Abstract
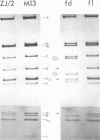
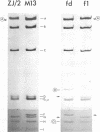
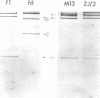
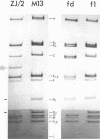
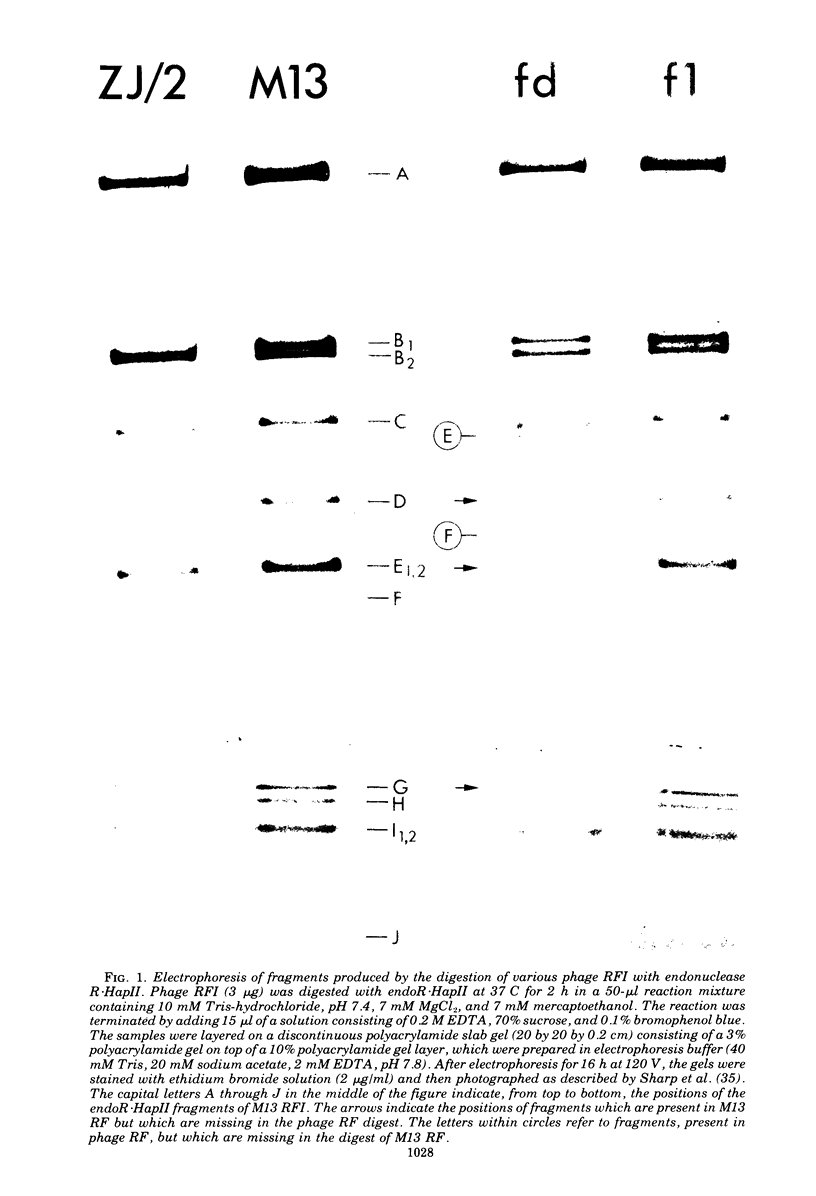
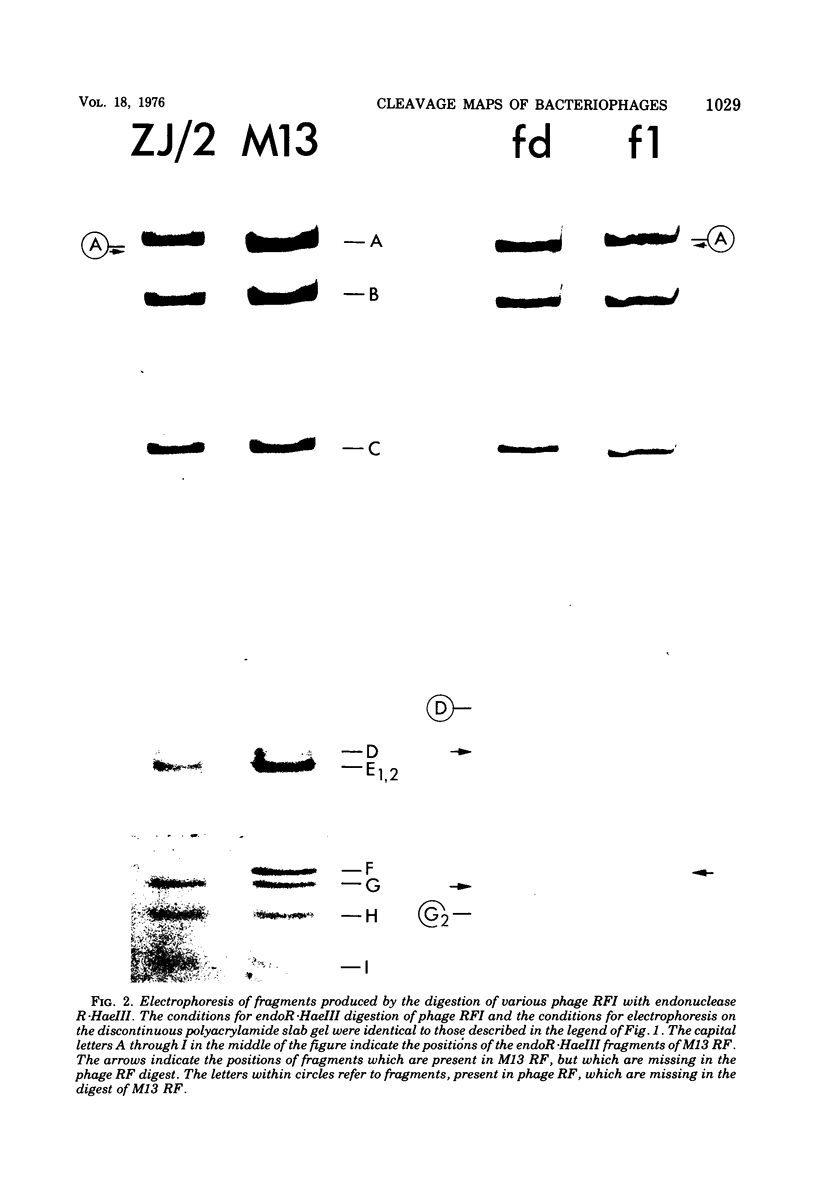
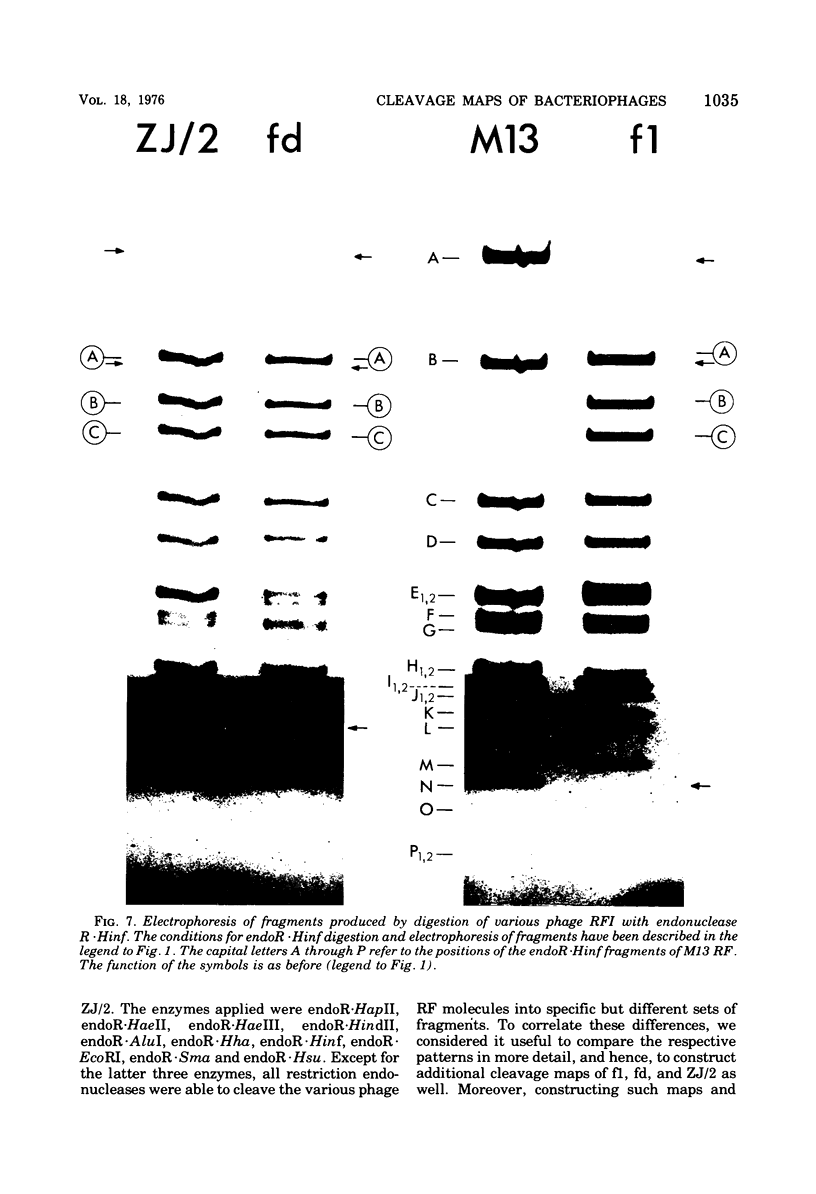
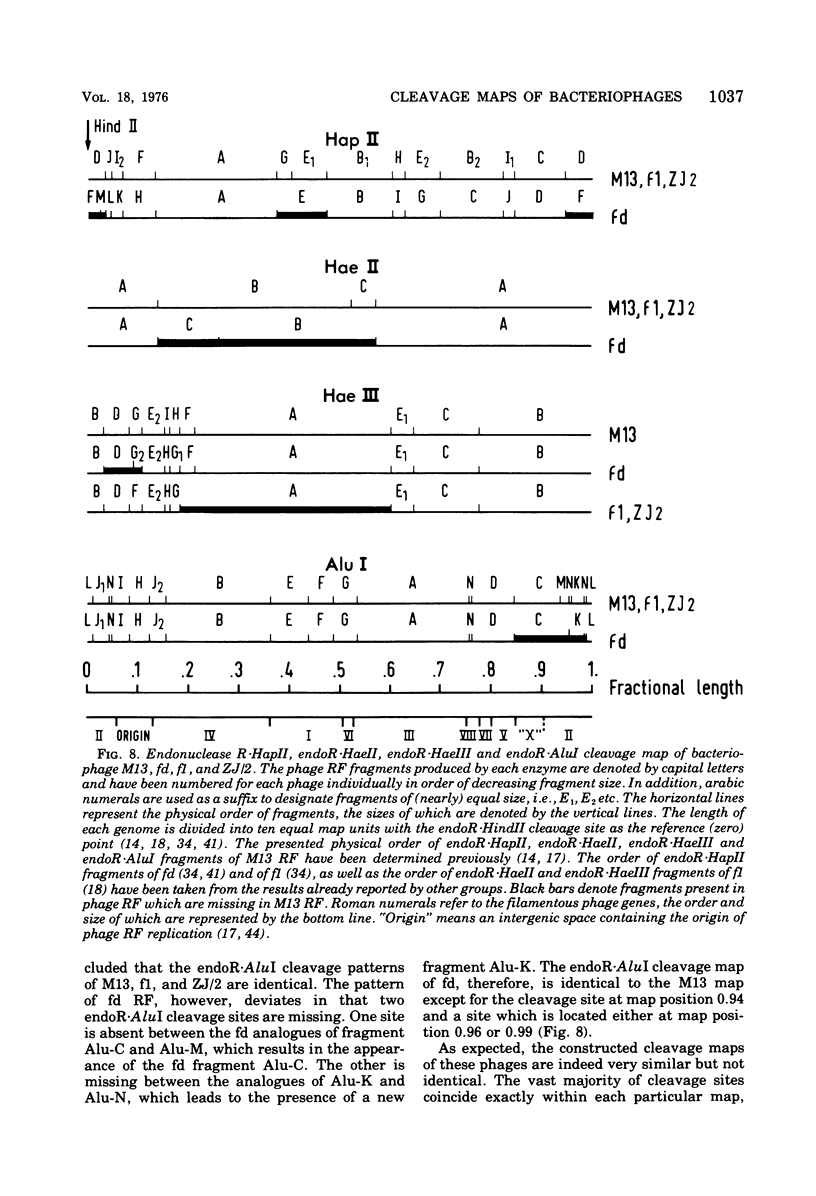
The replicative form DNAs of bacteriophage M13, fd, f1, and ZJ/2 were found to be sensitive to cleavage by the restriction endonucleases endoR-HapII, endoR-HaeII, endoR-HaeIII, endoR-HindII, endoR-AluI, endoR-Hha, and endoR-Hinf. With respect to M13 DNA the number of cleavage sites varied from 21 for endoR-Hinf, 18 for endoR-AluI, 15 for endoR-Hha, 13 for endoR-HapII, 10 for endoR-HaeIII, 3 for endoR-HaeII, to only a single site for endoR-HindII. In contrast to M13, fd and f1, the ZJ/2 DNA molecule was not cleaved by the endoR-HindII endonuclease. No cleavage site on either phage DNA was detected for the endonucleases endoR-Hsu, endoR-EcoRI and endoR-Sma. When compared with M13 DNA, several differences were noted in the number and size of cleavage products obtained with DNA of phage fd, f1, and ZJ/2. From the results of these analyses, using the M13 enzyme cleavage maps as a reference, the endoR-HapII, endoR-HaeII, endoR-HaeIII, endoR-HindII and endoR-AluI maps of phage fd, f1, and ZJ/2 could be constructed. As is expected for very closely related phages, the enzyme cleavage patterns exhibit a high degree of homology. Phage f1 and ZJ/2 are most related since an identical pattern was obtained with seven different restriction endonucleases. Evidence is provided also that f1 is more similar to M13 than to fd. Furthermore, characteristic differences exist within the endoR-Hinf enzyme cleavage pattern of all the four phages tested. Digestion of phage DNA with this enzyme, therefore, provides a new and sensitive method of distinguishing these closely related filamentous coliphages .
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B., Frey L., Delius H. Isolation and characterization of gene 5 protein of filamentous bacterial viruses. J Mol Biol. 1972 Jul 14;68(1):139–152. doi: 10.1016/0022-2836(72)90269-0. [DOI] [PubMed] [Google Scholar]
- DeFilippes F. M. A new method for isolation of a restriction enzyme from Hemophilus parainfluenzae. Biochem Biophys Res Commun. 1974 Jun 4;58(3):586–596. doi: 10.1016/s0006-291x(74)80460-2. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Marvin D. A. Altered coding in single stranded DNA viruses? Nature. 1969 Feb 22;221(5182):769–770. doi: 10.1038/221769a0. [DOI] [PubMed] [Google Scholar]
- Edens L., Konings R. N., Schoenmakers J. G. Physical mapping of the central terminator for transcription on the bacteriophage M13 genome. Nucleic Acids Res. 1975 Oct;2(10):1811–1820. doi: 10.1093/nar/2.10.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea V., Vovis G. F., Zinder N. D. Genetic studies with heteroduplex DNA of bacteriophage fl. Asymmetric segregation, base correction and implications for the mechanism of genetic recombination. J Mol Biol. 1975 Aug 15;96(3):495–509. doi: 10.1016/0022-2836(75)90175-8. [DOI] [PubMed] [Google Scholar]
- Garfin D. E., Goodman H. M. Nucleotide sequences at the cleavage sites of two restriction endonucleases from Hemophilus parainfluenzae. Biochem Biophys Res Commun. 1974 Jul 10;59(1):108–116. doi: 10.1016/s0006-291x(74)80181-6. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Evolution of phi chi 174. II. A cleavage map of the G4 phage genome and comparison with the cleavage map of phi chi 174. Virology. 1975 Feb;63(2):320–325. doi: 10.1016/0042-6822(75)90306-2. [DOI] [PubMed] [Google Scholar]
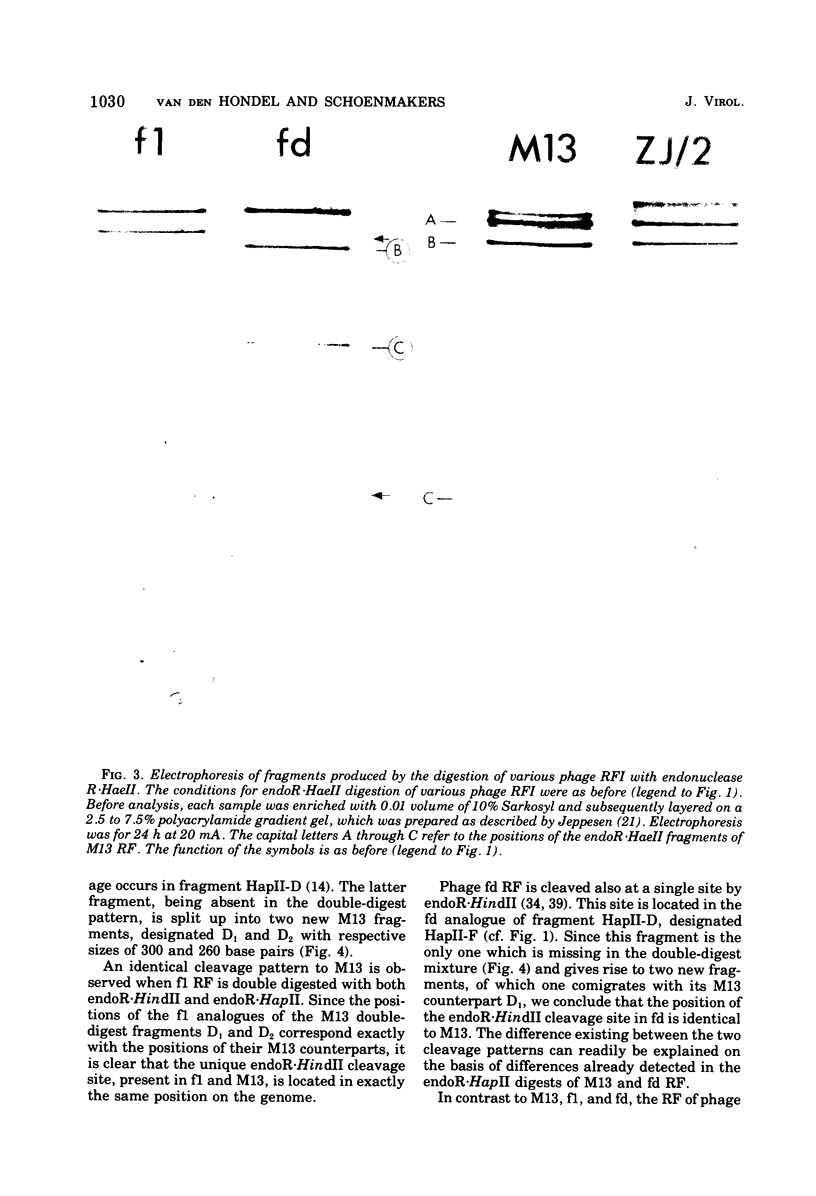
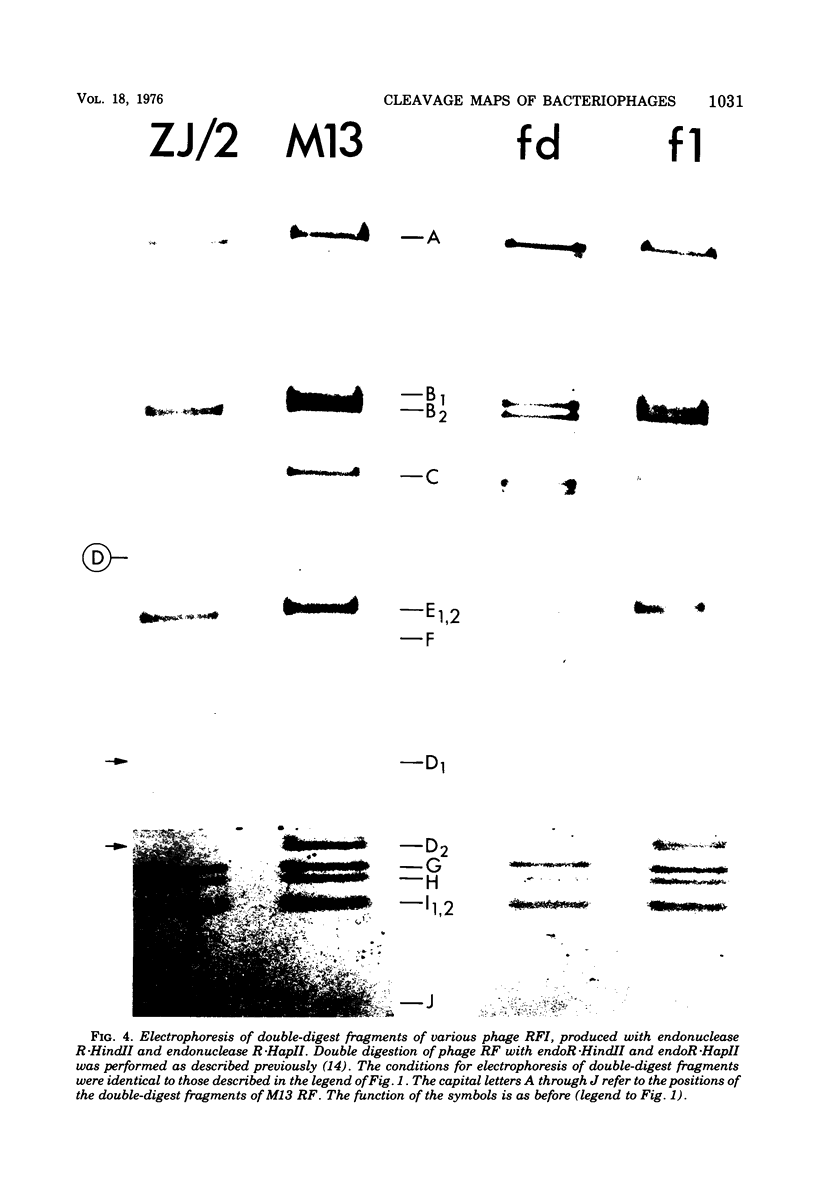
- Griffith J., Kornberg A. Mini M13 bacteriophage: circular fragments of M13 DNA are replicated and packaged during normal infections. Virology. 1974 May;59(1):139–152. doi: 10.1016/0042-6822(74)90211-6. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Fragment maps of phiX-174 replicative DNA produced by restriction enzymes from haemophilus aphirophilus and haemophilus influenzae H-I. J Virol. 1974 Nov;14(5):1142–1151. doi: 10.1128/jvi.14.5.1142-1151.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T. J., Pratt D. The proteins of bacteriophage M13. Proc Natl Acad Sci U S A. 1969 Mar;62(3):800–807. doi: 10.1073/pnas.62.3.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondel C. A., Konings R. N., Schoenmakers J. G. Regulation of gene activity in bacteriophage M13 DNA: Coupled transcription and translation of purified genes and gene-fragments. Virology. 1975 Oct;67(2):487–497. doi: 10.1016/0042-6822(75)90449-3. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Enea V., Zinder N. D. Cleavage map of bacteriophage f1: location of the Escherichia coli B-specific modification sites. J Mol Biol. 1975 Jun 25;95(2):147–165. doi: 10.1016/0022-2836(75)90388-5. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Zinder N. D. Effect of deoxyribonucleic acid length on the adenosine triphosphatase activity of Escherichia coli restriction endonuclease B. J Biol Chem. 1974 Jan 25;249(2):543–552. [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
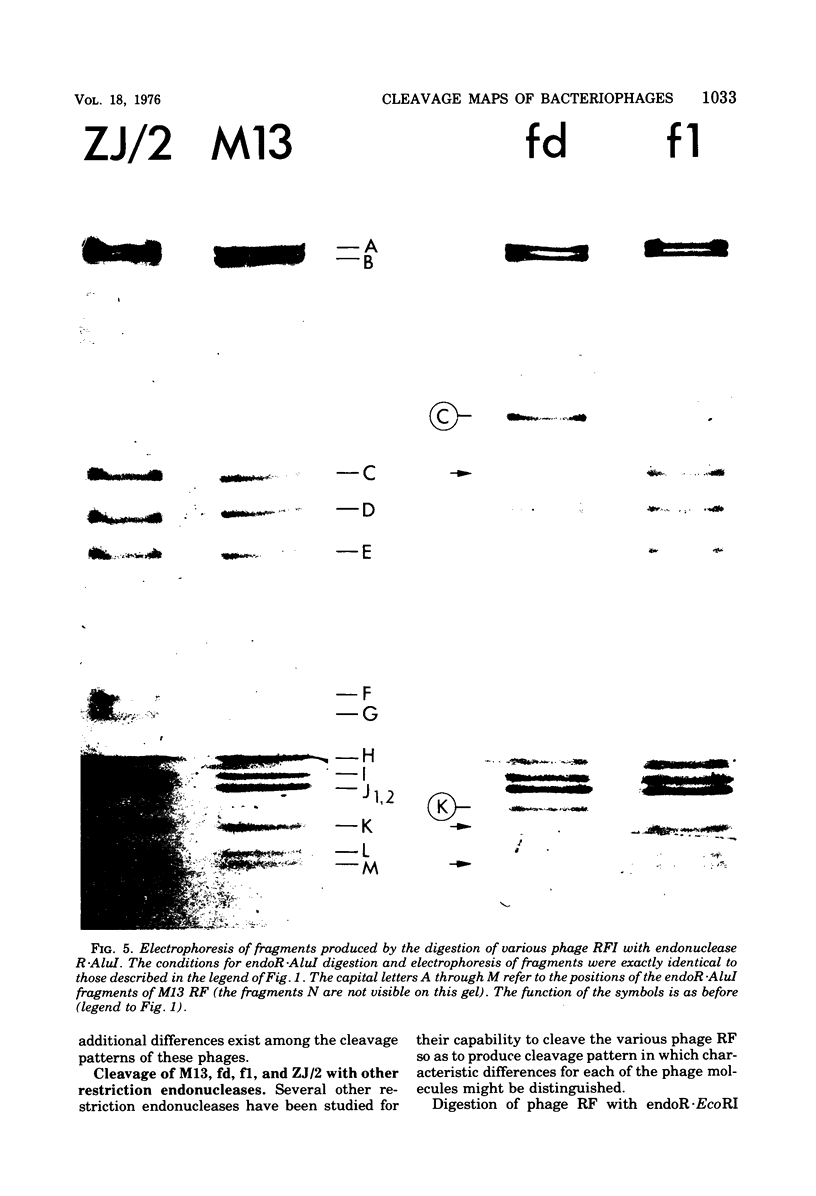
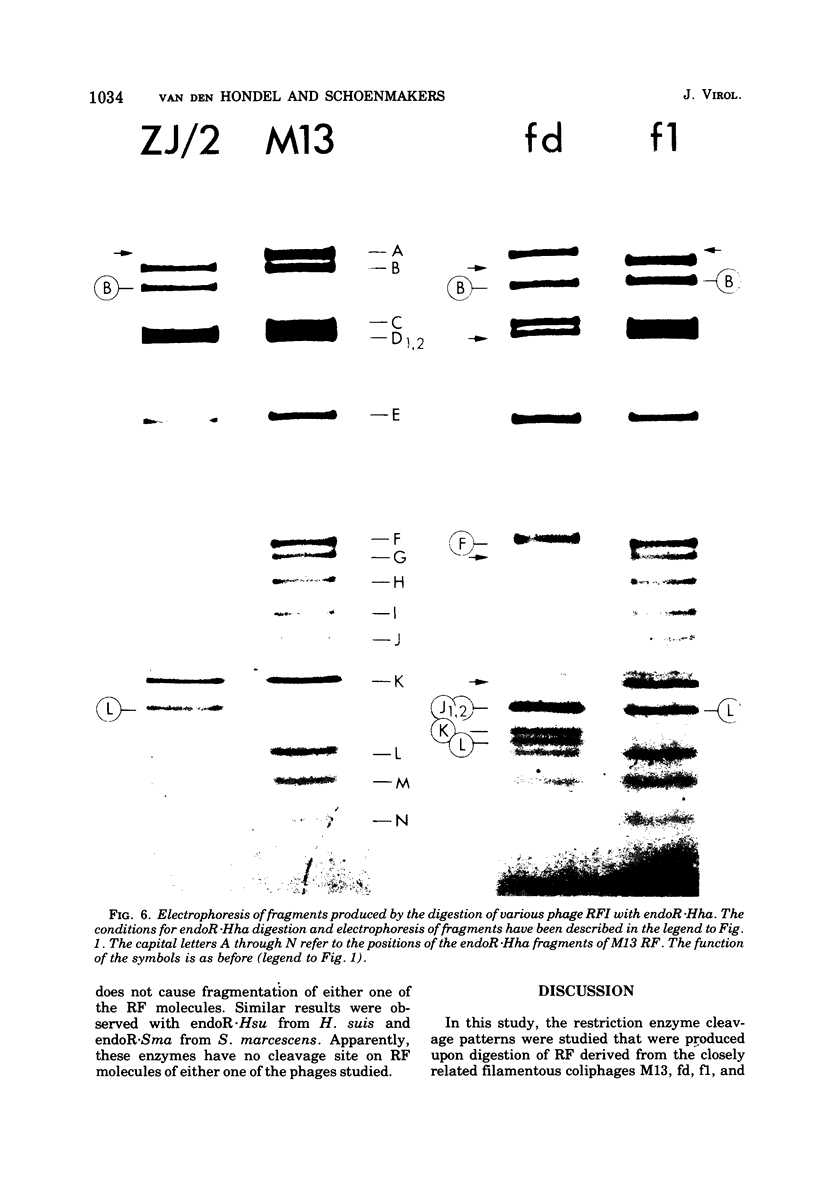
- Jeppesen P. G. A method for separating DNA fragments by electrophoresis in polyacrylamide concentration gradient slab gels. Anal Biochem. 1974 Mar;58(1):195–207. doi: 10.1016/0003-2697(74)90458-8. [DOI] [PubMed] [Google Scholar]
- Konings R. N., Hulsebos T., Van den Hondel C. A. Identification and characterization of the in vitro synthesized gene products of bacteriophage M13. J Virol. 1975 Mar;15(3):570–584. doi: 10.1128/jvi.15.3.570-584.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings R. N., Schoenmakers J. G. Bacteriophage M13 DNA-directed in vitro synthesis of gene 5 protein. Mol Biol Rep. 1974 Feb;1(5):251–256. doi: 10.1007/BF00417579. [DOI] [PubMed] [Google Scholar]
- Konings R. N. Synthesis of phage M13 specific proteins in a DNA-dependent cell-free system. FEBS Lett. 1973 Sep 1;35(1):155–160. doi: 10.1016/0014-5793(73)80600-3. [DOI] [PubMed] [Google Scholar]
- Lin N. S., Pratt D. Bacteriophage M 13 gene 2 protein: increasing its yield in infected cells, and identification and localization. Virology. 1974 Oct;61(2):334–342. doi: 10.1016/0042-6822(74)90271-2. [DOI] [PubMed] [Google Scholar]
- Linn S., Lautenberger J. A., Eskin B., Lackey D. Host-controlled restriction and modification enzymes of Escherichia coli B. Fed Proc. 1974 May;33(5):1128–1134. [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- Marvin D. A., Hohn B. Filamentous bacterial viruses. Bacteriol Rev. 1969 Jun;33(2):172–209. doi: 10.1128/br.33.2.172-209.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin D. A., Schaller H. The topology of DNA from the small filamentous bacteriophage fd. J Mol Biol. 1966 Jan;15(1):1–7. doi: 10.1016/s0022-2836(66)80204-8. [DOI] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Oey J. L., Knippers R. Properties of the isolated gene 5 protein of bacteriophage fd. J Mol Biol. 1972 Jul 14;68(1):125–138. doi: 10.1016/0022-2836(72)90268-9. [DOI] [PubMed] [Google Scholar]
- Okamoto T., Sugimoto K., Sugisaki H., Takanami M. Studies on bacteriophage fd DNA. II. Localization of RNA initiation sites on the cleavage map of the fd genome. J Mol Biol. 1975 Jun 15;95(1):33–44. doi: 10.1016/0022-2836(75)90333-2. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Studies on bacteriophage fd DNA I. A cleavage map of the fd genome. J Mol Biol. 1975 Jun 15;95(1):21–31. [PubMed] [Google Scholar]
- Sugisaki H., Takanami M. DNA sequence restricted by restriction endonuclease AP from Haemophilus aphirophilus. Nat New Biol. 1973 Dec 5;246(153):138–140. doi: 10.1038/newbio246138a0. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Griffith J., Geider K., Schaller H., Kornberg A. Initiation of deoxyribonucleic acid synthesis. VII. A unique location of the gap in the M13 replicative duplex synthesized in vitro. J Biol Chem. 1974 May 25;249(10):3049–3054. [PubMed] [Google Scholar]
- Takanami M., Kojo H. Cleavage site specificity of an endonuclease prepared from Heamophilus influenzae strain H-I. FEBS Lett. 1973 Feb 1;29(3):267–270. doi: 10.1016/0014-5793(73)80035-3. [DOI] [PubMed] [Google Scholar]
- Tate W. P., Petersen G. B. The pyrimidine oligodeoxyribonucleotides from the DNA molecules of bacteriophages f1, fd, and M13. Virology. 1974 Jan;57(1):77–84. doi: 10.1016/0042-6822(74)90109-3. [DOI] [PubMed] [Google Scholar]
- Van Den Hondel C. A., Schoenmakers J. G. Studies on bacteriophage M13 DNA. 1. A cleavage map of the M13 genome. Eur J Biochem. 1975 May 6;53(2):547–558. doi: 10.1111/j.1432-1033.1975.tb04098.x. [DOI] [PubMed] [Google Scholar]
- Van Den Hondel C. A., Weijers A., Konings R. N., Schoenmakers J. G. Studies on bacteriophage M13 DNA. 2. The gene order of the M13 genome. Eur J Biochem. 1975 May 6;53(2):559–567. doi: 10.1111/j.1432-1033.1975.tb04099.x. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Endonuclease R-EcoRII restriction of bacteriophage f1 DNA in vitro: ordering of genes V and VII, location of an RNA promotor for gene VIII. J Virol. 1975 Sep;16(3):674–684. doi: 10.1128/jvi.16.3.674-684.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Kinetics of methylation of DNA by a restriction endonuclease from Escherichia coli B. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3810–3813. doi: 10.1073/pnas.71.10.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Wright M., Wickner S., Hurwitz J. Conversion of phiX174 and fd single-stranded DNA to replicative forms in extracts of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3233–3237. doi: 10.1073/pnas.69.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., Kornberg A. DNA polymerase 3 star requires ATP to start synthesis on a primed DNA. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3679–3683. doi: 10.1073/pnas.70.12.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman R. L., Dunker A. K., Marvin D. A. Filamentous bacterial viruses. 3. Physical and chemical characterization of the If1 virion. Virology. 1972 Apr;48(1):230–244. doi: 10.1016/0042-6822(72)90130-4. [DOI] [PubMed] [Google Scholar]