Abstract
Hematopoietic tyrosine phosphatase (HePTP) regulates orthogonal MAP kinase signaling cascades by dephosphorylating both ERK and p38. HePTP recognizes a docking site (D-recruitment site, DRS) on its targets using a conserved N-terminal sequence motif (D-motif). Using solution NMR spectroscopy and isothermal titration calorimetry (ITC), we compare, for the first time, the docking interactions of HePTP with ERK2 and p38α. Our results demonstrate that ERK2/HePTP interactions primarily involve the D-motif, while a contiguous region called the kinase specificity motif (KIS) also plays a key role in p38α/HePTP interactions. D-motif/DRS interactions for the two kinases, while similar overall, do show some specific differences.
Keywords: MAP kinase, ERK, p38, tyrosine phosphatase, HePTP, D-recruitment site, TROSY
In higher eukaryotes the pleiotropic MAP kinases (ERK1-5; p38α,β,γ,δ; JNK1-3) phosphorylate specific serine/threonine residues on their target proteins, leading to cellular responses as diverse as proliferation, differentiation, survival and apoptosis.1 MAP kinase signaling cascades are mediated by “docking” interactions of the kinases with their substrates, phosphatases and adapter proteins.2 These docking interactions involve specific structural motifs on the MAP kinases and conserved sequence motifs on their interaction partners. Docking interactions with hematopoietic tyrosine phosphatase (HePTP, a class-I protein tyrosine phosphatase) enables the deactivation of both ERK1/2 and p38α by selective dephosphorylation of their activation loop tyrosines and also helps to sequester the kinases in their resting state in the cytosol.3, 4 These interactions require a region located on the disordered N-terminus of HePTP containing a consensus sequence (R/K)2-3-X2-6-φA-X-φB (φA,B are hydrophobic residues) known as a kinase interaction motif (KIM),5 a DEJL-motif,6 a D-site or a D-motif.7 This sequence motif (we use D-motif here) is recognized by the D-recruitment site (DRS) on MAP kinases.8, 9 The MAPK DRS consists of two distinct sub-sites, one (Φchg) forms electrostatic, and the other (Φhyd), forms hydrophobic interactions with D-motif sequences.10 Crystallographic analyses largely involving short peptide ligands have revealed the versatility of D-motif/DRS interactions2, 8, 9, 11 but attempts to crystallize complexes formed by full-length kinases with regulatory proteins have mostly been unsuccessful, likely due to the inherent flexibility of elements outside the canonical docking regions. Recently we used a combination of SAXS and NMR spectroscopy to study docking interactions involving ERK212 and p38α13 utilizing both peptide ligands as well as full-length HePTP. For p38α in the resting state,13 the NMR spectral perturbations in the presence of either a 17-residue peptide encoding the D-motif (KIM) sequence of HePTP (KIM15-31) or full-length HePTP, revealed engagement of both Φchg and Φhyd sub-sites of the DRS. This contrasts available crystallographic data for p38α in complex with short D-motif peptides that appear to indicate a pronounced engagement only of the Φhyd sub-site.11, 14 Crystal structures of ERK2/D-motif peptide complexes8, 9 exhibit engagement of both DRS sub-sites suggesting possible differences in D-motif/DRS interactions between ERK2 and p38α. However, engagement of the Φchg sub-site was seen in a crystal structure of the complex of p38α with a substrate, full-length MAPKAP kinase 2 (MK2),15 and also in a complex with the MAP kinase binding domain of the dual-specificity phosphatase MKP5.16 Given these contrasting results, we used solution NMR spectroscopy to compare D-motif/DRS interactions involving p38α and ERK2 in their resting states using the same peptide ligand, KIM15-31, and full-length HePTP (called HePTP from hereon). Our results provide for the first time a comparison of the interactions of two MAP kinases with the same D-motif peptide and the same full-length phosphatase. We show that HePTP recognizes ERK2 mainly through the D-motif and p38α through both the D-motif and a contiguous kinase specificity (KIS)17 motif. The catalytic PTP domain of HePTP does not appear to impart any significant structural stability in binding either ERK2 or p38α.
Focusing first on the spectral perturbations (Figure 1) induced on the ERK2 DRS by KIM15-31, we noted an excellent agreement with those expected from the crystal structure (with KIM16-31).9 Several Φchg sub-site residues on Loop16 (Y315, D316 and E320) and on Loop4 (H78, N80) were affected. Unexpectedly, the common-docking (CD) residue (D319 in ERK2), whose sidechain was deemed critical for binding, is minimally perturbed (D316 on p38α is also minimally perturbed). This may be explained by the complex dependence of backbone spectral perturbations in the case of interactions involving sidechains, on the induced conformational changes affecting the residue in question as well as the neighboring ones. In principle, different contributions may cancel each other; hence for isolated perturbations (or lack thereof) the possibility of false negatives has to be accounted for. The fact that both residues flanking the CD Asp are perturbed suggests that this is likely the case here. Similarly, all of the structural elements comprising the Φhyd sub-site (Loop8, Loop11, helices αD and αE) were perturbed. This includes Y126 that contacts the peptide L27’ (φA, peptide residues primed) sidechain, and both L113 and L155, which form the binding pocket for L29’ (φB). We use the original definition9 of the hydrophobic residues based on sequence (contrast Figure 4 of Francis et. al. based on structure13). The sidechains of the perturbed H123 (αE) and Q117 (Loop8) form hydrogen bonds with the peptide backbone in the crystal structure.
FIGURE 1.
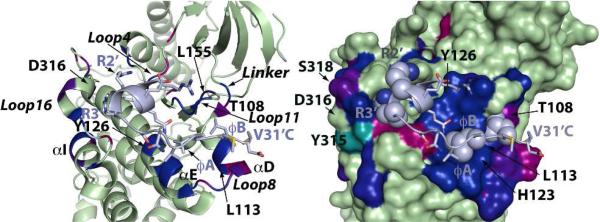
Close-up view of spectral perturbations induced by KIM15-31, plotted using a red (minimum perturbation) to blue (maximum perturbation) gradient (the scale is optimized for maximum visual contrast), on the ribbon (left) and surface (right) representations of an ERK2 mutant bound to KIM16-31m (light blue). Residues with perturbations below the 0.041ppm threshold are colored green. Resonances that disappear upon binding are colored cyan. On the right panel, sidechains belonging to the consensus D-motif sequence: 20R-21R-X5 -27L-X-29L are shown as spheres.
V31’, the terminal residue of KIM16-31 was found to be disordered in the initial crystal structures of the ERK2/KIM complex.9 However, a V31'C mutation on KIM16-31 (KIM16-31m) and a corresponding T116C mutation (at the end of helix αD) on ERK2 led to a high-resolution structure where an intermolecular disulfide bridge was observed between the two newly introduced residues.9 In p38α this same region hosts a well-defined hydrophobic binding pocket that, in co-crystals with D-motif peptides derived from either a substrate MEF2A or an activator MKK3b, docks the sidechain belonging to the φB residue.11 In fact, these structures exhibit an overall register shift of the entire consensus motif, with the φA sidechain occupying the pocket normally recognized by φB in the complexes involving ERK2. However our previous NMR results indicate that this is not the case when p38α binds KIM15-31, with φA and φB recognizing the homologous ERK2 pockets and V31’ entering the cavity occupied by φB in the p38α-peptide co-crystals.13 It should be noted that this particular pocket in ERK2 is partially occluded by the T108 (perturbed) sidechain making it less able to host bulky sidechains from the ligands. A direct comparison of the perturbations induced by KIM15-31 on ERK2 and p38α does in fact show a very similar pattern of perturbations at the Φhyd sub-site (Figure S2). The extensive perturbations seen for Loop8, Loop11, αD and αE hosting the critical residues of the Φhyd sub-site indicates its full engagement for both kinases. We may therefore conclude that, at saturating concentrations, the overall interaction topology of KIM15-31 with the DRS of both kinases is conserved in solution. A closer comparison of the spectral perturbations on a per-residue basis does nevertheless reveal differences (Figure S2). On the ERK2 Φhyd sub-site, KIM15-31 causes the largest changes on helix αE and on Loop11 (the binding pocket hosting the φA i.e. L27’ sidechain), while perturbations on helix αD (hosting the φB i.e. L29’, and the V31’ sidechains) are less prominent; spectral perturbations are more uniform on the p38α Φhyd sub-site. Differences between the two kinases can also be seen at their Φchg sub-sites. For instance, in p38α, E81, located on Loop4, is perturbed while the corresponding ERK2 residue E79 is not (however the flanking H78 and N80 are perturbed). On the other hand KIM15-31 induces more significant changes on Loop16 in ERK2 compared to p38α. The ERK2 residues E312, S318, I322 and A323 are perturbed but the corresponding positions on p38α are not. Also perturbations on helix αI are seen only for ERK2 (V302, E303, Q304). It has been suggested that some of these ERK2 residues (E303, S318) are part of an elaborate ERK2-specific network of hydrogen bonds and electrostatic interactions that are remodeled by ligand binding.9 Thus, our data indicates that while KIM15-31 engages the Φchg sub-site in both proteins, the subsequent reorganization of the electrostatics in this region is distinct in each case. The comparatively reduced spectral perturbations observed for the p38α Φchg sub-site may indicate that this region is less involved in KIM15-31 binding than in the case of ERK2, a possible reason for the difference in affinity as measured using ITC (Table S3).
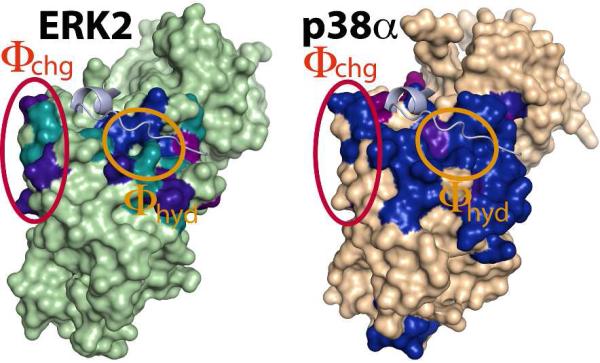
A more substantial difference between the two kinases is observed upon binding of HePTP (Figures 2, S3). ITC measurements indicate that HePTP binds both kinases with higher affinity than KIM15-31 (Table S3). However, HePTP induces a significantly larger number of spectral perturbations only on p38α when compared with KIM15-31, including a larger involvement of the Φchg sub-site, as well as a number of distal residues located on the C-lobe.13 Both NMR and ITC data showed that these latter perturbations were the result of binding the KIS motif on HePTP. In fact p38α binds the KIMKIS peptide with about a 7-fold higher affinity than KIM15-31 (Table S3). In contrast, the binding of HePTP to ERK2 produces a set of perturbations not dissimilar to those resulting from KIM15-31, with only a few additional perturbations observed in areas distal to the DRS. This is consistent with roughly a 3-fold increase in affinity for HePTP compared with KIM15-31 (Table S3). Further, the affinity of the KIMKIS peptide toward ERK2 is similar to KIM15-31 alone (1.8-fold increase, Table S3). These observations indicate that the KIS element is less critical for the ERK2/HePTP interaction than for the p38α/HePTP interaction. This observation is in line with an earlier report that demonstrated that various members of the KIM-PTP family (of which HePTP is a member) use their variable KIS sequences to differentially target MAP kinases.17 With HePTP, the majority of the backbone resonances belonging to the ERK2 DRS move to positions identical to those observed in the presence of KIM15-31, indicating a similar engagement of the D-motif for the two cases. However, a subset of residues located on the Φhyd sub-site, specifically on Loop8, Loop11 and helix αD, experiences smaller overall spectral perturbations in the presence of the full-length phosphatase (Figure S4). In the case of the resonance corresponding to the αD residue T116 (discussed above), the direction of shift during the titration course is opposite of that for KIM15-31. This suggests a different, perhaps weaker contacts with the C-terminus of the D-motif, and more likely of V31’, in the case of full-length HePTP. For p38α, the additional contacts with KIMKIS compared with HePTP that lead to a higher affinity (Table S3) in the former case has been discussed previously.13 In a SAXS study,12 we had shown that the catalytic (PTP) domain of HePTP was delocalized in a region below the ERK2 activation loop. The lack of significant spectral perturbations in this region for ERK2 suggests that the catalytic PTP domain of HePTP does not make appreciable contact with ERK2, similar to the case of p38α demonstrated previously.13
FIGURE 2.
Spectral perturbations induced by HePTP on ERK2 (left) and p38α (right). Color-coding as in Figure 1 (ERK2 threshold - 0.052 ppm; p38α residues with perturbations below the threshold are colored brown). The KIM16-31m peptide (light blue) bound to ERK2 has been modeled onto the p38α surface to aid visualization of the DRS.
We may conclude that D-motif/DRS interactions are essential for binding of HePTP to both p38α and ERK2 in their resting states. However, unlike p38α, where interactions involving the KIS motif of HePTP significantly enhance binding, for ERK2 these interactions are less important. The catalytic PTP domain of HePTP contributes to the binding of neither p38α nor ERK2. D-motif/DRS recognition modes for ERK2 and p38α are topologically similar overall and consistent with that depicted by the crystal structure of KIM16-31m bound to ERK29. The specific variations of perturbations for the two kinases likely result from different contributions of the individual portions of the DRS and its constituent sub-sites in determining the details of their respective binding modes. Interestingly, no other distal perturbations were observed on the N-lobes of ERK2 or p38α upon binding either KIM15-31 or HePTP. A number of crystallographic studies on both these MAP kinases9, 14 showed that the binding of various ligands to the DRS induces a number of distal structural changes that, triggered by conformational rearrangements on Loop4 and Loop16, and through the mediation of helix αC, leads to changes in the Gly-rich loop, the N-terminus and the activation loop. The activation loop, in particular, was shown to adopt a new well-defined conformation in the case of ERK2,9 and became more flexible for p38α.14 While resonances belonging to the activation loop of ERK2 in its resting state are currently unassigned, these assignments are complete for p38α. Resonances corresponding to all the other areas mentioned above have been largely identified for both kinases.10, 13, 18, 19 The lack of spectral perturbations indicates the absence of significant conformational differences between the free and bound states in solution in these regions of either kinase both in the presence of the short D-motif peptide or full-length HePTP. Clearly, more work is required to establish if the discrepancy between the solution and crystallographic studies is due to a curiously consistent interaction in crystallo, or the existence of bound-like states in the solution ensemble.
Supplementary Material
Acknowledgments
Funding Sources
Supported by grants: NIH: GM084278 (to R.G.), GM059802, CA167505 (to K.N.D.), 8G12MD007603 (partial support of the CCNY NMR facility); American Cancer Society: RSG-08-067-01-LIB (to R.P.) and the Welch Foundation F-1390 (to K.N.D.).
ABBREVIATIONS
- MAP kinase
mitogen activated protein kinase
- ERK
extracellular signal-regulated kinase
- DRS
D-recruitment site
- HePTP
hematopoietic protein tyrosine phosphatase
- KIM
kinase interaction motif
- KIS
kinase specificity motif
- KIM15-31
encodes HePTP residues 15-31
- KIM16-31
KIM15-31 missing V15
- KIM16-31m
KIM16-31 with a V31'C mutation
- KIMKIS
encodes HePTP residues 15-56 with a C42'S mutation
Footnotes
Supporting Information
Supporting methods, NMR and ITC data. This material is available free of charge via the Internet at http://pubs.acs.org.
Authors declare no competing financial interests.
REFERENCES
- 1.Dhillon AS, Hagan S, Rath O, Kolch W. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 2.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Chem. Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena M, Williams S, Gilman J, Mustelin T. J. Biol. Chem. 1998;273:15340–15344. doi: 10.1074/jbc.273.25.15340. [DOI] [PubMed] [Google Scholar]
- 4.Saxena M, Williams S, Brockdorff J, Gilman J, Mustelin T. J. Biol. Chem. 1999;274:11693–11700. doi: 10.1074/jbc.274.17.11693. [DOI] [PubMed] [Google Scholar]
- 5.Pulido R, Zuniga A, Ullrich A. EMBO J. 1998;17:7337–7350. doi: 10.1093/emboj/17.24.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell L. Biochem. Soc. Trans. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Sun JP, Zhou B, Zhang ZY. Proc. Natl. Acad. Sci. USA. 2006;103:5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Structure. 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Piserchio A, Warthaka M, Devkota AK, Kaoud TS, Lee S, Abramczyk O, Ren P, Dalby KN, Ghose R. Biochemistry. 2011;50:3660–3672. doi: 10.1021/bi2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Mol. Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 12.Francis DM, Rozycki B, Tortajada A, Hummer G, Peti W, Page R. J. Am. Chem. Soc. 2011;133:17138–17141. doi: 10.1021/ja2075136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis DM, Rozycki B, Koveal D, Hummer G, Page R, Peti W. Nature Chem. Biol. 2011;7:916–924. doi: 10.1038/nchembio.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akella R, Min X, Wu Q, Gardner KH, Goldsmith EJ. Structure. 2010;18:1571–1578. doi: 10.1016/j.str.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 15.ter Haar E, Prabhakar P, Liu X, Lepre C. J. Biol. Chem. 2007;282:9733–9739. doi: 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YY, Wu JW, Wang ZX. Science Signal. 2011;4:ra88. doi: 10.1126/scisignal.2002241. [DOI] [PubMed] [Google Scholar]
- 17.Munoz JJ, Tarrega C, Blanco-Aparicio C, Pulido R. Biochem. J. 2003;372:193–201. doi: 10.1042/BJ20021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piserchio A, Dalby KN, Ghose R. Meth. Mol. Biol. 2012;831:359–368. doi: 10.1007/978-1-61779-480-3_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogtherr M, Saxena K, Grimme S, Betz M, Schieborr U, Pescatore B, Langer T, Schwalbe H. J. Biomol. NMR. 2005;32:175. doi: 10.1007/s10858-005-2449-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.