atrophy (98). Proteins encoded by these disease causing genes, including FMRP, TDP-43, FUS and SMN, are components of transport mRNP granules and likely play important roles in mRNA granules assembly, sorting and/or mRNA localization. The continued study of mRNA localization and the function of local protein synthesis will further benefit our understanding of human development, disease mechanism and contribute to the discovery of therapeutic strategies that modulate mRNA localization and local translation.
Acknowledgments
We thank Drs. Christina Gross, Wilfried Rossell and Sharon Swanger for their critical reading of this manuscript. This work was supported by funding from NIH (to GB).
References
- 1.Lawrence JB, Singer RH. Intracellular localizationof messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 2.Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci. 2005;118(Pt 11):2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, King ML. Sending RNAs into the future: RNA localization and germ cell fate. IUBMB Life. 2004;56(1):19–27. doi: 10.1080/15216540310001658886. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 6.Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly (Austin) 2009;3(1):15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- 7.Yisraeli J, Melton D. The maternal mRNA Vg1 is correctly localized following injection. Nature. 1988;336:592–595. doi: 10.1038/336592a0. [DOI] [PubMed] [Google Scholar]
- 8.Deshler DO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1130. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 9.Litman P, Barg J, Rindzoonski L, Ginzburg I. Subcellular localization of tau mRNA in differentiating neuronal cell culture : Implications for neuronal polarity. Neuron. 1993;10:627–638. doi: 10.1016/0896-6273(93)90165-n. [DOI] [PubMed] [Google Scholar]
- 10.Garner CC, Tucker RP, Matus A. Selective localization of mRNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–679. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 11.Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKIIa is a cis acting signal for the localization and translation of mRNA in dendrites. PNAS. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassell GJ, Zhang HL, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Koisk KS. Sorting of beta actin mRNA and protein to neurites and growth cones in culture. Journal of Neuroscience. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23(32):10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123(2):431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51(6):685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453(7191):115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29(15):4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13(3):291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 20.Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30(46):15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25(4):778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cripe L, Morris E, Fulton AB. Vimentin mRNA location changes during muscle development. Proc Natl Acad Sci U S A. 1993;90(7):2724–2728. doi: 10.1073/pnas.90.7.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cajigas IJ, Tushev G, Will TJ, Tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74(3):453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26(51):13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 28.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27(20):5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J Neurobiol. 2004;61(2):222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- 30.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca(2+)-dependent growth cone guidance. Nat Neurosci. 2006;9(10):1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 31.Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9(10):1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. J Neurosci. 2010;30(28):9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci. 2011;31(27):9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31(2):261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 35.Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11(8):1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49(2):215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin Neurobiol. 2008;18(5):524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Swanger SA, Bassell GJ. Making and breaking synapses through local mRNA regulation. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127(2):441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13(5):625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Current Bio. 1999;(9):333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 43.Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176(1):36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 44.Macdonald PM, Kerr K, Smith JL, Leask A. RNA regulatory element Ble1 directs the early step on bicoid RNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- 45.Macdonald PM, Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 46.Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36(3):507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- 47.Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3(11):1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- 48.Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13(10):1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- 49.Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17(5):638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12(7):697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biological Chemistry. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- 52.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9(6):333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 53.Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. Embo J. 1997;16(7):1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17(4):2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24(2):148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19(23):6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25(11):4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X Mental Retardation Protein targets G Quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 59.Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB, Patel DJ. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18(7):796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14(6):926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bullock SL. Translocation of mRNAs by molecular motors: think complex? Semin Cell Dev Biol. 2007;18(2):194–201. doi: 10.1016/j.semcdb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 63.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13(2):161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19(20):5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci U S A. 2000;97(10):5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23(13):1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 2001;20(15):4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6(5):427–435. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- 69.Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12(3):571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 70.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. Journal of Neuroscience. 1996;16(24):7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43(4):513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 72.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell. 2009;20(8):2265–2275. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24(17):1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18(12):1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol. 2001;153(2):307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13(3):199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol Cell Biol. 2007;27(23):8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19(24):6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427(6976):753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 81.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428(6986):959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 82.di Penta A, Mercaldo V, Florenzano F, Munck S, Ciotti MT, Zalfa F, Mercanti D, Molinari M, Bagni C, Achsel T. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J Cell Biol. 2009;184(3):423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106(5):607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 84.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21(13):3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130(1):179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 86.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134(5):843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156(1):41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Glinka M, Herrmann T, Funk N, Havlicek S, Rossoll W, Winkler C, Sendtner M. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum Mol Genet. 2010;19(10):1951–1966. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- 89.Goetze B, Tuebing F, Xie Y, Dorostkar MM, Thomas S, Pehl U, Boehm S, Macchi P, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J Cell Biol. 2006;172(2):221–231. doi: 10.1083/jcb.200509035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma S, Liu G, Sun Y, Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim Biophys Acta. 2007;1773(6):912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163(4):801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, Bassell GJ. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31(10):3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Piazzon N, Rage F, Schlotter F, Moine H, Branlant C, Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem. 2008;283(9):5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 94.Tadesse H, Deschenes-Furry J, Boisvenue S, Cote J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet. 2008;17(4):506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H, Xing L, Singer RH, Bassell GJ. QNQKE targeting motif for the SMN-Gemin multiprotein complexin neurons. J Neurosci Res. 2007;85(12):2657–2667. doi: 10.1002/jnr.21308. [DOI] [PubMed] [Google Scholar]
- 96.Zhang H, Xing L, Rossoll W, Wichterle H, Singer RH, Bassell GJ. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J Neurosci. 2006;26(33):8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossoll W, Bassell GJ. Spinal muscular atrophy and a model for survival of motor neuron protein function in axonal ribonucleoprotein complexes. Results Probl Cell Differ. 2009;48:289–326. doi: 10.1007/400_2009_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10(8):597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res. 2012 doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Darnell JC. Defects in translational regulation contributing to human cognitive and behavioral disease. Curr Opin Genet Dev. 2011;21(4):465–473. doi: 10.1016/j.gde.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5(4):635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6(5):798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 103.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32(4):683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 104.Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19(5):2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mikl M, Vendra G, Kiebler MA. Independent localization of MAP2, CaMKIIalpha and beta-actin RNAs in low copy numbers. EMBO Rep. 2011;12(10):1077–1084. doi: 10.1038/embor.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A. 2012;109(12):4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tubing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci. 2010;30(11):4160–4170. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies. Cell. 2012;149(4):768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 109.Kwan AC, Dombeck DA, Webb WW. Polarized microtubule arrays in apical dendrites and axons. Proc Natl Acad Sci U S A. 2008;105(32):11370–11375. doi: 10.1073/pnas.0805199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85(21):8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Latham VM, Yu EH, Tullio AN, Adelstein RS, Singer RH. A rho-dependent signaling pathway operating through myosin localizes b-actin mRNA in fibroblasts. Current Biology. 2001;11:1010–1016. doi: 10.1016/s0960-9822(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 112.Yoshimura A, Fujii R, Watanabe Y, Okabe S, Fukui K, Takumi T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr Biol. 2006;16(23):2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 113.Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15(6):587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 114.Krauss J, Lopez de Quinto S, Nusslein-Volhard C, Ephrussi A. Myosin-V regulates oskar mRNA localization in the Drosophila oocyte. Curr Biol. 2009;19(12):1058–1063. doi: 10.1016/j.cub.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 115.Latham VM, Jr, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126(5):1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kislauskis EH, Zhu X, Singer RH. Beta-actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136(6):1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta -actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci U S A. 2001;98(13):7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438(7067):512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 119.Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Willis DE, Xu M, Donnelly CJ, Tep C, Kendall M, Erenstheyn M, English AW, Schanen NC, Kirn-Safran CB, Yoon SO, Bassell GJ, Twiss JL. Axonal Localization of transgene mRNA in mature PNS and CNS neurons. J Neurosci. 2011;31(41):14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30(22):4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kilchert C, Spang A. Cotranslational transport of ABP140 mRNA to the distal pole of S. cerevisiae. EMBO J. 2011;30(17):3567–3580. doi: 10.1038/emboj.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liao G, Ma X, Liu G. An RNA-zipcode-independent mechanism that localizes Dia1 mRNA to the perinuclear ER through interactions between Dia1 nascent peptide and Rho-GTP. J Cell Sci. 2011;124(Pt 4):589–599. doi: 10.1242/jcs.072421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 125.Daigle N, Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods. 2007;4(8):633–636. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 126.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Slobodin B, Gerst JE. A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes. RNA. 2010;16(11):2277–2290. doi: 10.1261/rna.2091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Santangelo PJ, Lifland AW, Curt P, Sasaki Y, Bassell GJ, Lindquist ME, Crowe JE., Jr Single molecule-sensitive probes for imaging RNA in live cells. Nat Methods. 2009;6(5):347–349. doi: 10.1038/nmeth.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lifland AW, Zurla C, Yu J, Santangelo PJ. Dynamics of native beta-actin mRNA transport in the cytoplasm. Traffic. 2011;12(8):1000–1011. doi: 10.1111/j.1600-0854.2011.01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15(3):426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hengst U, Jaffrey SR. Function and translational regulation of mRNA in developing axons. Semin Cell Dev Biol. 2007;18(2):209–215. doi: 10.1016/j.semcdb.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Murashov AK, Chintalgattu V, Islamov RR, Lever TE, Pak ES, Sierpinski PL, Katwa LC, Van Scott MR. RNAi pathway is functional in peripheral nerve axons. FASEB J. 2007;21(3):656–670. doi: 10.1096/fj.06-6155com. [DOI] [PubMed] [Google Scholar]
- 133.Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13(8):1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16(8):1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 136.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42(5):673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28(30):7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136(6):1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]