Abstract
While initiation of cannabis use is around 40% heritable, not much is known about the underlying genetic etiology. Here, we meta-analysed two genome-wide association studies of initiation of cannabis use with (>10,000 individuals). None of the genetic variants reached genome-wide significance. We also performed a gene-based association test, which also revealed no significant effects of individual genes. Finally, we estimated that only approximately 6.0% of the variation in cannabis initiation is due to common genetic variants. Future genetic studies using larger sample sizes and different methodologies (including sequencing) might provide more insight in the complex genetic etiology of cannabis use.
Keywords: genetics, cannabis, heritability, association
Cannabis is the most widely used illicit drug worldwide, and numerous population based family and twin studies indicate that risk of cannabis use runs in families. A recent meta-analysis of existing twin studies reported heritability estimates of 45% for males and 39% for females for cannabis use initiation (Verweij, Zietsch, Lynskey, et al., 2010). Not much is known about the genetic variants and biological mechanisms underlying this heritability. The few linkage studies that examined cannabis use phenotypes reported non-overlapping linkage peaks that did not meet genome-wide significance (see Agrawal and Lynskey, 2009). Candidate gene association studies for cannabis use have mainly focussed on genes known to be involved in the endogenous cannabinoid system: the cannabinoid receptor 1 gene (CNR1) and the fatty acid amide hydrolase gene (FAAH). But again, while some nominally significant associations were reported, others were unable to replicate these (see Agrawal and Lynskey, 2009). A recent large scale study testing for association of these two and eight other candidate genes with lifetime frequency of cannabis use did not support association with any of the genes (Verweij, Zietsch, Liu, et al., 2011; NB: this study used the same Australian subsample as used in the present study). The lack of replication may point to potential publication bias and false-positive findings in candidate gene association studies and also highlights the limited understanding of the neurobiology of cannabis use. Other, hypothesis-free, approaches are therefore needed to further unravel the genetic etiology of cannabis use.
Recently, Agrawal et al. (2011) performed the first genome-wide association study for cannabis use phenotypes, using a sample of 708 cannabis-dependent cases and 2,346 controls. They did not identify any genetic variants significantly associated with cannabis dependence, potentially because of a lack of statistical power. The present study has two components. First, we meta-analysed results from two genome-wide association analyses of initiation of cannabis use (ever versus never) in order to identify genetic variants underlying cannabis use. With a combined sample size of over 10,000 individuals (from 4,622 independent families) and a substantially higher number of cases, this study provided more power to identify genetic variants of small effect size than the study by Agrawal et al. (2011). Secondly, we used a recently developed method utilising genome-wide SNP data to estimate the overall percentage of variance in initiation of cannabis use that is due to common genetic variants, which will provide insight into the genetic etiology underlying cannabis use.
Data used in this study come from Australian and UK (TwinsUK, http://www.twinsuk.ac.uk; Spector and Williams, 2006) twin registries (see Table 1 for sample details). As part of larger questionnaires, individuals were asked whether they had ever used cannabis. Table 1 shows the prevalence of cannabis use initiation for individuals included in the present study. It should be noted that the prevalence is substantially higher in the Australian sample. Higher levels of cannabis use have generally been reported in Australia than in the UK (UNODC, 2010), but the difference may be larger due to age differences between the two samples and because a subset of the Australian sample has been ascertained for familial alcohol and nicotine use.
Table 1.
Sample descriptives.
| Australia (N=2,538 independent families) |
UK (N=2084 independent families) |
|||||
|---|---|---|---|---|---|---|
| Males | Females | Total | Males | Females | Total | |
| Sample Size | 3292 | 3883 | 7175 | 216 | 2700 | 2916 |
| Age (M±SD) | 45.2 ± 10.8 | 44.5 ± 11.1 | 44.8 ± 11.0 | 58.7 ± 13.5 | 58.7 ± 12.2 | 58.7 ± 12.3 |
| Percentage of individuals that have used cannabis |
58.4% | 47.4% | 52.4% | 18.5% | 11.3% | 11.8% |
Note that the Australian sample includes 403 and the UK sample 224 identical twin pairs
Genotype data were obtained using different Illumina SNP platforms (317K, HumanCNV370-Quadv3, HumanCNV370v1, and Human610-Quad). Standard quality control procedures were applied as outlined previously (Medland, Nyholt, Painter, et al., 2009), including checks for ancestry outliers, Hardy Weinberg Equilibrium, Mendelian errors, call rate, and minor allele frequency. Subsequently, both datasets were imputed separately using MACH (Li, Willer, Ding, et al., 2010) using reference data from the European HapMap I + II samples, (Release 22 Build 36). Only SNPs with an imputation quality score (r2) greater than 0.3 were retained, and SNPs were filtered on allele frequency. In total, ~2,4 million SNPs were available for the association analyses.
For each sample separately, the dosage score at each SNP was tested for association with cannabis use initiation using the family-based association test implemented in Merlin-offline (Chen and Abecasis, 2007), correcting for sex, age, age2, and sex*age effects. We performed a total test of association which employs a linear regression method while explicitly correcting for the relatedness between family members (including MZ twins) within the variance/covariance model. The strongest associations were verified using the Generalized Disequilibrium Test (GDT, Chen, Manichaikul, and Rich, 2009) which is specifically designed for use with dichotomous data. However, unlike Merlin-offline, which can analyse dosage data, GDT can only be used with hard-call genotypes, which are less informative than dosage data thus resulting in a less powerful test. A meta-analysis of the association results from both studies was conducted in METAL (Abecasis, 2009) using the effect sizes of the SNPs along with their standard errors. Next, we conducted a gene-based test (VEGAS, Liu, McRae, Nyholt, et al., 2010) in order to determine if particular genes harbour an excess of associated variants. This test summarises evidence for association on a per gene basis by considering the p-value of all SNPs within the gene, while accounting for linkage disequilibrium and number of SNPs in the gene.
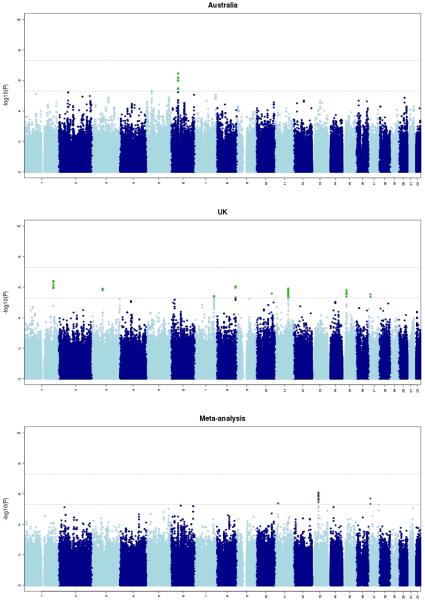
Results from the meta-analysis are summarised in Figure 1 (Manhattan plot), Supplementary Figure 1 (Q-Q plot), and Supplementary Table 1 (strongest associations; note that the results were similar with Merlin-offline and GDT), and the strongest gene-based associations are shown in Supplementary Table 2. We did not identify any SNP or gene that was significantly associated with cannabis use initiation, after correcting for multiple testing. An approximate power calculation (Purcell, Cherny, and Sham, 2003) indicates the combined samples provided 65% and 98% power to detect a genetic variant (with a minor allele frequency of 0.25) with a relative risk of 1.15 and 1.2, respectively. This indicates that individual common genetic variants of this size or greater do not contribute to individual differences in initiation of cannabis use. The power calculation shows that our sample is underpowered to detect common genetic variants of smaller effect sizes, indicating the need for larger sample sizes to enable the detection of these variants.
Figure 1.
Results of the genome-wide association analyses for lifetime cannabis use. The x-axis shows the chromosome numbers and the y-axis the significance of the association signals (i.e. −log10(P) value).
We then estimated the proportion of variance in initiation of cannabis use that could be explained by the aggregate effect of all SNPs using GCTA (Yang, Lee, Goddard, et al., 2011). This estimate is obtained by correlating the genetic similarity (i.e. the identity by state at the SNP level) between individuals (relatedness < 0.025) with their phenotypic similarity. The methodology is explained in more detail in the Supplementary material. We estimated that only 6.0% (SE=10.2, p=0.28) of the variance in cannabis use initiation is due to the aggregated effect of common variants. The relatively large standard error indicates that this estimate is somewhat imprecise, mostly due to the limited power of the sample (N=4612 unrelated individuals), but also by the measurement of the phenotype (binary, as opposed to continuous) and potentially by the heterogeneity of the phenotype between the two samples.
This SNP-based heritability estimate is substantially lower than the heritability estimate of ~40% obtained from twin studies (Verweij, et al., 2010). The discrepancy between twin and SNP-based heritability estimates is larger than for some other phenotypes, such as height and intelligence (Davies, Tenesa, Payton, et al., 2011; Yang, Benyamin, McEvoy, et al., 2010), but similar to for example personality, where the SNP-based estimates for various personality traits are between 4 and 12%, while the twin and family-based heritability estimates are between 30 and 60% (Verweij, Yang, Lahti, et al., 2012; Vinkhuyzen, Pedersen, Yang, et al., 2012). Note that heritability estimates from twin studies include the effects of all causal genetic variants, while the heritability estimated using GCTA includes only the effects of variants that are in linkage disequilibrium with the SNPs included in the analyses. These SNPs do not capture all genetic variants, especially not rare variants or variants with low minor allele frequencies. Although the SNP-based heritability estimate is somewhat imprecise due to limited power, the result raises the possibility that the role of common genetic variants in the heritability of initiation of cannabis use is low, which could help to explain why we were unable to find any genetic association. It may also suggest that non-additive genetic effects (dominance and/or epistasis), interactions with the environment, and/or rare mutations also play a role. Alternatively, twin studies may have overestimated the relative contribution of genetic influences (see Vinkhuyzen, et al., 2012). As with other complex traits, future genetic studies using larger sample sizes and different methodologies (including sequencing) might provide more insight in the complex genetic etiology of cannabis use.
Supplementary Material
Acknowledgements
We greatly thank the twins for their participation. This study was supported in part by grants AA013326, AA014041, AA13320, AA013321, DA23668, and DA12854 from the National Institutes of Health, Bethesda, Md. We would also like to thank the staff of the Twin Research Unit (KCL) for their help and support in undertaking this project. The Wellcome Trust provides core support for Department of Twin Research. KJHV is supported by an ANZ Trustees PhD scholarship in Medical Research. The authors declare no conflict of interests. BB is the recipient of an NHMRC Biomedical Postdoctoral Fellowship (552498).
Footnotes
Author contributions:
KJHV and SEM were responsible for the study design. MTL, AA, GWM, PAFM, ACH, TDS, and NGM contributed to the acquisition of the data. KJHV performed most analyses and was responsible for writing up the paper. AAEV, BB, SDG, and SEM assisted in the data analysis. All authors provided critical revision of the manuscript and approved final version for publication.
References
- Abecasis GR. METAL. 2009 from http://www.sph.umich.edu/csg/abecasis/Metal/ [Google Scholar]
- Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction. 2009;104(4):518–532. doi: 10.1111/j.1360-0443.2009.02504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, Neuman R, Howells W, Fisher S, Fox L, Cloninger R, Dick DM, Doheny KF, Edenberg HJ, Goate AM, Hesselbrock V, Johnson E, Kramer J, Kuperman S, Nurnberger JI, Pugh E, Schuckit M, Tischfield J, Rice JP, Bucholz KK, Bierut LJ, Consortium G. A genome-wide association study of DSM-IV cannabis dependence. Addiction Biology. 2011;16(3):514–518. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. American Journal of Human Genetics. 2007;81(5):913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Manichaikul A, Rich SS. A Generalized Family-Based Association Test for Dichotomous Traits. American Journal of Human Genetics. 2009;85(3):364–376. doi: 10.1016/j.ajhg.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, McGhee K, Lopez L, Gow AJ, Corley J, Redmond P, Fox HC, Haggarty P, Whalley LJ, McNeill G, Goddard ME, Espeseth T, Lundervold AJ, Reinvang I, Pickles A, Steen VM, Ollier W, Porteous DJ, Horan M, Starr JM, Pendleton N, Visscher PM, Deary IJ. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011;16(10):996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, AMFS Investigators. Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, Gordon SD, Ferreira MAR, Wright MJ, Henders AK, Campbell MJ, Duffy DL, Hansell NK, Macgregor S, Slutske WS, Heath AC, Montgomery GW, Martin NG. Common variants in the trichohyalin gene are associated with straight hair in Europeans. American Journal of Human Genetics. 2009;85(5):750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Spector TD, Williams FMK. The UK Adult Twin Registry (TwinsUK) Twin Research and Human Genetics. 2006;9(6):899–906. doi: 10.1375/183242706779462462. [DOI] [PubMed] [Google Scholar]
- UNODC . World Drug Report 2010. United Nations Publication; 2010. [Google Scholar]
- Verweij KJH, Yang J, Lahti J, Veijola J, Hintsanen M, Pulkki-Råback L, Heinonen K, Pouta A, Pesonen AK, Widen E, Taanila A, Isohanni M, Miettunen J, Palotie A, Penke L, Service SK, Heath AC, Montgomery GW, Raitakari O, Kähönen M, Viikari J, Räikkönen K, Eriksson JG, Keltikangas-Järvinen L, Lehtimäki T, Martin NG, Järvelin MR, Visscher PM, Keller MC, Zietsch BP. Maintenance of genetic variation in human personality: Testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evolution. 2012 doi: 10.1111/j.1558-5646.2012.01679.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Liu JZ, Medland SE, Lynskey MT, Madden PAF, Agrawal A, Montgomery GW, Heath AC, Martin NG. No association of candidate genes with cannabis use in a large sample of Australian twin families. Addiction Biology. 2011;17(3):687–690. doi: 10.1111/j.1369-1600.2011.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG, Boomsma DI, Vink JM. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105(3):417–430. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkhuyzen AAE, Pedersen N, Yang J, Lee SH, Magnussen P, Iacono WG, McGue M, Madden PAF, Heath AC, Luciano M, Payton A, Horan M, Ollier W, Pendleton N, Deary IJ, Montgomery GW, Martin NG, Visscher PM, Wray NR. Common SNPs explain some of the variation in the personality dimensions of Neuroticism and Extraversion. Translational Psychiatry. 2012 doi: 10.1038/tp.2012.27. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42(7):565–U131. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.