Autism spectrum disorders (ASD) are an early onset, heterogeneous group of heritable neuropsychiatric disorders with symptoms that include deficits in social interaction skills, impaired communication ability, and ritualistic-like repetitive behaviors1,2. One of the hypotheses for a common molecular mechanism underlying ASD is altered translational control resulting in exaggerated protein synthesis3. Genetic variants in chromosome 4q, which contains the EIF4E locus, have been described in autistic patients4,5. Importantly, a rare single nucleotide polymorphism has been identified in autism that is associated with increased promoter activity in the EIF4E gene6. Herein we show that genetically increasing the levels of eIF4E in mice7 results in exaggerated cap-dependent translation and aberrant behaviors reminiscent of autism, including repetitive/perseverative behaviors and deficits in social interactions. Moreover, these autistic-like behaviors are accompanied by synaptic pathophysiology in the medial prefrontal cortex, striatum, and hippocampus. The autistic-like behaviors displayed by the eIF4E transgenic mice are corrected by intracerebroventricular (ICV) infusions of the cap-dependent translation inhibitor 4EGI-1. Our findings demonstrate a causal relationship between exaggerated cap-dependent translation, synaptic dysfunction, and aberrant behaviors associated with autism.
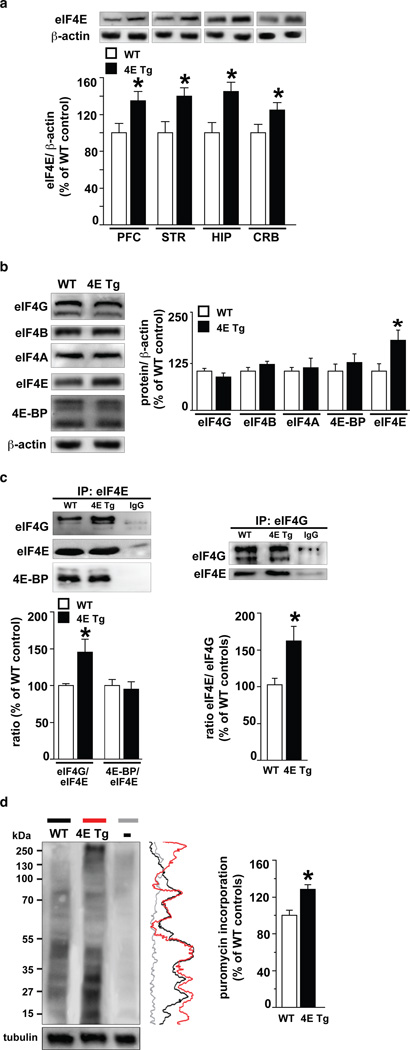
eIF4E transgenic mice7 exhibited increased levels of eIF4E across brain regions (Fig. 1a) without compensatory changes in levels of other translational control proteins (Fig. 1b). We investigated whether eIF4E was bound preferentially to either 4E-BP or eIF4G, which repress and promote, respectively, the initiation of cap-dependent translation8,9. We found significantly higher levels of eIF4E/eIF4G interactions in the brains of eIF4E transgenic mice (Fig. 1c and Supplementary Fig. 1a) with no alterations in the interaction between eIF4E and 4E-BP (Fig. 1c, left and Supplementary Fig. 1a). To confirm that the increased eIF4E/eIF4G interactions resulted in increased protein synthesis, we infused puromycin into the lateral ventricle of cannulated mice and labeled newly synthesized proteins using SUnSET10,11 and observed increased de novo cap-dependent translation (Fig. 1d and Supplementary Fig. 1b-g). Overall, our results indicate that overexpression of eIF4E results in exaggerated cap-dependent translation in the brains of eIF4E transgenic mice.
Figure 1. eIF4E transgenic mice exhibit increased eIF4E/eIF4G interactions and exaggerated cap-dependent translation.
a, eIF4E transgenic mice (4E Tg) exhibit increased eIF4E expression in multiple brain regions. n=4 mice/genotype, *p<0.05 vs wild-type (WT), Student's t-test. b, 4E Tg mice exhibit normal expression of other translational control proteins. n=4 mice/genotype. *p<0.05 vs WT, Student's t-test. c, 4E Tg mice exhibit increased eIF4E/eIF4G interactions. Immunoprecipitation of eIF4E (left panel) and eIF4G (right panel). n=3 mice/genotype, *p<0.05 vs WT, Student's t-test. d, 4E Tg mice exhibit increased translation as measured with SUnSET (see Supplementary Methods). Vertical line traces of each autoradiogram are shown on the right. n=3 mice/genotype, *p<0.05 vs WT, Student's t-test. – represents a control sample without puromycin. All data are shown as mean ± SEM.
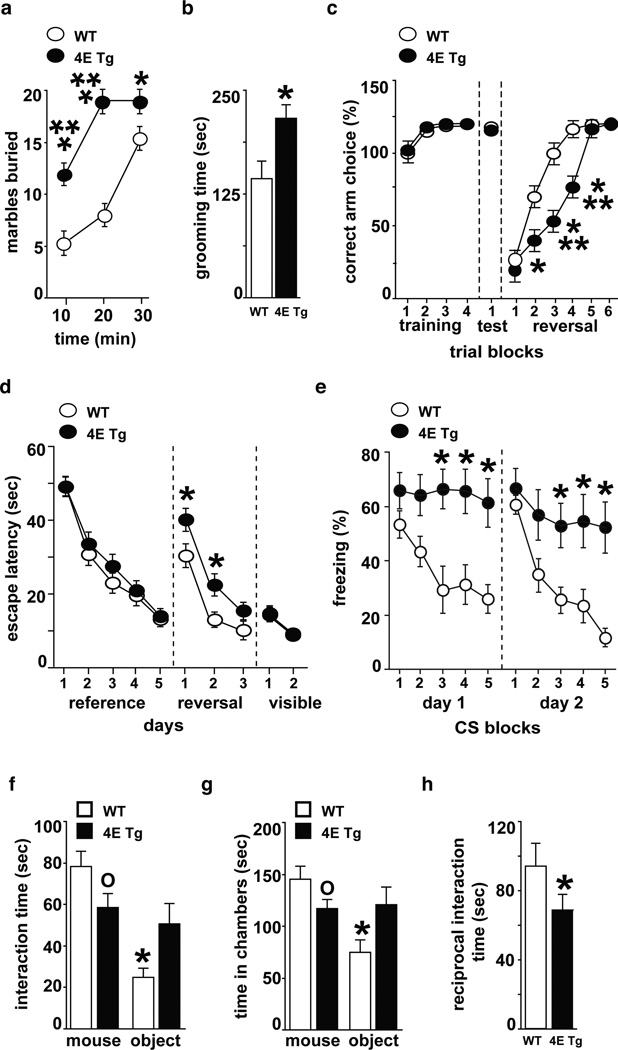
We then determined whether eIF4E transgenic mice display repetitive and perseverative behaviors, which are behavioral domains required for ASD diagnosis2. eIF4E transgenic mice exhibited repetitive digging behavior in the marble burying test12 and increased self-grooming13 compared to wild-type littermates (Fig. 2a, 2b). eIF4E transgenic mice also displayed cognitive inflexibility in both a water-based Y-maze task and a modified version of the Morris water maze14,15. Learning ability in the acquisition and memory phases of these tasks was intact; however, in the reversal phases, eIF4E transgenic mice were impaired in locating the new platform positions (Fig. 2c, 2d and Supplementary Fig. 2e-h). We tested an additional form of behavioral inflexibility by examining the eIF4E transgenic mice for extinction of cued fear conditioning and found that they did not exhibit a significant reduction in freezing responses following extinction training (Fig. 2e). These experiments suggest that excessive cap-dependent translation in the brain affects the ability to suppress previously codified response patterns and the ability to form new behavioral strategies in response to changed environmental circumstances.
Figure 2. eIF4E transgenic mice exhibit ASD-like behaviors.
eIF4E transgenic mice (4E Tg) were compared to wild-type (WT) littermates. a, Marble burying test: n=21–22 mice/genotype, ***p<0.001 and *p<0.05 vs WT, repeated measures (RM) ANOVA [time X genotype, F(2,46)=31.62, p< 0.001] followed by Bonferroni-Dunn test. b, Self-grooming test: n=12 mice/genotype, *p<0.05 vs WT, Student’s t-test. c, Y-maze reversal task: n=21–22 mice/genotype, *p<0.05 and ***p<0.001 vs WT, RM ANOVA [time X genotype, F(5,138)=16.74, p< 0.001] followed by Bonferroni-Dunn test. d, Morris water maze (MWM) reversal learning: n=12–13 mice/genotype, *p<0.05 vs WT, RM ANOVA [time X genotype, F(3,92)=6.1, p< 0.001] followed by Bonferroni-Dunn test. e, Extinction of cued fear memory (15 CS/day represented as 3 CS/block): n=6 mice/genotype, *p<0.05 vs WT, RM ANOVA [day1: time X genotype, F(4,40)=5.73, p< 0.001; day2: time X genotype, F(4,40)=4.81, p< 0.01] followed by Bonferroni-Dunn test. f,g, Social behavior test: time spent interacting with either a stranger mouse f, or in the chambers g. n=6 mice/genotype, *p< 0.05 and °p< 0.05 vs. WT, RM ANOVA [f: stimulus X genotype, F(1,10)=6.04, p< 0.05; g: stimulus X genotype, F(1,10)=6.12, p< 0.05] followed by Bonferroni-Dunn test. h, Reciprocal social interaction task: n=6 mice/genotype, *p<0.05 vs WT controls, Student's t-test. All data are shown as mean ± SEM.
Abnormalities in social interaction skills are another behavioral defect displayed by individuals with ASD2. In tests to examine social behavior16–18, the eIF4E transgenic mice did not show a preference for a nonspecific stranger versus a novel, inanimate object (Fig. 2f, 2g). Moreover, eIF4E transgenic mice exhibited diminished reciprocal interactions with a freely moving stranger mouse (Fig. 2h), further evidence of deficits in social behavior. The deficits in social behavior of the eIF4E transgenic mice are unlikely to be caused by a generalized increase in anxiety (Supplementary Fig. 2c, 2d and 2j). Moreover, the eIF4E transgenic mice exhibited mild hyperactivity (Supplementary Fig. 2a and 2b), but no impairments in motor coordination, motor learning and sensorimotor gating (Supplementary Fig 2i and 2k, 2l). Taken together, our behavioral analysis of the eIF4E transgenic mice indicates that increased cap-dependent translation in the brain results in a distinct pattern of behavioral abnormalities consistent with ASD.
Previous studies suggest that ASD symptoms such as cognitive inflexibility and deficits in social behavior are generated by abnormalities in prefrontal and/or striatal circuits19. Consistent with this idea, the medial prefrontal cortex (mPFC) is implicated in the modulation of social behaviors and social skills20, whereas motor, social, and communication impairments in boys with ASD are associated with anatomical abnormalities in the striatum21. Therefore, we next examined whether the eIF4E transgenic mice exhibited specific synaptic pathophysiologies in the mPFC and striatum.
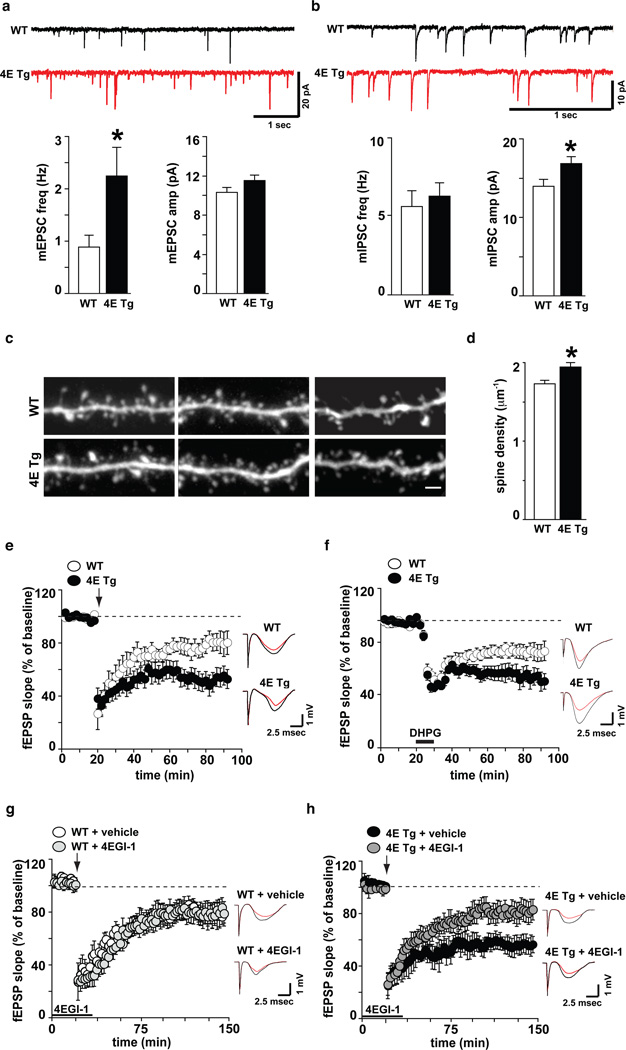
In the eIF4E transgenic mice, examination of spontaneous synaptic “mini” events in layers 2/3 of acute mPFC slices revealed an increase in the frequency but not amplitude of excitatory events (mEPSC; Fig 3a), and an increase in the amplitude, but not frequency, of inhibitory events (mIPSC; Fig 3b). No changes were observed in layer 5 (Supplementary Fig. 3a, 3b). Thus, our data suggests an enhancement of excitatory input and post-synaptic sensitivity for inhibitory events onto layer 2/3 pyramidal neurons, consistent with the hypothesis that autism may arise from an imbalance between excitatory and inhibitory synaptic transmission22.
Figure 3. eIF4E transgenic mice exhibit alterations in synaptic function, dendritic spine density, and synaptic plasticity.
a, eIF4E transgenic mice (4E Tg) exhibit increased mEPSC frequency and b, increased mIPSC amplitude in layer 2/3 mPFC pyramidal neurons. n=27–30 neurons/genotype, *p<0.05 vs WT, Student's t-test. c,d, 4E Tg mice exhibit increased dendritic spine density in layer 2/3 mPFC pyramidal neurons. High-magnification images c, and quantification d, of spiny dendrites from wild-type (WT) and 4E Tg mice. n= 12 neurons/genotype, *p<0.05 vs WT, Student's t-test. Scale bar = 2 µm. e, 4E Tg mice exhibit enhanced striatal LTD. n=13 slices from 8 mice/genotype. f, 4E Tg mice exhibit enhanced hippocampal mGluR-LTD. n=15 slices from 8 mice/genotype. g, h, 4EGI-1 normalizes enhanced striatal LTD displayed by 4E Tg mice h, without impacting LTD in WT mice g. n=18 slices from 9 mice/genotype/treatment. All field recordings were analyzed with repeated measure ANOVAs. Arrows indicate delivery of high-frequency stimulation (HFS). Solid bars indicate the duration of bath application of DHPG (10 µM, 10 min) and 4EGI-1 (100 µM, 45 min). Representative traces (right panels) showing fEPSP before (black) and 60 min after (red) HFS. All data are shown as mean ± SEM.
To determine whether the increased frequency of spontaneous mEPSCs might result from an enhanced number of synaptic contacts, we imaged dendritic spines using two-photon laser-scanning microscopy (Fig. 3c, 3d and Supplementary Fig. 3c, 3d). We found a significant increase (~12%) in spine density and observed a significantly smaller spine volume in the eIF4E transgenic mice compared to wild-type littermates (WT=0.123±0.004 um3 and 4E Tg=0.110±0.004 um3, p=0.01 vs. WT, Student’s t-test).
Next, we examined whether increased expression of eIF4E also resulted in synaptic pathophysiology in the striatum. We used high-frequency stimulation (HFS) to induce long-term depression (LTD) in acute striatal slices23 and found that eIF4E transgenic mice exhibited enhanced LTD compared to wild-type littermates (Fig. 3e, Supplementary Fig. 3e, 3f). We hypothesize that the enhanced LTD in eIF4E transgenic mice results in altered efficiency of striatal information storage and processing, culminating in the inability to form new motor patterns and/or to disengage from previously learned motor behaviors.
To determine whether the synaptic alterations described in the eIF4E transgenic mice were selective for the fronto-striatal circuit, we examined synaptic plasticity in the hippocampus24. We found that eIF4E transgenic mice exhibited enhanced mGluR-LTD compared to wild-type littermates (Fig. 3f, Supplementary Fig. 3g, 3h), consistent with previous studies showing that changes in brain protein synthesis are accompanied by altered (enhanced or reduced) hippocampal mGluR-LTD25,26. Thus, consistent with the ubiquitous increase in brain expression of eIF4E, the eIF4E transgenic mice display altered synaptic function and plasticity in several brain regions (mPFC, striatum and hippocampus) implicated in behavioral abnormalities associated with ASD.
Finally, we asked whether exaggerated cap-dependent translation was responsible for the synaptic alterations and ASD-like behaviors displayed by the eIF4E transgenic mice. We took advantage of 4EGI-1, an inhibitor of eIF4E/eIF4G interactions8,11, to block the synaptic and behavioral consequences of increased eIF4E expression. Bath application of 4EGI-1 normalized the enhanced striatal LTD observed in the eIF4E transgenic mice (Fig. 3g and 3h), suggesting that exaggerated striatal LTD (Fig. 3h) is a direct consequence of increased binding of eIF4E to eIF4G (Supplementary Fig. 3i-k).
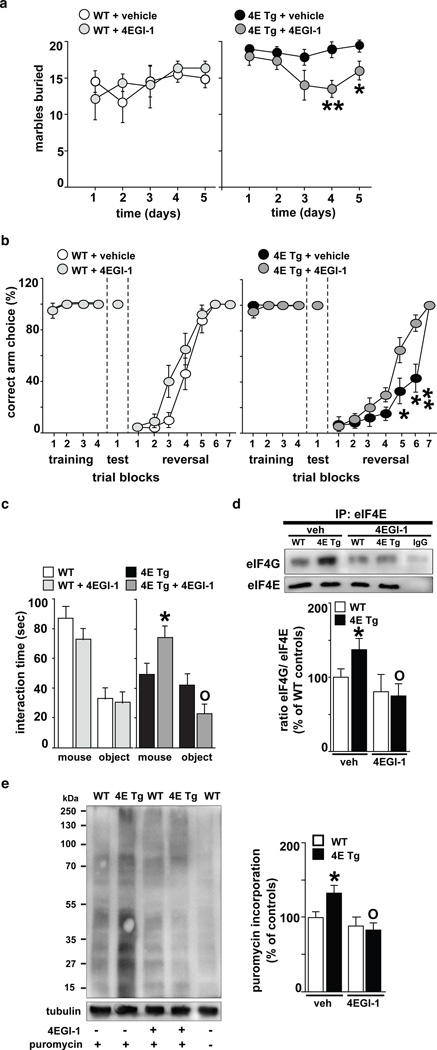
Next, we employed a subthreshold dose of 4EGI-111 to normalize the behavioral abnormalities in eIF4E transgenic mice without impairing their wild-type littermates. eIF4E transgenic mice treated with 4EGI-1 exhibited a decrease in repetitive behavior during the marble burying task, which started on day four and persisted throughout day five (Fig. 4a). Moreover, we found that 4EGI-1 maintained the behavioral effects observed in the marble-burying task (Supplementary Fig. 4a and 4b). We also found that blockade of eIF4E/eIF4G interactions with 4EGI-1 significantly improved the performance of eIF4E transgenic mice in the reversal phase of the Y-maze test (Fig. 4b). These findings indicate that chronic treatment of eIF4E transgenic mice with 4EGI-1 reverses their repetitive and perseverative behaviors. We also found that infusions of 4EGI-1 rescued the social behavior deficits displayed by the eIF4E transgenic mice in the three-chamber arena test, as they exhibited an increased preference for a non-specific stranger compared to a novel object (Fig. 4c).
Figure 4. The cap-dependent translation inhibitor 4EGI-1 reverses ASD-like behaviors displayed by eIF4E transgenic mice.
a, Treatment of eIF4E transgenic mice (4E Tg) with 4EGI-1 reduces the marble-burying behavior n=6 mice/genotype/treatment, **p< 0.01, *p< 0.05 vs. 4E Tg + vehicle, two-way repeated measures (RM) ANOVA [treatment X genotype, F(1,20)= 4.21, p< 0.05] followed by Bonferroni-Dunn test. b, 4EGI-1 improves the cognitive flexibility of 4E Tg mice in the Y-maze test. n=6–7 mice/genotype/treatment, **p< 0.01, *p< 0.05 vs. 4E Tg + vehicle, two-way RM ANOVA [treatment X genotype, F(1,21)=4.61, p< 0.05] followed by Bonferroni-Dunn test. c, 4EGI-1 improves social behavior of 4E Tg mice in the three-chamber arena test. n=6 mice/genotype/treatment, *p< 0.05 and °p< 0.05 vs. 4E Tg + vehicle, two-way RM ANOVA [treatment X genotype, F(1,20)=6.26, p< 0.05] followed by Bonferroni-Dunn test. d, 4EGI-1decreases the enhanced eIF4E/eIF4G interactions in 4E Tg mice. Immunoprecipitation (IP) of eIF4E in the striatum. n=4 mice/genotype, *p<0.05 and °p<0.05 vs vehicle-treated wild-type (WT) and 4E Tg, respectively, two-way ANOVA, followed by Bonferroni-Dunn test. e, 4EGI-1 normalizes the exaggerated cap-dependent translation in 4E Tg mice as measured with SUnSET. The last WT sample represents a control without puromycin. *p<0.05 and °p<0.05 vs vehicle-treated WT and 4E Tg, respectively, two-way ANOVA, followed by Bonferroni-Dunn test. All data are shown as mean ± SEM.
At the completion of the behavioral studies with 4EGI-1, we performed co-immunoprecipitation experiments, confirming that 4EGI-1 reduced the increased eIF4E/eIF4G interactions exhibited by the eIF4E transgenic mice (Fig. 4d and Supplementary Fig. 4c-e). Furthermore, puromycin-labeling of newly synthesized proteins was reduced to wild-type levels, indicating that 4EGI-1 was effective in attenuating the increased cap-dependent translation in the eIF4E transgenic mice (Fig. 4e and Supplementary Fig. 4f and 4g). Together, these results indicate that repeated treatment of eIF4E transgenic mice with 4EGI-1 reverses the increased binding of eIF4E to eIF4G, exaggerated cap-dependent translation, and reversal of ASD-like behaviors.
Herein we have demonstrated that increased eIF4E expression and, consequently, dysregulated translational control at the initiation phase of protein synthesis in mice results in the appearance of synaptic dysfunction and aberrant behaviors consistent with ASD. Based on our observations, we hypothesize that exaggerated cap-dependent protein synthesis in the eIF4E transgenic mice and FXS model mice27,28 results in enhanced translation of a specific subset of mRNAs. Thus, the identity of both these mRNAs and the cis-acting elements in the 5’ UTR responsible for eIF4E-dependent protein synthesis and their possible overlap with fragile X mental retardation protein target mRNAs will be important investigations in future studies.
In closing, our studies with eIF4E transgenic mice indicate that ASD-like behaviors can be induced by exaggerated cap-dependent translation in the brain. Moreover, we demonstrated that aberrant repetitive, perseverative, and social behaviors displayed by eIF4E transgenic mice are reversed by reducing eIF4E/eIF4G interactions, thereby restoring translational homeostasis. Thus, our findings establish a causal link between exaggerated cap-dependent translation and behaviors associated with autism. Finally, our findings indicate that behavioral defects caused by exaggerated cap-dependent translation, which also occurs in FXS29,30, a disorder with a high incidence of ASD, are not irrevocable and can be corrected well into adulthood.
Methods Summary
All procedures involving animals were approved by the New York University Animal Care and Use Committee and followed the NIH Guidelines for the use of animals in research. For a detailed description of all the techniques used in this study, please refer to Methods section. All the experiments were performed with the examiners blind to genotype.
Methods
Housing
Generation of βT-Eif4e transgenic mice (eIF4E transgenic mice) has been described previously7.
For all the experiments, we made use of littermates derived from crossing heterozygotes. Mice were back-crossed to the N10 generation in C57BL/6J mice. Overall, eIF4E transgenic mice were viable, fertile, and showed no gross anatomical abnormalities in the age range used for this study. eIF4E transgenic mice and their wild-type littermates were housed in groups of 3–4 animals per cage and kept on a regular 12 h light/dark cycle (lights on at 7:00 am). Food and water were available ad libitum.
Surgery and drug infusion
Mice were anesthetized [ketamine (100 mg/kg) and xylazine (10 mg/kg)] and mounted onto a stereotaxic apparatus. Cannulae (26-gauge) were implanted unilaterally at the following coordinates (−0.22 mm anterioposterior, +1 mm mediolateral, and −2.4 mm dorsoventral)31. Mice were allowed one week to recover after the surgery.
The infusions of the eIF4E/eIF4G inhibitor 4EGI-1 were performed as described previously11. 4EGI-1 dissolved in 100% DMSO was diluted in vehicle [0.5% (2-hydroxypropyl)-β-cyclodextrin and 1% DMSO in ACSF]. 4EGI-1 (20 µM) or vehicle were infused over 1 min (0.5 µL/min; Harvard Apparatus). On the last day of treatment, mice received infusion of 4EGI-1 alone or puromycin (25 µg in 0.5 µL) prior to 4EGI-1 infusions. All behavior and tissue dissection occurred 1 hr after 4EGI-1 infusions.
Behavior
The following behavioral tests were performed on male eIF4E transgenic mice and their wild-type littermates (2–6 months of age) as described previously: novelty-induced locomotor activity32, open field33, elevated plus maze33, rotarod34, prepulse inhibition33, marble burying14, social behavior16, direct social interaction35,36, y-maze and the Morris water maze7,35.
For all experiments, mice were acclimated to the testing room 30 min prior to behavioral training and all behavior apparatuses were cleaned between each trial with 30% ethanol. The experimenter was blind to genotype and drug treatment while performing and scoring all behavioral tasks. All behavioral tests were performed starting with the least aversive task first (locomotor activity) and ending with the most aversive (either water-based mazes or extinction of fear memory).
Western blots
Mice were killed by decapitation 1 hr after the infusion with either 4EGI-1 alone or 4EGI-1 and puromycin. The striatum and prefrontal cortex were rapidly dissected, placed on an ice-cold surface, and sonicated in 1% SDS and boiled for 10 min. Aliquots (2 µl) of the homogenate were used for protein determination with a BCA (bicinchoninic acid) assay kit (Pierce, Thermo Scientific, Rockford, USA). Equal amounts of protein (20 µg) for each sample were loaded onto 10% polyacrylamide gels. Proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred overnight to polyvinylidene difluoride membranes (Immobilon-Psq, Millipore Corporation, Billerica, USA). The membranes were immunoblotted with antibodies against eIF4E (1:1000), eIF4G (1:1000), eIF4B (1:1000), eIF4A (1:1000), 4E-BP (1:1000) (Cell Signaling Technology, MA, USA). Antibodies against β-actin and tubulin (1:5000, Cell Signaling Technology, MA, USA) were used to estimate the total amount of proteins. Detection was based on HRP-conjugated secondary antibody (Promega, WI, USA) and chemiluminescence reagent (ECL or ECL plus; GEHealthcare, Buckinghamshire, UK), and visualized using a Kodak 4000MM imager to obtain pixel density values for the band of interest (Carestream, NY, USA). All images were obtained using maximum sensitivity settings with no binning (0–65 K signal range). No images analyzed presented saturating signals for the bands of interest (>65 K grayscale value). The amount of each protein was normalized for the amount of the corresponding β-actin or tubulin detected in the sample.
Immunoprecipitation
Tissue was homogenized in ice-cold lysis immunoprecipitation buffer containing (in mM): 40 hepes (pH 7.5), 150 NaCl, 10 pyrophosphate, 10 glycerophosphate, 1 EDTA and 0.1% CHAPS, Protease Inhibitor II, Phosphatase Inhibitor Mixture I, II (Sigma-Aldrich, MO, USA). Cleared homogenate (500 µg) was incubated with either anti-eIF4G (2.5 µg) or eIF4E (2.5 µg) (Bethyl Laboratories, TX, USA) and gently shaken overnight at 4°C. The antibody/lysate mix was incubated with 75 µL IgG bound to agarose-beads (Thermo Scientific, IL, USA). The bead/sample slurry was incubated while rocking at 4°C overnight. Supernatant was removed and saved, and immunoprecipitates were washed three times in lysis buffer, and once in wash buffer (50 mM hepes pH 7.5, 40 mM NaCl, 2 mM EDTA). SDS/PAGE buffer was added to the washed immunoprecipitates, which then were resolved on 4 to 12% gradient gels. Efficiency of the immunoprecipitation was determined by examining the supernatant and wash fractions obtained from the procedure on images obtained from Kodak 4000MM imager (see Western Blots). Band density values for coimmunoprecipitated eFI4E, eIF4G and 4E-BP were normalized to immunoprecipitated eIF4G or eIF4E.
SuNSET
A protein synthesis assay was performed as previously described using the SuNSET method 11. Puromycin-treated samples were identified on blots using the mouse monoclonal antibody 12D10 (1:5000 from a 5-mg/mL stock). Because only a small fraction of the brain proteins were labeled, signal from blots was identified using ECL-Advance (GEHealthcare, Buckinghamshire, UK).
Electrophysiology
Hippocampal (400 µm), prefrontal and striatal slices (300 µm) for electrophysiology were prepared as described previously24.
Solution to maintain slices
Cutting solution (CS; in mM): 110 Sucrose, 60 NaCl, 3 KCl, 1.25 NaH2PO4, 28 NaHCO3, 0.5 CaCl2, 7 MgCl2, 5 Glucose, 0.6 Ascorbate. Artificial cerebrospinal fluid (ACSF; in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 D-glucose, 2 CaCl2, and 1 MgCl2. Slices were incubated at room temperature and then were placed in the recording chamber for additional recovery time of 60 min at 33°C.
Extracellular recordings
Extracellular field EPSPs were recorded as described previously23,24. In all the experiments, baseline synaptic transmission was monitored for at least 20 min before long-term depression (LTD) induction. Three trains of high-frequency stimulation (3 sec duration, 100 Hz frequency at 20 sec intervals) were used to induced LTD in striatal slices23 while 10 min of incubation with DHPG (50 µM) was used to induce mGluR-dependent LTD in hippocampal slices24. The slope of fEPSP was expressed as percent of the baseline average before LTD induction.
Intracellular recordings
Medial prefrontal pyramidal cells were illuminated and visualized using a x60 water-immersion objective mounted on a fixed-stage microscope (BX61-WI, Olympus, Center Valley, PA), and the image was displayed on a video monitor using a charge-coupled device camera (Hamamatsu, Bridgewater, NJ). Recordings were amplified by multiclamp 700B and digitized by Digidata 1440 (Molecular Devices, Sunnyvale, CA). The recording electrode was pulled from a borosilicate glass pipette (3–5 MΩ) using an electrode puller (P-97, Sutter Instruments, Navato, CA), was filled with an internal solution according to the specific experimental requirement, and was patched onto the soma. Rs was compensated ~70% and was readjusted before each experiment. A measured liquid junction potential was corrected by adjusting the pipette offset. All voltage-clamp recordings were low-pass filtered at 10 kHz and sampled at 50 kHz.
Internal solution for mEPSC (in mM): 120 cesium methane-sulfonate, 10 HEPES, 10 EGTA, 4 MgCl2, 0.4 NaGTP, 4 MgATP, 10 phosphocreatine, 5 QX-314 (pH adjusted to 7.3 with CsOH, 290 mOsm). Bicuculline 50 µM and tetrodotoxin (TTX) 1 µM (Tocris, Ellisville, MI) were added to the external ACSF bath solution.
Internal solution for mIPSC (in mM): 140 CsCl, 10 EGTA, 10 HEPES, 2 MgCl2, 2.0 Mg-ATP, 4 Na2-ATP, 0.4 Na2-GTP, 5 QX-314 (pH adjusted to 7.3 with CsOH, 290 mOsm), thus yielding a chloride reversal potential of around 2 mV for the chloride currents. 40 µM 6,7-dinitroquinoxaline-2,3-dione (DNQX), 50 µM 2-amino-5-phosphonopentanoate (D-AP-5) and 1 µM TTX were added to the ACSF bath solution.
In these conditions, mEPSC and mIPSC were recorded in voltage clamp at −70 mV and measured for 120 sec or 60 sec, respectively.
Dendritic spine morphology
Dendritic spine density experiments were performed as previously described37,38. Briefly, two-photon imaging was accomplished with a custom microscope and high-resolution stacks (x = 0.13 µm, y = 0.13 µm, z = 0.2 µm per voxel) of dendritic segments throughout the entire cell were taken for morphological analysis in NeuronStudio. Spine head volume was calculated using a rayburst algorithm. Images were deconvolved prior to volume measurements using custom routines written in MATLAB (Mathworks).
Supplementary Material
Acknowlegements
We would like to thank Dr. Joseph LeDoux and members of his laboratory for their technical support and suggestions. We would also like to thank Drs. David St. Clair and Dr. Zosia Miedzybrodzka for their comments on the manuscript. This research was supported by NIH grants NS034007, NS047384, and NS078718, and Department of Defense award W81XWH-11-1-0389 (E.K.), NIH grant CA154916 (D.R.), and the Wellcome Trust (A.F.M.).
Footnotes
Author Contributions
The study was directed by E.K. and conceived and designed by E.S. and E.K. E.S. performed the molecular, behavioral, and electrophysiogical experiments. T.N.H. performed behavioral experiments. A.F.M. and A.G.C. performed the dendritic spine density experiments. P.P. contributed the anti-puromycin (12D10) antibody. D.R. contributed with reagents and expertise concerning translation control by eIF4E. H.K. performed the cortical whole-cell electrophysiolgical experiments. The manuscript was written by E.S. and E.K. and edited by all of the authors.
The authors declare no competing financial interests.
References
- 1.Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119:747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rapin I, Tuchman RF. Autism: definition, neurobiology, screening, diagnosis. Pediatric Clinics of NA. 2008;55:1129–1146. doi: 10.1016/j.pcl.2008.07.005. viii. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher RJ, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Autism Genome Project Consortium, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonan AL, et al. A genomewide screen of 345 families for autismsusceptibility loci. Am J Hum Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neves-Pereira M, et al. Deregulation of EIF4E: a novel mechanism for autism. Journal of Medical Genetics. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- 7.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 8.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 9.Gingras AC, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Meth. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 11.Hoeffer CA, et al. Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proceedings of the National Academy of Sciences. 2011;108:3383–3388. doi: 10.1073/pnas.1013063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas A, et al. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology. 2009;204:361–373. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeffer CA, et al. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60:832–845. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehninger D, et al. Reversal of learning deficits in a Tsc2 mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moy SS, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou J, et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon C-H, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fineberg NA, et al. Probing compulsive and impulsive behaviors, from 10 animal models to endophenotypes: a narrative review. Neuropsychopharmacology. 2010;35:591–604. doi: 10.1038/npp.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu A, Adler M, Crocetti D, Miller MI, Mostofsky SH. Basal ganglia shapes predict social, communication, and motor dysfunctions in boys with autism spectrum disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:539–551. 551.e1–551.e4. doi: 10.1016/j.jaac.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Tabuchi K, et al. A Neuroligin-3 Mutation Implicated in Autism Increases Inhibitory Synaptic Transmission in Mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calabresi P, Maj R, Pisani A, Mercuri NB, Bernardi G. Longterm synaptic depression in the striatum: physiological and pharmacological characterization. J Neurosci. 1992;12:4224–4233. doi: 10.1523/JNEUROSCI.12-11-04224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou L, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent longterm depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Huber KM, (null), Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 26.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronesi JA, et al. Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat Neurosci. 2012;15:431–440. doi: 10.1038/nn.3033. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dölen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. Third Edition. Academic Press; 2007. [Google Scholar]
- 32.Errico F, et al. The GTP-binding protein Rhes modulates dopamine signalling in striatal medium spiny neurons. Mol Cell Neurosci. 2008;37:335–345. doi: 10.1016/j.mcn.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Banko JL. Behavioral alterations in mice lacking the translation repressor 4E-BP2. Neurobiology of Learning and Memory. 2007;87:248–256. doi: 10.1016/j.nlm.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Borgkvist A, et al. Altered dopaminergic innervation and amphetamine response in adult Otx2 conditional mutant mice. Mol Cell Neurosci. 2006;31:293–302. doi: 10.1016/j.mcn.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 35.Chévere-Torres I, Maki JM, Santini E, Klann E. Impaired social interactions and motor learning skills in tuberous sclerosis complex model mice expressing a dominant/negative form of tuberin. Neurobiology of Disease. 2011 doi: 10.1016/j.nbd.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumitriu D, Rodriguez A, Morrison JH. High-throughput, detailed, cell-specific neuroanatomy of dendritic spines using microinjection and confocal microscopy. Nat Protoc. 2011;6:1391–1411. doi: 10.1038/nprot.2011.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chalifoux JR, Carter AG. GABAB receptors modulate NMDA receptor calcium signals in dendritic spines. Neuron. 2010;66:101–113. doi: 10.1016/j.neuron.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.