Abstract
The past 25 years have seen significant advances in understanding the diversity and functions of glycoprotein glycans in Drosophila melanogaster. Genetic screens have captured mutations that reveal important biological activities modulated by glycans, including protein folding and trafficking, as well as cell signaling, tissue morphogenesis, fertility, and viability. Many of these glycan functions have parallels in vertebrate development and disease, providing increasing opportunities to dissect pathologic mechanisms using Drosophila genetics. Advances in the sensitivity of structural analytic techniques have allowed the glycan profiles of wild-type and mutant tissues to be assessed, revealing novel glycan structures that may be functionally analogous to vertebrate glycans. This review describes a selected set of recent advances in understanding the functions of N-linked and O-linked (non-glycosaminoglycan) glycoprotein glycans in Drosophila with emphasis on their relatedness to vertebrate organisms.
Keywords: Drosophila, glycosylation, N-linked, O-linked
Introduction and Scope
Given the small size of the organism, it might be expected that Drosophila melanogaster would be a less than optimal system in which to pursue structural and functional glycomics. However, the opportunity to address glycan function through genetic manipulation is attracting a growing number of glycobiologists to the field, in addition to mainstream Drosophilists that have come upon glycobiology through unbiased genetic screens or the discovery of unexpected functional connections. Gains in analytic sensitivity realized over the past decade, primarily driven by mass spectrometry, have also enhanced the accessibility of glycan characterization following experimental perturbation or genetic manipulation for many small organisms. Thus, Drosophila glycobiology has begun to address major questions of broad relevance for all animals and, importantly, for human diseases. In this review, we focus on a selected handful of such recent advances in understanding the functional impact of N-linked and O-linked glycoprotein glycans in Drosophila and point out similarities and differences associated with analogous vertebrate structures. We apologize to our fellow Drosophila glycobiologists whose significant results we are unable to discuss due to the constraints imposed by the format of this review. For example, early appreciation for the essential role of glycosaminoglycan-based modulation of morphogen signaling arose from Drosophila genetic screens and many fine reviews cover this area very well [1,2]. Likewise, appreciation for the participation of glycosphingolipids in cell signaling and cell-cell interactions is growing rapidly and, despite the significant structural differences between Drosophila and vertebrate glycosphingolipids, functional and pathophysiological corollaries have emerged [3–12]. Drosophila glycosphingolipid glycobiology, Drosophila GAG functions, as well as nucleocytoplasmic glycosylation [13,14] are beyond the scope of this review.
The Glycobiology of Drosophila N-linked glycans
Mass spectrometry-based techniques have produced total N-linked glycan profiles for glycoproteins harvested from Drosophila embryos [15–17]. Unlike vertebrates, Drosophila N-linked glycan profiles display an overwhelming abundance of high and paucimannose structures, and a relatively low amount of hybrid and complex glycans [18]. As is also the case in vertebrates, N-linked glycosylation in Drosophila is initiated by adding a conserved 14-mer oligosaccharide (Glc3Man9GlcNAc2) en bloc onto NXS/T/C motifs within newly synthesized polypeptides. Subsequent trimming and processing of this oligosaccharide in the ER/Golgi drives protein maturation and trafficking [19,20]. Drosophila has provided a platform to study protein maturation associated with some human diseases. For example, N-linked glycosylation of rhodopsin is critical for its proper trafficking in photoreceptor neurons [21,22]. Mutations that remove the N-linked glycosylation site of human rhodopsin cause autosomal dominant retinitis pigmentosa, a retinal degenerative disorder [23]. The N-linked glycan of Drosophila rhodopsin (Rh1) is essential for its exit from the ER [24] but is extensively trimmed or completely removed during its maturation and transport to the light sensing organelle, the rhabdomere [25,26]. Although much of the deglycosylation pathway for Rh1 is unknown, the process has recently been shown to be regulated by a phosphatase (Drosophila Metallophosphoesterase, dMPPE) [27]. In dmppe mutants Rh1 is incompletely deglycosylated, which renders it more sensitive to endocytic degradation, thereby leading to morphological and functional defects in photoreceptors in aged animal. The relevant target of dMPPE is phosphorylated α-mannosidase II (α-Man-II); dephosphorylated α-Man-II does not act on rhodopsin, thereby blocking full deglycosylation and transport of Rh1 to the rhabdomere. While this extreme processing may be a special case, in which stringent limitations on glycan size are imposed by the constraints of packing protein into a specific subcellular compartment, it remains to be determined how many other glycoproteins are similarly processed and how broadly phosphorylaton/dephosphorylation cycles influence glycoprotein glycan processing.
The glycans on many glycoproteins facilitate protein folding in the ER through interactions with lectinic chaperones and also serve as markers that divert misfolded glycoproteins into the ER-associated degradation (ERAD) pathway. Cytoplasmic peptide:N-glycanase (PNGase) participates in ERAD by catalyzing the cleavage of N-linked glycans from misfolded glycoproteins that have been extruded from the ER. Glycan removal facilitates degradation of the polypeptide by the ubiquitin-proteasome system [28–30]. PNGase is conserved among eukaryotes from yeast to human and it has been demonstrated that its deglycosylation activity is essential for degradation of most target glycoproteins [31,32]. Recently, a Drosophila ortholog of cytoplasmic PNGase, PNGase-like (Pngl), was shown to be enzymatically inactive while still retaining its carbohydrate binding activity [33]. The loss of deglycosylation activity of Pngl is due to the lack of the second CXXC motif, which binds a zinc atom essential for enzymatic activity and is also lacking in other dipteran insects. Drosophila pngl mutants display severe developmental defects, reduced viability, and adult sterility. These phenotypes were fully rescued by transgenic expression of wild-type Pngl. However, a mutant form of Pngl, in which a key Cys residue of the catalytic triad was converted to Ala, could not rescue the developmental defect. This finding indicates that a biochemical activity other than deglycosylation, which is yet to be determined, is important for at least some of the biological functions of Pngl.
Once successfully folded and transported along the secretory pathway, the most abundant N-linked glycan structures acquired by Drosophila glycoproteins are paucimannosidic. The abundance of these highly trimmed glycans is attributable to the activity of a processing β-N-acetylglucosaminidase called fused lobes (fdl). The Fdl enzyme specifically cleaves off the terminal GlcNAc residue added by GlcNAcT1 from the branch point intermediate structure, GlcNAcMan3GlcNAc2, thereby blocking further elongation of the α3 arm or elaboration of additional branches [34,35]. The “fused lobes” name comes from the observation that fdl mutants exhibit morphological changes in the adult brain, where the bilateral mushroom body β-lobes are found aberrantly fused at the midline [36]. The fused lobe phenotype was also observed in Drosophila mgat1 (GnT1) mutants, which lack the enzyme responsible for transferring GlcNAc onto Man5GlcNAc2, thereby initiating the formation of hybrid and complex glycans [37]. Drosophila mgat1 null mutants also exhibited locomotor defects and reduced life span, demonstrating the biological impact of altered N-linked glycan complexity for Drosophila central nervous system development and function. More recently, it was reported that tissue-specific knockdown of Drosophila mgat1 in the nervous system decreased locomotor activity and life span [38]. Furthermore, the reduced life span associated with Drosophila mgat1 null mutants was not just rescued by neuron-specific expression of Mgat1, even better, their life span was increased significantly compared to control. While these important findings clearly demonstrated that Mgat1-dependent glycan processing in the nervous system contributes to Drosophila longevity, it still remains to be understood how one mutation that decreases glycan complexity (Mgat1/GnT1) yields phenotypes so similar to another mutation that increases glycan complexity (Fdl).
Beneath the large shadow cast by the highly abundant paucimanose glycans, in-depth mass spectrometric analysis of the minor glycans in Drosophila tissues has revealed the presence of di-, mono-, and non-fucosylated hybrid/complex N-linked glycans with Gal or GalNAc extenstions, forming LacNAc or LacdiNAc termini, respectively [15–17]. Terminal LacNAc groups are found to be capped by acidic sugars, either sialic acid or glucuronic acid, but similarly capped LacdiNAc termini have not yet been detected [39,40]. The identity of the Drosophila enzyme responsible for LacNAc formation on N-linked glycans is still unresolved. Three orthologs of mammalian β4GalT-1 have been annotated in the Drosophila genome and their enzymatic properties have been investigated [41–44]. Of these three candidates, the enzyme with the least similarity to mammalian β4GalT-1 is more closely related to mammalian β4GalT-7, which transfers Gal to Xyl in the production of the glycosaminoglycan linker tetrasaccharide [41]. The other two genes appear to encode invertebrate β4GalNAcTs (β4GalNAcTA and B) rather than β4GalTs since they exhibit in vitro donor preference for UDP-GalNAc over UDP-Gal [44,45]. Based on the sequence differences between invertebrate β4GalNAcTs and mammalian β4GalTs and the crystal structure of bovine β4GalT-1, it has been proposed that a single amino acid mutation at the nucleotide sugar binding site (isoleucine/leucine in invertebrate β4GalNAcTs to tyrosine in mammalian β4GalTs) occurred over 500 million years ago, shifting the ancestral specificity from UDP-GalNAc to UDP-Gal. Indeed, switching the corresponding isoleucine residue of Drosophila β4GalNAcTA to tyrosine dramatically changed the donor preference of the enzyme [46]. Nevertheless, these observations do not exclude the possibility that invertebrate β4GalNAcTs transfer Gal onto GlcNAc in vivo. Such a low efficiency enzyme activity might partially explain why Drosophila N-linked glycan profiles contain much less LacNAc-containing glycans than those of mammalian species. The relative paucity of LacdiNAc terminated N-linked glycans may reflect differential compartmentalization of enzyme, acceptor glycoprotein, and UDP-GalNAc/UDP-Gal. Consistent with this proposal, β4GalNAcTB appears to be primarily responsible for generating LacdiNAc groups on Drosophila glycosphingolipids, but requires an escort protein (GABPI) to pilot it towards its functionally appropriate Golgi localization [47].
Unlike the putative Drosophila galactosyltransferase activities, little ambiguity surrounds the identity of the sialyltransferase that caps hybrid and complex glycans in Drosophila. A single sialyltransferase gene (DSiaT) has been identified in Drosophila [48]. Consistent with glycomic characterization of the sialylated glycans detected in Drosophila embryos, DSiaT is most homologous to the vertebrate ST6Gal1 enzyme [15]. DSiaT expression is restricted to a subset of central nervous system neurons where its activity is proposed to modulate neuronal excitability. Therefore, DSiaT mutants display various abnormalities including reduced life span, locomotor defects, temperature-sensitive paralysis, and altered neuromuscular junction morphology [49]. Interestingly, in vitro characterization of DSiaT enzyme activity indicated that LacdiNAc is a better acceptor than LacNAc, but sialylated LacdiNAc N-linked glycans have not yet been detected in Drosophila tissues [15,16,48]. LacNAc groups on N-linked glycans are also modified by addition of GlcA [16]. However, the relevant biosynthetic machinery and biological function(s) of glycoprotein glucuronylation are yet to be identified.
N-linked glycan processing occurs in a stepwise fashion, dictated by the topographic distribution of processing enzymes across the Golgi apparatus. Many longstanding questions remain regarding the trafficking and regulation of glycan processing enzymes in this organelle. Unlike the classic depictions of stacked cisternae that are characteristic of the Golgi found in most vertebrate cells, the Golgi of Drosophila cells consists of discrete puncta distributed throughout the cytoplasm, implying the existence of robust intraorganellar targeting mechanisms [50,51]. Cellular and genetic analysis has demonstrated the existence of discrete trafficking pathways that independently support glycoprotein or glycosaminoglycan processing through the Drosophila Golgi apparatus [52]. Recently, RNAi and classic mutagenesis screens have taken advantage of the tissue specific expression of a unique difucosylated N-linked glycan to characterize genes that regulate the glycosylation machinery. These approaches have revealed the participation of specific RNA binding proteins in regulating the expression of glycosyltransferases [53] and identified a role for protein phosphorylation in Golgi trafficking [54]. Similarly, RNAi screens targeting a broad range of protein kinases in vertebrate cells have also implicated phosphorylation as a central regulator of Golgi trafficking during interphase, in addition to the previously identified importance of cell cycle kinases for redistribution of the Golgi during mitosis [55,56]. Further characterization of Golgi-directed kinases and their substrates will likely provide new mechanistic links between cell signaling and glycomic plasticity for all classes of glycans.
The Glycobiology of Drosophila O-linked glycans
While the first characterizations of Drosophila N-linked glycans began to appear in the literature during the mid-1980s[57,40], it was another decade before lectin histochemistry and biochemical analysis documented the distribution and biosynthesis of O-linked glycoprotein glycosylation in the organism [39,58,59]. The passing of yet another decade, the first of this century, witnessed evidence for the inducible biosynthesis of core 1 disaccharide, the spatial and temporal regulation of polypeptide GalNAcT expression, and novel influences of specific O-glycan structures on cell signaling [60–63]. In particular, the demonstration that modification of the Drosophila Notch receptor protein with O-linked Fuc affects its signaling, and that extension of O-Fuc with GlcNAc modulates Notch’s ability to discriminate between ligands, opened fertile new ground for coupling genetic analysis with functional glycomics [64]. Analogous modifications of vertebrate Notch proteins were identified contemporaneously with the Drosophila work, raising awareness of how little we really understood about the functions of O-linked glycosylation in any organism [65,66]. In the opening years of the second decade of this century, Drosophila O-linked glycoprotein glycobiology continues to draw parallels and contrasts with vertebrate O-glycan functions in developmental and disease contexts.
As in other animals, the Drosophila genome contains multiple polypeptide GalNAcT genes, called PGANTs in Drosophila and ppGalNAcTs in vertebrates, which encode candidate enzymes for the initiation of mucin-type O-linked glycan formation. Currently, 9 PGANT gene products have been shown to have enzymatic activity, while another 4 candidates remain to be validated. As in other organisms, the Drosophila PGANTs fall into two broad families, those that transfer GalNAc to naked peptide and those that prefer to transfer GalNAc to peptides that are already modified by glycan. Systematic analysis has demonstrated that individual PGANTs are expressed in characteristic, but dynamic and frequently overlapping patterns during development [62]. In some Drosophila tissues only one PGANT is expressed or is highly dominant over others, providing unique opportunities to assess the function of mucin-type glycosylation through RNAi or mutagenesis strategies. These approaches have demonstrated that in various tissue contexts specific PGANTs regulate cell adhesion, modulate extracellular matrix composition, participate in epithelial morphogenesis, or are otherwise essential for viability [67–69]. These phenotypes resonate well with the pathophysiology of human diseases, such as autoimmunity, cancer progression, and congenital heart disease, in which altered mucin type O-glycosylation has been implicated [70–75]. Therefore, the accessibility of the Drosophila system for further mechanistic characterization of phenotypes may yield new insight and new targets for understanding several disease processes.
The O-linked modifications of Notch continue to engage the interest of glycobiologists for their novelty, diversity, and functional significance. In both vertebrates and Drosophila, O-linked Fuc on EGF repeats was the first functional glycan modification of Notch to be described. The loss of O-Fuc was demonstrated genetically and biochemically to decrease Notch signaling, leading to the hypothesis that the O-Fuc moiety contributed to receptor activation. Subsequent work revealed that enzymatically inactive O-fucosyltransferase (OFUT-1 in Drosophila) rescued Notch signaling and reversed intracellular Notch accumulation associated with an enzyme null mutant. These findings led to the proposal that OFUT-1 acts as a chaperone, ensuring the proper folding and/or trafficking of the Notch receptor and that this activity did not require transfer of O-Fuc to Notch [76,77]. Rather, the addition of O-Fuc to Notch primarily serves to provide a substrate for glycan elongation by Fringe, producing GlcNAcβ3Fuc, which establishes the ligand binding specificity of Notch [64]. In vertebrates, the disaccharide generated by Fringe is rapidly elongated by the addition of a Gal and sialic acid (SA) residue, generating a tetrasaccharide that has not been detected in Drosophila. In fact, the major O-Fuc glycan in Drosophila is a branched trisaccharide, GlcNAcβ3(GlcAβ4)Fuc, carrying GlcA rather than SA as its acidic residue [78]. The GlcAT responsible for branching the Fringe product is yet to be identified.
O-Fucosylation of Notch requires GDP-Fuc and is thought to occur in the endoplasmic reticulum, although direct evidence for ER transport of this nucleotide sugar has been lacking. Mutations in two Drosophila genes, now known as efr and gfr, have identified transporters with overlapping influence on Notch signaling. Efr transports GDP-Fuc, as well as UDP-GlcNAc and UDP-Xyl, into the ER, providing what should be an essential pool of GDP-Fuc for O-fucosylation of Notch. Gfr transports GDP-Fuc into the Golgi, where it can serve as donor substrate for fucosylation of N-linked glycans [79]. Loss of Gfr results in significant underfucosylation of N-linked glycoprotein glycans, but also produces mild, temperature sensitive Notch phenotypes [80]. Thus, it is proposed that GDP-Fuc taken up into the Golgi by Gfr may be delivered to the ER by retrograde transport where it makes an important contribution to the total pool of nucleotide sugar required for O-fucosylation of Notch [79,81]. The Drosophila Gfr gene is homologous to a human GDP-Fuc transporter (solute carrier family member 35C1, SLC35C1), which is mutated in a congenital disorder of glycosylation (CDG type IIc, also known as Leukocyte Adhesion Deficiency Type II or LADII) that is characterized by growth retardation, intellectual disability, and immune deficiency [82,83]. While the immune deficiency of this disorder arises primarily from loss of fucosylated glycans that mediate leukocyte trafficking, the developmental phenotypes may reflect altered Notch signaling [81]. In contrast to Gfr, Efr transports GDP-Fuc directly into the ER pool expected to contribute to O-fucosylation of Notch. However, Efr mutants do not display Notch phenotypes, indicating that retrograde transport of GDP-Fuc from the Golgi is sufficient to maintain Notch signaling. In the Efr/Gfr double mutant, Notch phenotypes more severe than those seen in the Gfr single mutant are detected. The clear demonstration of genetic interactions between Gfr and Efr may reveal the existence of feedback mechanisms that regulate the distribution of nucleotide sugars across the secretory pathway or may suggest that a significant portion of Notch O-fucosylation actually occurs in the Golgi. The development of better subcellular markers and sensitive methods to determine the distribution of GDP-Fuc (and other nucleotide sugars) would help to clarify the mechanisms that regulate Notch O-fucosylation.
Ongoing studies of Drosophila and vertebrate Notch signaling continue to reveal the importance of novel O-linked glycan modifications [84]. Molecular characterization of a temperature sensitive mutation, called rumi, that produces Notch phenotypes at the restrictive temperature identified a novel protein with a candidate glycosyltransferase domain [85]. Biochemical characterization of recombinant Rumi revealed that the protein possesses an O-glucosyltransferase activity specific for EGF domains, such as those in the Notch extracellular domain. Addition of O-Glc to Ser/Thr residues by Rumi occurs at a consensus that is distinct from O-Fuc addition and individual Notch EGF repeats can carry both O-Fuc and O-Glc, or either alone [86]. Three rumi candidate homologues were identified in vertebrates, but only one retains enzymatic activity [87]. Knockout of the enzymatically active mouse rumi, called a POGLUT (protein O-glucosyltransferase), results in lethality at embryonic day 9.5 with accompanying defects in neural tube development, somitogenesis, and cardiovascular elaboration [88]. While some aspects of the mouse knockout phenotypes can be attributed to altered Notch signaling, the full range of defects are broader than can be explained by loss of Notch, suggesting additional POGLUT targets in vertebrates. Additional targets for Rumi have not yet been identified in Drosophila. Interestingly, unlike O-Fuc addition, ligand binding to Notch is not affected in rumi mutants but proteolytic cleavage of Notch is impaired, blocking propagation of the intracellular signal normally transmitted by liberation of the Notch cytoplasmic domain [85,88]. Further characterization of Drosphila Rumi and vertebrate POGLUT has revealed that these enzymes possess two activities. In addition to transferring Glc, they also transfer Xyl to Ser/Thr residues [87]. In theory, the addition of O-Xyl provides a substrate for glycosaminoglycan elongation, but it is currently unclear whether O-Xyl addition results in elaboration of a GAG chain on Notch or any other protein substrate, or whether O-Xyl by itself can modulate Notch function. O-Glc residues on Notch EGF repeats in Drosophila are extended by the addition of a Xyl to form the Xylα3Glc disaccharide [89]. In vertebrates, an additional Xyl is added to form the Xylα3Xylα3Glc trisaccharide, which has not been detected in Drosophila [78,90]. While the vertebrate XylT enzymes have been identified, Drosophila homologues have not been pursued and phenotypes associated with loss of the Drosophila or vertebrate enzymes remain to be explored [89].
Further expanding the diversity of O-linked glycans on Notch, recent reports have placed O-GlcNAc on EGF repeats of Drosophila (and mouse) Notch protein [91,92]. While O-linked GlcNAc has been extensively studied as a modification of nuclear and cytoplasmic proteins, these recent findings are the first to place O-GlcNAc on the extracellular domain of a transmembrane protein [93]. Extracellular O-GlcNAc modification is achieved by an enzyme (EOGT) that is distinct from the enzyme responsible for nucleocytoplasmic additon of O-GlcNAc (OGT) and loss of EOGT results in phenotypes that indicate a role for extracellular O-GlcNAc in cell-matrix interactions [94,95].
Are all of the O-linked glycan modifications of Notch now identified? The answer to this question is not clear and its ultimate resolution will require a substantial advance in current glycoproteomic analytic techniques. As mentioned previously, whole embryo glycomic analysis detects the presence of a branched O-Fuc trisaccharide carrying a GlcA residue. This O-Fuc trisaccharide is a major O-glycan of the Drosophila embryo and is, therefore, likely to be found on proteins other than just Notch [78]. Does this glycan even exist in linkage to Notch? An answer to this question will only come from analysis of the glycosylation of Notch protein isolated from developing tissues. Constructs expressed in cell culture are unlikely to reproduce the genetic, epigenetic, and cell-cell interactions that drive endogenous glycosylation of vitally important receptor proteins.
Similar considerations pertain for understanding the full diversity of glycoprotein acceptors that carry other O-glycan modifications. Protein O-mannosylation in Drosophila is carried out by the coordinated activity of two proteins, POMT-1 and POMT-2 (also known as rotated and twisted abdomen, respectively), which modify the Drosophila dystroglycan protein [96–98]. Loss of O-mannosylation in Drosophila results in muscle degeneration which phenocopies the pathophysiology of human congenital muscular dystrophies [99,96]. But unlike O-mannosylation of vertebrate alpha-dystroglycan, Drosophila O-Man has not yet been shown to be elongated in a POMGNT-1 or Large dependent manner [100]. The existence of other O-Man modified proteins in Drosophila and vertebrates is predicted by several lines of evidence but comprehensive glycoproteomic identification of these targets remains to be achieved in either organism [101,102]. Additionally, a second protein O-fucosyltransferase (POFUT-2 in vertebrates) links Fuc to Ser/Thr residues of thrombospondin repeats (TSRs) but not to EGF repeats; a Drosophila homolog (OFUT-2) has been annotated but not characterized [103,104]. In vertebrates, this O-Fuc on TSR repeats can be extended by addition of a Glc residue, generating a Glcβ3Fuc disaccharide [103]. Loss of this glucosyltransferase activity, which has not yet been identified in Drosophila, results in Peters Plus syndrome, characterized by eye defects, short stature, developmental delay, and cleft lip/palate [105]. An essential goal for future glycomic and glycoproteomic analysis of O-linked glycosylation in Drosophila is to establish the endogenous diversity of glycoproteins carrying specific glycan structures. In so doing, glycoproteomic analysis can identify new targets for genetic and glycobiological analysis that may be relevant for understanding the function of analogous glycans in vertebrate systems.
Concluding remarks
More than a century of random mutagenesis screens in Drosophila have pursued the identification of developmentally important genes in an unbiased manner. These approaches have repeatedly opened new frontiers for understanding biological processes that are broadly conserved across animal species. Among these important paradigms, several functional consequences of protein glycosylation have been revealed through both forward and reverse genetic strategies. The contribution of N-linked glycans to protein folding and stability, the impact of altered glycosylation on cell signaling, neural activity, adult longevity and fertility, as well as the connection between well-defined glycans and specific disease pathologies have emerged as glycan functions in Drosophila with clear parallels in vertebrates. Continued application of genetic strategies to identify new genes, new interactions, and new functions related to Drosophila glycans promises to enhance all of glycobiology. But, unlike proteins, glycans are not the translated products of a predictive linear code. Therefore, rapid genetic advances in Drosophila glycobiology will mandate the development of tools for glycan structural analysis that can keep pace with the need to assign biochemical context to new mutants.
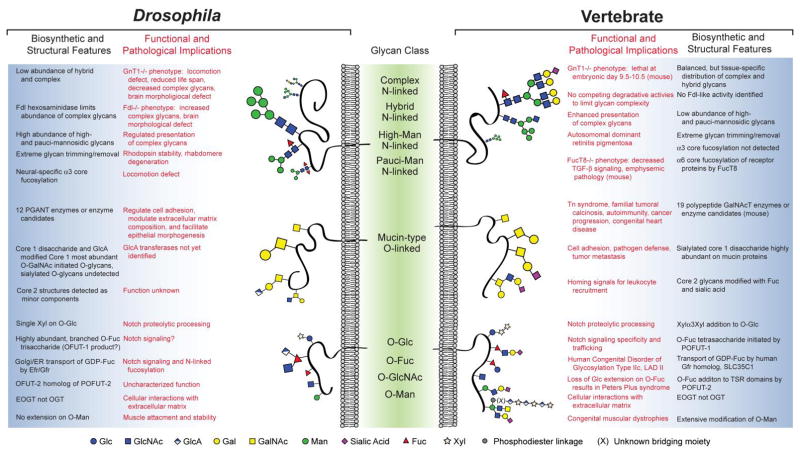
Figure 1. Comparative glycomics of Drosophila and vertebrate glycoprotein glycosylation.
Representations of N-linked and O-linked glycans are scaled proportionally to their relative abundance in Drosophila and vertebrate tissues. Monosaccharide designations are in accordance with the guidelines proposed by the Consortium for Functional Glycomics.
Acknowledgments
The authors acknowledge the support of grant R01-GM072839 (to MT) from the National Institutes of Health/National Institute of General Medicine.
References
- 1.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131(24):6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 2.Selleck SB. Genetic dissection of proteoglycan function in Drosophila and C. elegans. Seminars in cell & developmental biology. 2001;12(2):127–134. doi: 10.1006/scdb.2000.0242. [DOI] [PubMed] [Google Scholar]
- 3.Chen YW, Pedersen JW, Wandall HH, Levery SB, Pizette S, Clausen H, Cohen SM. Glycosphingolipids with extended sugar chain have specialized functions in development and behavior of Drosophila. Dev Biol. 2007;306(2):736–749. doi: 10.1016/j.ydbio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Dahlgaard K, Jung A, Qvortrup K, Clausen H, Kjaerulff O, Wandall HH. Neurofibromatosis-like phenotype in Drosophila caused by lack of glucosylceramide extension. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1115453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller R, Altmann F, Zhou D, Hennet T. The Drosophila melanogaster brainiac protein is a glycolipid-specific beta 1,3N-acetylglucosaminyltransferase. The Journal of biological chemistry. 2002;277(36):32417–32420. doi: 10.1074/jbc.C200381200. [DOI] [PubMed] [Google Scholar]
- 6.Pizette S, Rabouille C, Cohen SM, Therond P. Glycosphingolipids control the extracellular gradient of the Drosophila EGFR ligand Gurken. Development. 2009;136(4):551–561. doi: 10.1242/dev.031104. [DOI] [PubMed] [Google Scholar]
- 7.Schwientek T, Keck B, Levery SB, Jensen MA, Pedersen JW, Wandall HH, Stroud M, Cohen SM, Amado M, Clausen H. The Drosophila gene brainiac encodes a glycosyltransferase putatively involved in glycosphingolipid synthesis. The Journal of biological chemistry. 2002;277(36):32421–32429. doi: 10.1074/jbc.M206213200. [DOI] [PubMed] [Google Scholar]
- 8.Seppo A, Moreland M, Schweingruber H, Tiemeyer M. Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. European journal of biochemistry/FEBS. 2000;267(12):3549–3558. doi: 10.1046/j.1432-1327.2000.01383.x. [DOI] [PubMed] [Google Scholar]
- 9.Seppo A, Tiemeyer M. Function and structure of Drosophila glycans. Glycobiology. 2000;10(8):751–760. doi: 10.1093/glycob/10.8.751. [DOI] [PubMed] [Google Scholar]
- 10.Stolz A, Haines N, Pich A, Irvine KD, Hokke CH, Deelder AM, Gerardy-Schahn R, Wuhrer M, Bakker H. Distinct contributions of beta 4GalNAcTA and beta 4GalNAcTB to Drosophila glycosphingolipid biosynthesis. Glycoconj J. 2008;25(2):167–175. doi: 10.1007/s10719-007-9069-5. [DOI] [PubMed] [Google Scholar]
- 11.Wandall HH, Pedersen JW, Park C, Levery SB, Pizette S, Cohen SM, Schwientek T, Clausen H. Drosophila egghead encodes a beta 1,4-mannosyltransferase predicted to form the immediate precursor glycosphingolipid substrate for brainiac. The Journal of biological chemistry. 2003;278(3):1411–1414. doi: 10.1074/jbc.C200619200. [DOI] [PubMed] [Google Scholar]
- 12.Wandall HH, Pizette S, Pedersen JW, Eichert H, Levery SB, Mandel U, Cohen SM, Clausen H. Egghead and brainiac are essential for glycosphingolipid biosynthesis in vivo. The Journal of biological chemistry. 2005;280(6):4858–4863. doi: 10.1074/jbc.C400571200. [DOI] [PubMed] [Google Scholar]
- 13.Park S, Park SH, Baek JY, Jy YJ, Kim KS, Roth J, Cho JW, Choe KM. Protein O-GlcNAcylation regulates Drosophila growth through the insulin signaling pathway. Cellular and molecular life sciences: CMLS. 2011;68(20):3377–3384. doi: 10.1007/s00018-011-0640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekine O, Love DC, Rubenstein DS, Hanover JA. Blocking O-linked GlcNAc cycling in Drosophila insulin-producing cells perturbs glucose-insulin homeostasis. The Journal of biological chemistry. 2010;285(49):38684–38691. doi: 10.1074/jbc.M110.155192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. The Journal of biological chemistry. 2007;282(12):9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 16.Aoki K, Tiemeyer M. The glycomics of glycan glucuronylation in Drosophila melanogaster. Methods Enzymol. 2010;480:297–321. doi: 10.1016/S0076-6879(10)80014-X. [DOI] [PubMed] [Google Scholar]
- 17.North SJ, Koles K, Hembd C, Morris HR, Dell A, Panin VM, Haslam SM. Glycomic studies of Drosophila melanogaster embryos. Glycoconj J. 2006;23(5–6):345–354. doi: 10.1007/s10719-006-6693-4. [DOI] [PubMed] [Google Scholar]
- 18.ten Hagen KG, Zhang L, Tian E, Zhang Y. Glycobiology on the fly: developmental and mechanistic insights from Drosophila. Glycobiology. 2009;19(2):102–111. doi: 10.1093/glycob/cwn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annual review of biochemistry. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 20.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291(5512):2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 21.Hargrave PA. The amino-terminal tryptic peptide of bovine rhodopsin. A glycopeptide containing two sites of oligosaccharide attachment. Biochimica et biophysica acta. 1977;492(1):83–94. doi: 10.1016/0005-2795(77)90216-1. [DOI] [PubMed] [Google Scholar]
- 22.Murray AR, Fliesler SJ, Al-Ubaidi MR. Rhodopsin: the functional significance of asn-linked glycosylation and other post-translational modifications. Ophthalmic Genet. 2009;30(3):109–120. doi: 10.1080/13816810902962405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dryja TP, Li T. Molecular genetics of retinitis pigmentosa. Hum Mol Genet. 1995;4(Spec No):1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 24.Webel R, Menon I, O’Tousa JE, Colley NJ. Role of asparagine-linked oligosaccharides in rhodopsin maturation and association with its molecular chaperone, NinaA. The Journal of biological chemistry. 2000;275(32):24752–24759. doi: 10.1074/jbc.M002668200. [DOI] [PubMed] [Google Scholar]
- 25.Katanosaka K, Tokunaga F, Kawamura S, Ozaki K. N-linked glycosylation of Drosophila rhodopsin occurs exclusively in the amino-teminal domain and function in rhodopsin maturation. FEBS letters. 1998;424:149–154. doi: 10.1016/s0014-5793(98)00160-4. [DOI] [PubMed] [Google Scholar]
- 26.Satoh A, Tokunaga F, Kawamura S, Ozaki K. In situ inhibition of vesicle transport and protein processing in the dominant negative Rab1 mutant of Drosophila. Journal of cell science. 1997;110 (Pt 23):2943–2953. doi: 10.1242/jcs.110.23.2943. [DOI] [PubMed] [Google Scholar]
- 27.Cao J, Li Y, Xia W, Reddig K, Hu W, Xie W, Li HS, Han J. A Drosophila metallophosphoesterase mediates deglycosylation of rhodopsin. EMBO J. 2011;30(18):3701–3713. doi: 10.1038/emboj.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T, Park H, Hollingsworth NM, Sternglanz R, Lennarz WJ. PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. The Journal of cell biology. 2000;149(5):1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T. Cytoplasmic peptide:N-glycanase and catabolic pathway for free N-glycans in the cytosol. Seminars in cell & developmental biology. 2007;18(6):762–769. doi: 10.1016/j.semcdb.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Spiro RG. Role of N-linked polymannose oligosaccharides in targeting glycoproteins for endoplasmic reticulum-associated degradation. Cellular and molecular life sciences: CMLS. 2004;61(9):1025–1041. doi: 10.1007/s00018-004-4037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altrich-VanLith ML, Ostankovitch M, Polefrone JM, Mosse CA, Shabanowitz J, Hunt DF, Engelhard VH. Processing of a Class I-Restricted Epitope from Tyrosinase Requieres Peptide N-Glycanase and the Cooperative Action of Endoplasmic Reticulum Aminopeptidase 1 and Cytosolic Protease. J Immunol. 2006 doi: 10.4049/jimmunol.177.8.5440. [DOI] [PubMed] [Google Scholar]
- 32.Kario E, Tirosh B, Ploegh HL, Navon A. N-linked glycosylation does not impair proteasomal degradation but affects class I major histocompatibility complex presentation. The Journal of biological chemistry. 2008;283(1):244–254. doi: 10.1074/jbc.M706237200. [DOI] [PubMed] [Google Scholar]
- 33.Funakoshi Y, Negishi Y, Gengen JP, Seino J, Ishii K, Lennarz WJ, Matsuo I, Ito Y, Taniguchi N, Suzuki T. Evidence for an Essential Deglycosylation-Independent Activity of PNGase in Drosophila melanogaster. PLoS One. 2010;5(5):e10545. doi: 10.1371/journal.pone.0010545.g001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonard R, Rendic D, Rabouille C, Wilson IB, Preat T, Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. The Journal of biological chemistry. 2006;281(8):4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- 35.Geisler C, Aumiller JJ, Jarvis DL. A fused lobes gene encodes the processing beta-N-acetylglucosaminidase in Sf9 cells. The Journal of biological chemistry. 2008;283(17):11330–11339. doi: 10.1074/jbc.M710279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boquet I, Hitier R, Dumas M, Chaminade M, Preat T. Central brain postembryonic development in Drosophila: implication of genes expressed at the interhemispheric junction. J Neurobiol. 2000;42(1):33–48. [PubMed] [Google Scholar]
- 37.Sarkar M, Leventis PA, Silvescu CI, Reinhold VN, Schachter H, Boulianne GL. Null mutations in Drosophila N-acetylglucosaminyltransferase I produce defects in locomotion and a reduced life span. The Journal of biological chemistry. 2006;281(18):12776–12785. doi: 10.1074/jbc.M512769200. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar M, Iliadi KG, Leventis PA, Schachter H, Boulianne GL. Neuronal expression of Mgat1 rescues the shortened life span of Drosophila Mgat11 null mutants and increases life span. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(21):9677–9682. doi: 10.1073/pnas.1004431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fredieu JR, Mahowald AP. Glycoconjugate expression during Drosophila embryogenesis. Acta anatomica. 1994;149(2):89–99. doi: 10.1159/000147562. [DOI] [PubMed] [Google Scholar]
- 40.Williams PJ, Wormald MR, Dwek RA, Rademacher TW, Parker GF, Roberts DR. Characterisation of oligosaccharides from Drosophila melanogaster glycoproteins. Biochimica et biophysica acta. 1991;1075(2):146–153. doi: 10.1016/0304-4165(91)90245-c. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura Y, Haines N, Chen J, Okajima T, Furukawa K, Urano T, Stanley P, Irvine KD, Furukawa K. Identification of a Drosophila gene encoding xylosylprotein beta4-galactosyltransferase that is essential for the synthesis of glycosaminoglycans and for morphogenesis. The Journal of biological chemistry. 2002;277(48):46280–46288. doi: 10.1074/jbc.M203873200. [DOI] [PubMed] [Google Scholar]
- 42.Takemae H, Ueda R, Okubo R, Nakato H, Izumi S, Saigo K, Nishihara S. Proteoglycan UDP-galactose:beta-xylose beta 1,4-galactosyltransferase I is essential for viability in Drosophila melanogaster. The Journal of biological chemistry. 2003;278(18):15571–15578. doi: 10.1074/jbc.M301123200. [DOI] [PubMed] [Google Scholar]
- 43.Vadaie N, Hulinsky RS, Jarvis DL. Identification and characterization of a Drosophila melanogaster ortholog of human beta1,4-galactosyltransferase VII. Glycobiology. 2002;12(10):589–597. doi: 10.1093/glycob/cwf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haines N, Irvine KD. Functional analysis of Drosophila beta1,4-N-acetlygalactosaminyltransferases. Glycobiology. 2005;15(4):335–346. doi: 10.1093/glycob/cwi017. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki N, Yoshida H, Fuwa TJ, Kinoshita-Toyoda A, Toyoda H, Hirabayashi Y, Ishida H, Ueda R, Nishihara S. Drosophila beta 1,4-N-acetylgalactosaminyltransferase-A synthesizes the LacdiNAc structures on several glycoproteins and glycosphingolipids. Biochemical and biophysical research communications. 2007;354(2):522–527. doi: 10.1016/j.bbrc.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Ramakrishnan B, Qasba PK. Role of a single amino acid in the evolution of glycans of invertebrates and vertebrates. J Mol Biol. 2007;365(3):570–576. doi: 10.1016/j.jmb.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johswich A, Kraft B, Wuhrer M, Berger M, Deelder AM, Hokke CH, Gerardy-Schahn R, Bakker H. Golgi targeting of Drosophila melanogaster beta4GalNAcTB requires a DHHC protein family-related protein as a pilot. The Journal of cell biology. 2009;184(1):173–183. doi: 10.1083/jcb.200801071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koles K, Irvine KD, Panin VM. Functional characterization of Drosophila sialyltransferase. The Journal of biological chemistry. 2004;279(6):4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- 49.Repnikova E, Koles K, Nakamura M, Pitts J, Li H, Ambavane A, Zoran MJ, Panin VM. Sialyltransferase regulates nervous system function in Drosophila. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30(18):6466–6476. doi: 10.1523/JNEUROSCI.5253-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ripoche J, Link B, Yucel JK, Tokuyasu K, Malhotra V. Location of Golgi membranes with reference to dividing nuclei in syncytial Drosophila embryos. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(5):1878–1882. doi: 10.1073/pnas.91.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto-Hino M, Abe M, Shibano T, Setoguchi Y, Awano W, Ueda R, Okano H, Goto S. Cisterna-specific Localization of Glycosylation-related Proteins to the Golgi Apparatus. Cell structure and function. 2012;37(1):55–63. doi: 10.1247/csf.11037. [DOI] [PubMed] [Google Scholar]
- 52.Yano H, Yamamoto-Hino M, Abe M, Kuwahara R, Haraguchi S, Kusaka I, Awano W, Kinoshita-Toyoda A, Toyoda H, Goto S. Distinct functional units of the Golgi complex in Drosophila cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(38):13467–13472. doi: 10.1073/pnas.0506681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto-Hino M, Kanie Y, Awano W, Aoki-Kinoshita KF, Yano H, Nishihara S, Okano H, Ueda R, Kanie O, Goto S. Identification of genes required for neural-specific glycosylation using functional genomics. PLoS genetics. 2010;6(12):e1001254. doi: 10.1371/journal.pgen.1001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baas S, Sharrow M, Kotu V, Middleton M, Nguyen K, Flanagan-Steet H, Aoki K, Tiemeyer M. Sugar-free frosting, a homolog of SAD kinase, drives neural-specific glycan expression in the Drosophila embryo. Development. 2011;138(3):553–563. doi: 10.1242/dev.055376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri HP. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. The Journal of cell biology. 2010;189(6):997–1011. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94(6):783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 57.Snow PM, Patel NH, Harrelson AL, Goodman CS. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embryos. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1987;7(12):4137–4144. doi: 10.1523/JNEUROSCI.07-12-04137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kramerov AA, Arbatsky NP, Rozovsky YM, Mikhaleva EA, Polesskaya OO, Gvozdev VA, Shibaev VN. Mucin-type glycoprotein from Drosophila melanogaster embryonic cells: characterization of carbohydrate component. FEBS letters. 1996;378(3):213–218. doi: 10.1016/0014-5793(95)01444-6. [DOI] [PubMed] [Google Scholar]
- 59.Kramerov AA, Mikhaleva EA, Rozovsky Ya M, Pochechueva TV, Baikova NA, Arsenjeva EL, Gvozdev VA. Insect mucin-type glycoprotein: immunodetection of the O-glycosylated epitope in Drosophila melanogaster cells and tissues. Insect biochemistry and molecular biology. 1997;27 (6):513–521. doi: 10.1016/s0965-1748(97)00026-x. [DOI] [PubMed] [Google Scholar]
- 60.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nature reviews. Molecular cell biology. 2003;4(10):786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 61.Theopold U, Dorian C, Schmidt O. Changes in glycosylation during Drosophila development. The influence of ecdysone on hemomucin isoforms. Insect biochemistry and molecular biology. 2001;31 (2):189–197. doi: 10.1016/s0965-1748(00)00117-x. [DOI] [PubMed] [Google Scholar]
- 62.Tian E, Ten Hagen KG. Expression of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology. 2006;16(2):83–95. doi: 10.1093/glycob/cwj051. [DOI] [PubMed] [Google Scholar]
- 63.Tian E, Ten Hagen KG. O-linked glycan expression during Drosophila development. Glycobiology. 2007;17(8):820–827. doi: 10.1093/glycob/cwm056. [DOI] [PubMed] [Google Scholar]
- 64.Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. The Journal of biological chemistry. 2003;278(43):42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- 65.Haltiwanger RS, Stanley P. Modulation of receptor signaling by glycosylation: fringe is an O-fucose-beta1,3-N-acetylglucosaminyltransferase. Biochimica et biophysica acta. 2002;1573(3):328–335. doi: 10.1016/s0304-4165(02)00400-2. [DOI] [PubMed] [Google Scholar]
- 66.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406(6794):369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 67.Tian E, Ten Hagen KG. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. The Journal of biological chemistry. 2007;282(1):606–614. doi: 10.1074/jbc.M606268200. [DOI] [PubMed] [Google Scholar]
- 68.Tran DT, Zhang L, Zhang Y, Tian E, Earl LA, Ten Hagen KG. Multiple members of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. The Journal of biological chemistry. 2012;287(8):5243–5252. doi: 10.1074/jbc.M111.306159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Zhang Y, Hagen KG. A mucin-type O-glycosyltransferase modulates cell adhesion during Drosophila development. The Journal of biological chemistry. 2008;283(49):34076–34086. doi: 10.1074/jbc.M804267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guda K, Moinova H, He J, Jamison O, Ravi L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, Lowe JB, Wiesner GL, Parmigiani G, Barnholtz-Sloan J, Dawson DW, Velculescu VE, Kinzler KW, Papadopoulos N, Vogelstein B, Willis J, Gerken TA, Markowitz SD. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(31):12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ichikawa S, Guigonis V, Imel EA, Courouble M, Heissat S, Henley JD, Sorenson AH, Petit B, Lienhardt A, Econs MJ. Novel GALNT3 mutations causing hyperostosis-hyperphosphatemia syndrome result in low intact fibroblast growth factor 23 concentrations. The Journal of clinical endocrinology and metabolism. 2007;92(5):1943–1947. doi: 10.1210/jc.2006–1825. [DOI] [PubMed] [Google Scholar]
- 73.Ju T, Cummings RD. Protein glycosylation: chaperone mutation in Tn syndrome. Nature. 2005;437(7063):1252. doi: 10.1038/4371252a. [DOI] [PubMed] [Google Scholar]
- 74.Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, Arnett DK, Peloso GM, Ordovas JM, Cupples LA. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC medical genetics. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V, Zelikovic I, Raimer S, Metzker A, Richard G, Sprecher E. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nature genetics. 2004;36(6):579–581. doi: 10.1038/ng1358. [DOI] [PubMed] [Google Scholar]
- 76.Okajima T, Reddy B, Matsuda T, Irvine KD. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC biology. 2008;6:1. doi: 10.1186/1741-7007-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasamura T, Ishikawa HO, Sasaki N, Higashi S, Kanai M, Nakao S, Ayukawa T, Aigaki T, Noda K, Miyoshi E, Taniguchi N, Matsuno K. The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of Notch in Drosophila. Development. 2007;134(7):1347–1356. doi: 10.1242/dev.02811. [DOI] [PubMed] [Google Scholar]
- 78.Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. The Journal of biological chemistry. 2008;283(44):30385–30400. doi: 10.1074/jbc.M804925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ishikawa HO, Ayukawa T, Nakayama M, Higashi S, Kamiyama S, Nishihara S, Aoki K, Ishida N, Sanai Y, Matsuno K. Two pathways for importing GDP-fucose into the endoplasmic reticulum lumen function redundantly in the O-fucosylation of Notch in Drosophila. The Journal of biological chemistry. 2010;285(6):4122–4129. doi: 10.1074/jbc.M109.016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishikawa HO, Higashi S, Ayukawa T, Sasamura T, Kitagawa M, Harigaya K, Aoki K, Ishida N, Sanai Y, Matsuno K. Notch deficiency implicated in the pathogenesis of congenital disorder of glycosylation IIc. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(51):18532–18537. doi: 10.1073/pnas.0504115102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamakawa T, Ayukawa T, Matsuno K. Metabolism and transportation pathways of GDP-fucose that are required for the O-fucosylation of Notch. Advances in experimental medicine and biology. 2012;727:37–46. doi: 10.1007/978-1-4614-0899-4_3. [DOI] [PubMed] [Google Scholar]
- 82.Lubke T, Marquardt T, Etzioni A, Hartmann E, von Figura K, Korner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nature genetics. 2001;28(1):73–76. doi: 10.1038/88299. [DOI] [PubMed] [Google Scholar]
- 83.Luhn K, Wild MK, Eckhardt M, Gerardy-Schahn R, Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nature genetics. 2001;28(1):69–72. doi: 10.1038/88289. [DOI] [PubMed] [Google Scholar]
- 84.Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Current topics in developmental biology. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- 85.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132(2):247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Current opinion in structural biology. 2011;21(5):583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takeuchi h, Fernandez-Valdivia RC, Caswell DS, Nita-Lazar A, Rana NA, Garner TP, Weldeghioeghis TK, Macnaughtan MA, Jafar-Nejad H, Haltiwanger RS. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1109696108/-/DCSupplemental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, Jafar-Nejad H. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138(10):1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. The Journal of biological chemistry. 2010;285(3):1582–1586. doi: 10.1074/jbc.C109.065409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sethi MK, Buettner FF, Ashikov A, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. The Journal of biological chemistry. 2012;287(4):2739–2748. doi: 10.1074/jbc.M111.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. The Journal of biological chemistry. 2008;283(51):35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 92.Sakaidani Y, Ichiyanagi N, Saito C, Nomura T, Ito M, Nishio Y, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochemical and biophysical research communications. 2012;419(1):14–19. doi: 10.1016/j.bbrc.2012.01.098. [DOI] [PubMed] [Google Scholar]
- 93.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual review of biochemistry. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. The Journal of biological chemistry. 1997;272(14):9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 95.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nature communications. 2011;2:583. doi: 10.1038/ncomms1591. [DOI] [PubMed] [Google Scholar]
- 96.Haines N, Seabrooke S, Stewart BA. Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Molecular biology of the cell. 2007;18(12):4721–4730. doi: 10.1091/mbc.E07-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lyalin D, Koles K, Roosendaal SD, Repnikova E, Van Wechel L, Panin VM. The twisted gene encodes Drosophila protein O-mannosyltransferase 2 and genetically interacts with the rotated abdomen gene encoding Drosophila protein O-mannosyltransferase 1. Genetics. 2006;172(1):343–353. doi: 10.1534/genetics.105.049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakamura N, Stalnaker SH, Lyalin D, Lavrova O, Wells L, Panin VM. Drosophila Dystroglycan is a target of O-mannosyltransferase activity of two protein O-mannosyltransferases, Rotated Abdomen and Twisted. Glycobiology. 2010;20(3):381–394. doi: 10.1093/glycob/cwp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. Journal of cell science. 2006;119(Pt 2):199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 100.Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 2010;327(5961):88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dino MR, Harroch S, Hockfield S, Matthews RT. Monoclonal antibody Cat-315 detects a glycoform of receptor protein tyrosine phosphatase beta/phosphacan early in CNS development that localizes to extrasynaptic sites prior to synapse formation. Neuroscience. 2006;142(4):1055–1069. doi: 10.1016/j.neuroscience.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 102.Stalnaker SH, Aoki K, Lim JM, Porterfield M, Liu M, Satz JS, Buskirk S, Xiong Y, Zhang P, Campbell KP, Hu H, Live D, Tiemeyer M, Wells L. Glycomic analyses of mouse models of congenital muscular dystrophy. The Journal of biological chemistry. 2011;286(24):21180–21190. doi: 10.1074/jbc.M110.203281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kozma K, Keusch JJ, Hegemann B, Luther KB, Klein D, Hess D, Haltiwanger RS, Hofsteenge J. Identification and characterization of abeta1,3-glucosyltransferase that synthesizes the Glc-beta1,3-Fuc disaccharide on thrombospondin type 1 repeats. The Journal of biological chemistry. 2006;281(48):36742–36751. doi: 10.1074/jbc.M605912200. [DOI] [PubMed] [Google Scholar]
- 104.Luo Y, Koles K, Vorndam W, Haltiwanger RS, Panin VM. Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. The Journal of biological chemistry. 2006;281(14):9393–9399. doi: 10.1074/jbc.M511975200. [DOI] [PubMed] [Google Scholar]
- 105.Hess D, Keusch JJ, Oberstein SA, Hennekam RC, Hofsteenge J. Peters Plus syndrome is a new congenital disorder of glycosylation and involves defective Omicron-glycosylation of thrombospondin type 1 repeats. The Journal of biological chemistry. 2008;283(12):7354–7360. doi: 10.1074/jbc.M710251200. [DOI] [PubMed] [Google Scholar]