Abstract
Objective
Protein inhibitor of activated STAT1 (PIAS1) is known to function as SUMO E3 ligase as well as transrepressor. The aim of the study is to elucidate the regulatory mechanisms for these two different functions, especially with respect to endothelial inflammation.
Methods and Results
The MAPK-activated protein kinase-2 (MK2) is pro-inflammatory kinase and phosphorylates PIAS1 at the Ser522 residue. Activation of MK2 enhances p53-SUMOylation, but a PIAS1 phosphorylation mutant, PIAS1-S522A, abolished this p53-SUMOylation, suggesting a critical role for PIAS1 S522 phosphorylation in its SUMO ligase activity. Since nuclear p53 can inhibit KLF2 promoter activity, we investigated the roles for PIAS1 phosphorylation and p53-SUMOylation in the KLF2 and eNOS expression. Both MK2 and PIAS1 overexpression increased KLF2 promoter activity and eNOS expression, which were inhibited by expressing a p53-SUMOylation defective mutant, p53-K386R, and PIAS1-S522A. PIAS1-S522A also abolished the anti-inflammatory effect of wild type (WT) PIAS1 in vitro and also in vivo which was examined by leukocyte rolling in microvessels of skin grafts transduced by adenovirus encoding WT PIAS1 or the S522A mutant.
Conclusion
Our study has identified a novel negative feedback regulatory pathway through which MK2 limits endothelial inflammation via the PIAS1 S522 phosphorylation-mediated increase in PIAS1 transrepression and SUMO ligase activity.
Keywords: Endothelial inflammation, MK2, NF-κB Transrepression, PIAS1, Vascular biology
Introduction
Endothelial dysfunction and subsequent atherosclerosis are characterized by chronic endothelial cell (EC) inflammation that is induced by a combination of risk factors such as hypertension, obesity, diabetes, smoking, hyperlipidemia, and genetics1, and by the accumulation of lipids, leukocytes, and fibrous elements to form arterial plaques2, 3. NF-κB is an important transcriptional regulator of many inflammatory genes such as those of chemokines, adhesion molecules, and cytokines that play major roles in endothelial inflammation and dysfunction. Protein modifications like phosphorylation, SUMOylation, and ubiquitination have come into focus as important regulators of signaling due to the transient nature of these modifications that affect EC phenotypes, a characteristic found in microvascular diseases where persistent activation of NF-κB contributes to inflammation and progressive compromise of the endothelial function4, 5. Although many studies have identified key positive regulators of NF-κB signaling, much less is understood about negative feedback mechanisms that down-regulate NF-κB-mediated endothelial inflammation in vascular pathologies.
Tumor necrosis factor-α (TNF) is a key pro-inflammatory cytokine involved in the progression of endothelial inflammation and dysfunction through the activation of NF-κB signaling6. Through the canonical NF-κB pathway, TNF activates IκB kinase (IKK) to phosphorylate and degrade IκB, releasing NF-κB into the nucleus where it can activate the transcription of inflammatory genes. Protein inhibitor of activated STAT-1 (PIAS1) negatively regulates this pathway by interacting with NF-κB to repress its transcriptional activity, affecting 48% of the TNF-induced genes7. Not surprisingly, PIAS1-deficient mice are hypersensitive to Lipopolysaccharides (LPS)-induced endotoxic shock7. Recently, Liu et al. have reported that phosphorylation of PIAS1 at Ser-90 (S90) by IKKα during TNF-induced inflammation blocks its NF-κB repressive function, which acts as a negative feedback mechanism against the TNF-IKKα-NF-κB signaling pathway8. However, the regulation of PIAS1’s transrepression of NF-κB in EC remains poorly understood.
In this study, we report that MAPK-activated protein kinase-2 (MK2) phosphorylates PIAS1 at Ser522 (S522) and promotes PIAS1 transrepression activity on NF-κB. In addition to the previously reported anti-inflammatory effect of IKKα-mediated PIAS1 S90 phosphorylation8, we found that phosphorylation of PIAS1 S522 increased its SUNO E3 ligase activity and subsequent p53-SUMOylation. It was reported that nuclear p53 promoted endothelial inflammation by down-regulating Kruppel-Like Factor 2 (KLF2)9, and we found that the MK2-mediated phosphorylation of PIAS1 S522 and subsequent p53-SUMOylation, which causes its nuclear export in EC10, increased KLF2 promoter activity as well as eNOS expression. The anti-inflammatory effect of PIAS1-WT was lost in ECs expressing the PIAS1-S522A mutant protein, suggesting that the MK2-mediated PIAS1 phosphorylation at S522 protects ECs against inflammation.
Materials and Methods
An expanded version of the Methods section is available in the online-only Data Supplement.
Generation of human anti-phospho-PIAS1-S522 antibody
A peptide corresponding to amino acids 512–532 of human PIAS1 (PAVDTSYINTS*LIQDYRHPFH) was synthesized (Peptibody Inc. 2216 Mermans Rd Charlotte, NC 28270, US) with an amino-terminus biotin label in both phosphorylated and unphosphorylated forms.
Cell Culture
Human umbilical vein endothelial cells (HUVECs) were obtained by collagenase digestion11 and cultured on 0.2% gelatin pre-coated dishes using M200 medium supplemented with LSGS (GIBCO) and 2% fetal calf serum (GIBCO). Mouse brain endothelial cells (bEND.3) were cultured in Dulbecco’s modified Eagle’s medium with 4 mM L-glutamine adjusted to contain 1.5 g/L sodium bicarbonate and 4.5 g/L glucose with 10% fetal bovine serum.
Endothelial cell transfection and luciferase reporter Assay
Fully confluent HUVECs were transfected with pNF-κB-luc reporter plasmid which contained five copies of the consensus NF-κB sequence linked to a minimal E1B promoter-luciferase gene (Stratagene) or KLF2-luc reporter plasmid with pRL-TK (encoding Renilla luciferase) plasmid which was used to normalize transfection efficiencies.
Adenovirus-mediated Gene Transfer
Fully confluent HUVECs were transduced with adenovirus vectors for LacZ (Ad-LacZ), wild-type MK2 (Ad-WT-MK2), and dominant-negative MK2 (Ad-DN-MK2) in fresh medium for 18 hrs before being treated with TNF.
Identification of MK2-mediated PIAS1 phosphorylation sites
PIAS1 phosphorylation sites were identified by a combination of NetPhos 2.012 analysis and in vitro kinase assays.
Real-time PCR assay
Total RNA was extracted using the TRIzol reagent according to the manufacturer’s instruction. Reverse transcriptions were performed in 20 μl mixture containing 1 μg of total RNA according to the manufacture’s protocol (Bio-Rad, #170-8890).
Immunoprecipitation (SUMO assay) and Western blot analysis
SUMOylation was detected by immunoprecipitation analysis as previously described10.
Skin transplants
Seven to Eight weeks old C57BL/6 (Jackson Labs) mice were anesthetized with ketamine and xylazine (80/13 mg/kg). Autologous skin transplants were performed by removing a skin piece from an ear and transplanting it to the flank of the same mouse.
Intravital microscopy
Intravital microscope analyses of the skin graft were carried out 1 week after transplants were made as previously described13 after some modification.
Statistical Analysis
Data are reported as mean ± S.D. as indicated. Statistical analysis was performed with the PRISM version 5.0 (GraphPad software). Differences were analyzed with a one-way or a two-way repeated–measure analysis of variance as appropriate, followed by Schéffe’s correction for multiple comparisons. *P < 0.05 and **P < 0.01.
Results
MK2 phosphorylates PIAS1 S522 and inhibits TNF-mediated NF-κB transactivation in endothelial cells
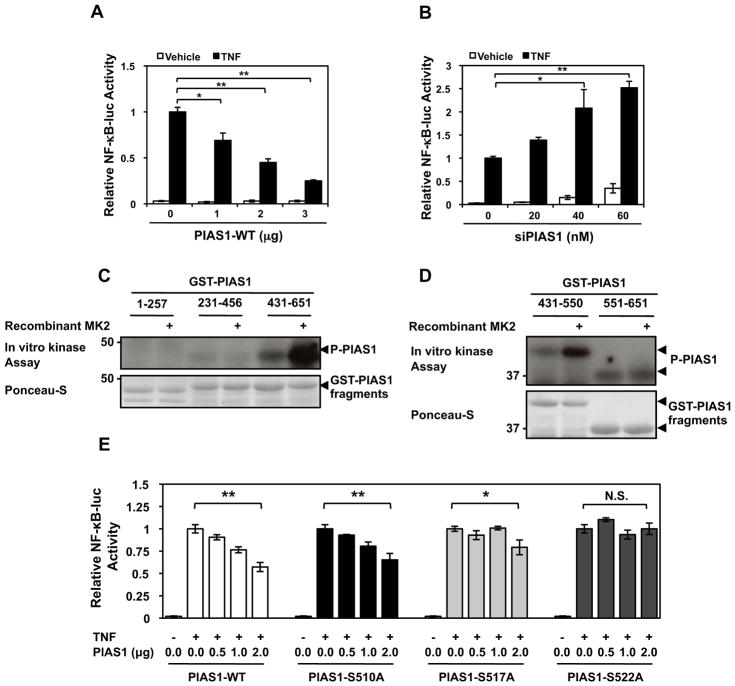
PIAS1 is a known negative regulator of NF-κB signaling as it interacts with p65 and represses the transcriptional activity of NF-κB7. However, the role of PIAS1 in endothelial inflammation remains unclear. We explored this question by expressing varying amounts of either PIAS1-WT or PIAS1 siRNA in HUVECs and then assaying for NF-κB reporter activity following TNF stimulation. Under these inflammatory conditions, expression of wild-type PIAS1 repressed NF-κB transactivation (Fig. 1A) while knockdown of endogenous PIAS1 with siRNA enhanced NF-κB transcriptional activity (Fig. 1B) in a dose-dependent manner.
Figure 1. MK2-mediated PIAS1 phosphorylation enhances NF-κB Transactivation under TNF stimulation.
HUVECs were transfected with an NF-κB-luc reporter construct and kinase dead wild-type PIAS1 (A) or PIAS1 siRNA20 (B), stimulated with TNF, and then assayed for firefly and Renilla luciferase activities. For experiments A–B, quantitative data are shown with n=3, values are mean ± S.D.; *P < 0.05, **P < 0.01. (C) PIAS1 was initially divided into three ~200 amino acid fragments (1–257, 231–456, 431–651) and analyzed by in vitro kinase assay. Note strong phosphorylation of the PIAS1 fragment 431–651 by MK2. A representative autoradiogram is shown along with a Ponceau stain to show GST-PIAS1 expression levels. (D) The PIAS1 431–651 fragment was further divided in half (431–550 and 551–651) and analysis was repeated. (E) PIAS1 phosphorylation mutants were constructed and used to evaluate their effects on NF-κB transactivation under TNF stimulation using dual-luciferase reporter assays. *P < 0.05, **P < 0.01 (n=3 mice; mean ± S.D.). (F) HUVECs were transfected for 48 hrs with either MK2 or control siRNA as indicated and then the cells were stimulated with TNF (10 ng/ml) as indicated. PIAS1 S522 phosphorylation was detected by immunoblotting with anti-phospho-PIAS1 S522 (top). The MK2 and PIAS1 expressions were detected by immnoblotting with anti-MK2 and -PIAS1, respectively. Values are mean ± S.D. (lower panel, n=3); **P < 0.01 compared to the sample without TNF stimulation, ##P < 0.01 compared to each concentration of TNF stimulation in the control siRNA. (G) Endogenous PIAS1 in HUVECs was depleted using siRNA, and after 48 hrs the cells were transduced with adenovirus containing PIAS1-WT or PIAS1- S522A mutant. After 16 hrs of transduction cells were stimulated by TNF for 10 min, and PIAS1 was immuno-precipitated by anti-PIAS1 and immunoblotted with anti-phospho-PIAS1 S522 (top). PIAS1 and tubulin expression were detected by immnoblotting with anti-PIAS1 and -tubulin, respectively. Values are mean ± S.D. (lower panel, n=3); *P < 0.05, **P < 0.01. (H) NF-κB transactivation under TNF stimulation was determined using dual-luciferase reporter assays in PIAS1 S522A mutant knock-in experiments. HUVECs were transfected with PIAS1 siRNA or control siRNA and after 48 hrs transduced with Ad-PIAS1-WT, Ad-PIAS1-S522A mutant, or Ad-LacZ. After 16 hrs of transduction the NF-κB-luc reporter was transfected, and cells were stimulated with TNF and then assayed for firefly and Renilla luciferase activities. Values are mean ± S.D. (n=3); *P < 0.05, **P < 0.01.
Both IKKα and MK2 are known inflammatory kinase14. We found that the canonical NF-κB pathway involving IKKα, IκBα, and p65 NF-κB phosphorylation was unaffected by the expression of the adenoviral wild-type MK2 (Ad-WT-MK2) or dominant-negative MK2 (Ad-DN-MK2) as seen in Supplemental Fig. I. Therefore, we hypothesized that MK2 could phosphorylate PIAS1 and inhibit its transcriptional repression on NF-κB, similar to TNF activating IKKα to phosphorylate PIAS18. As the first step to identify phosphorylation sites in PIAS112, we constructed glutathione S-transferase (GST) fused truncated PIAS1 fragments (AA1–257, 231–456, 431–651, 431–550, 551–651) and used them as substrates for in vitro kinase assays with recombinant active MK2. As shown in Figs. 1C–D, MK2 strongly phosphorylated the PIAS1 fragment containing amino acid residues 431–550. Using NetPhos 2.0, we then identified serine-510, -517, and -522 residues as candidates for MK2-mediated phosphorylation sites based on their high scores as potential phosphorylation sites12. As shown in Supplemental Fig. II, there is a significant reduction in PIAS1 AA431–651 phosphorylation in all three phosphorylation mutants.
Following the identification of MK2-mediated PIAS1 phosphorylation sites, we used the PIAS1 phosphorylation mutants to assay for NF-κB transactivation. The PIAS1-S510A mutant behaved similarly to PIAS1-WT and the PIAS1-S517A partially repressed NF-κB transcriptional activity at 2 μg/ml of cDNA (Fig. 1E). However, the PIAS1-S522A mutant lost its ability to repress NF-κB, suggesting that phosphorylation of PIAS1-S522 can functionally inhibit NF-κB transactivation.
PIAS1 S522 is phosphorylated by TNF treatment via endogenous MK2 activation and subsequently inhibits NF-kB activation
To determine the role of endogenous MK2-mediated PIAS1-S522 phosphorylation in NF-κB activation in ECs, we investigated whether TNF could increase endogenous PIAS1-S522 phosphorylation using a synthetic human antibody against phospho-PIAS1-S522 we generated by employing a phase display system. As shown in Fig. 1F, TNF increased endogenous PIAS1 S522 phosphorylation maximally within 10 min after TNF stimulation. Finally, to determine the role of endogenous MK2 in PIAS1 S522 phosphorylation inside cells, we used MK2 siRNA and found that endogenous PIAS1 S522 phosphorylation was completely inhibited by the depletion of MK2, supporting a crucial role of endogenous MK2 in TNF-initiated PIAS1-S522 phosphorylation.
Next to determine the role of endogenous PIAS1 S522 phosphorylation in TNF-initiated NF-κB activation, we performed knock-in experiments as shown in Fig. 1G. HUVECs were treated with PIAS1 siRNA for 48 hrs to deplete endogenous PIAS1 and transduced with adenovirus containing PIAS1-WT or PIAS1-S522A mutant. Expression levels of PIAS1 wild type and the PIAS1 S522A mutant were similar to that of endogenous PIAS1, and we also confirmed that PIAS1 S522 phosphorylation did not occur in the PIAS1 S522A mutant knocked-in cells (Fig. 1G upper panel). As shown in Fig. 1H, PIAS1 S522A mutation significantly enhanced TNF-induced NF-κB activation compared with wild type. These results suggest an inhibitory role of endogenous PIAS1 S522 phosphorylation in NF-κB activation.
Critical roles of MK2 and PIAS1 S522 phosphorylation in endogenous p53 SUMOylation
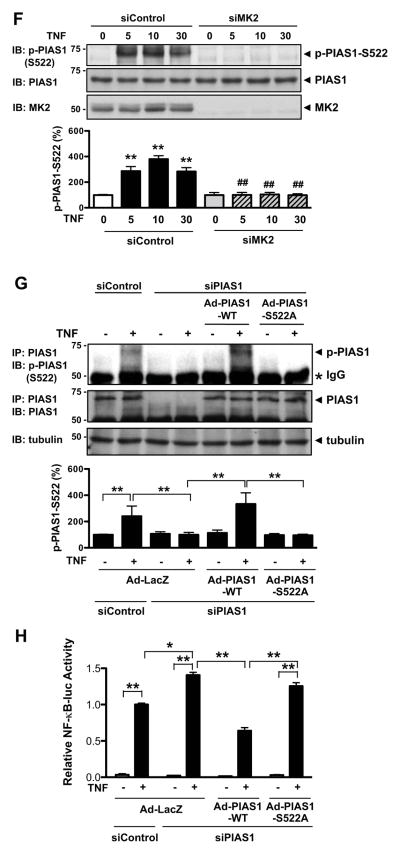
It has been reported that IKKα-mediated PIAS1 S90 phosphorylation inhibits NF-κB transactivation but that the PIAS1-S90A mutant retains its SUMO E3 ligase activity8. In Fig. 1 we demonstrate a role of MK2 on PIAS1 S522 phosphorylation, which inhibits NF-κB activation, but the role of MK2-mediated PIAS1 S522 phosphorylation in its SUMO E3 ligase activity remains unclear. Since we recently reported that disturbed flow-induced EC dysfunction may be mediated by p53-SUMOylation10, first we investigated whether MK2 kinase activity could regulate p53-SUMOylation. As shown in Fig. 2A, p53-SUMOylation induced by Ubc9 (SUMO E2 conjugating enzyme) and SUMO3 was significantly enhanced in cells co-transfected with MK2 but was completely abolished by kinase defective mutant DN-MK2. Next, to determine the role of endogenous MK2 in TNF-induced p53 SUMOylation, we depleted MK2 expression by using siRNA, and found that the depletion of MK2 significantly inhibited TNF-induced endogenous p53 SUMOylation in both HUVECs and bEND.3 (Fig. 2B and Supplemental Fig. IIIA), suggesting a key role of MK2 on TNF-induced p53 SUMOylation in endothelial cells.
Figure 2. MK2 increases p53-SUMOylation via PIAS1 phosphorylation.
(A) HUVECs were transduced with Ad-LacZ, Ad-MK2, or Ad-DN-MK2 for 18 hrs and then transfected for 24 hrs as indicated with Flag-tagged p53, HA-tagged SUMO3, and Ubc9. p53 SUMOylation was detected by immunoprecipitation with anti-Flag followed by Western blotting with anti-HA (top). Both protein expression and immunoprecipitated p53 were confirmed by anti-Flag and the MK2, Ubc9, and SUMO expressions were detected by indicated antibodies. Mono-SUMOylation band (≈74kDa) and poly-SUMOylation bands (> 78kDa) were detected. (B) HUVECs were transfected for 48 hrs with either MK2 or control siRNA as indicated and then the cells were stimulated with TNF (10 ng/ml) for 3 hrs. p53 SUMOylation was detected by immunoprecipitation with anti-p53 followed by Western blotting with anti-SUMO (top). The MK2, p53, and SUMO expressions were confirmed by immnoblotting with anti-MK2, - p53, and -SUMO, respectively. The quantitative data are shown with n=3 (A and B, lower panel), values are mean ± S.D.; **P < 0.01. (C) HUVECs were transduced for 18 hrs with Ad-PIAS1 or Ad-PIAS1-S522A with Ad-MK2 or Ad-LacZ as indicated, then the cells were stimulated with TNF for 3 hrs. p53 SUMOylation was detected by immunoprecipitation with anti-p53 followed by Western blotting with anti-SUMO (top). PIAS1, MK2, p53, and SUMO expression was confirmed by immnoblotting with anti-PIAS1, MK2, -p53, and -SUMO2/3, respectively. (D) Values are mean ± S.D. (Fig. 2C, n=3); *P < 0.05, **P < 0.01.
Next we examined the role of PIAS1-S522 phosphorylation in endogenoaus p53 SUMOylation in both HUVECs and bEND.3. We transduced adenovirus containing PIAS1 WT (Ad-PIAS1-WT) and S522A mutant (Ad-PIAS1-S522A) and examined endogenous p53 SUMOylation induced by TNF (Fig. 2C and Supplemental Fig. IIIC). In addition, to examine the role of MK2-mediated PIAS1-S522 phosphorylation in endogenous p53 SUMOylation, we also transduce Ad-MK2 and examined the effect of PIAS1-S522A mutant on MK2-mediated p53 SUMOylation. As shown in Figs. 2C–D and Supplemental Fig. IIIC, both TNF and MK2 transduction increased endogenous p53 SUMOylation in PIAS1-WT transduced endothelial cells. However, in PIAS1-S522A mutant transduced cells we did not find any increase of endogenous p53 SUMOylation induced by TNF, MK2, and the combination of MK2 transduction with TNF stimulation. Of note, PIAS1 wild type and S522A expression levels were similar. We also observed similar tendency that deletion of MK2 and PIAS1-S522A expression inhibited TNF-induced p53 SUMOylation in bEND.3 cells (Figs. 2B–D and Supplemental Fig. IIIA and C), demonstrating the importance of MK2 and PIAS1 S522 phosphorylation in p53 SUMOylation not only in HUVECs but also in bEND.3 cells.
Since PIAS1-S517 partially repressed NF-κB transcriptional activity, we also examined the effect of PIAS1 S517 phosphorylation on the p53 SUMOylation and found that PIAS1 S517 overexpression failed to inhibit TNF-mediated p53 SUMOylation (Supplemental Fig. IIIB). It suggests that TNF-induced PIAS1 S522 phosphorylation but not S517 phosphorylation can regulate p53 SUMOylation.
Roles of MK2-mediated PIAS1 S522 phosphorylation and subsequent p53-SUMOylation in KLF2 activity and eNOS expression
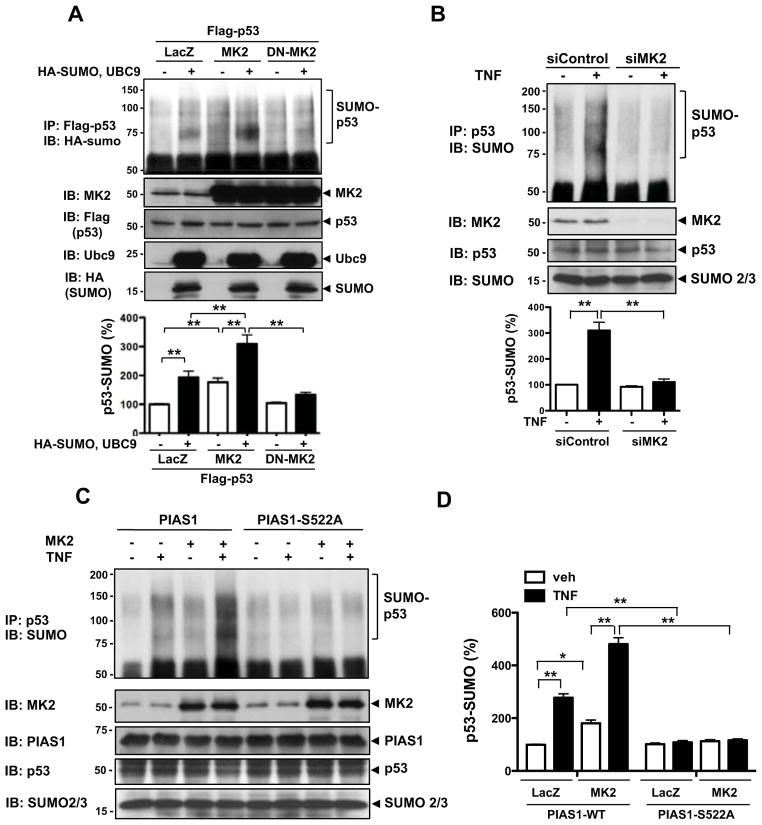
Recently Kumar et al. reported that KLF2 expression was suppressed by p53 via the conserved p53-binding repressor sequence in its promoter (−221 to −88)15. We have reported that p53-SUMOylation induces p53 nuclear export10. Therefore, we hypothesized that p53-SUMOylation induced by MK2-mediated PIAS1 S522 phosphorylation would increases KLF2 promoter activity as well as eNOS expression, which is regulated by KLF2. First, we investigated the role of MK2 and PIAS1 in KLF2 promoter activity using KLF2-luc promoter (−924 to +14) plasmid and pRL-TK plasmid used to normalize transfection efficiencies. As shown in Fig. 3A, overexpression of MK2 and PIAS1-WT significantly increased KLF2 promoter activity. Although co-transduction of p53-WT did not significantly change the MK2 and PIAS1-mediated increase of KLF2 promoter activity compared to Ad-LacZ (data not shown), the Ad-p53-KR mutant significantly inhibited KLF2 promoter activity, suggesting a critical role of p53-SUMOylation in the MK2- and PIAS1-mediated KLF2 promoter activity (Fig. 3A). Effects of p53-KR on KLF2 target genes were also examined. As shown in Fig. 3B, transduction of Ad-MK2 and Ad-PIAS1 increased eNOS expression. Interestingly, the p53-SUMOylation KR mutant significantly inhibited eNOS expression compared to p53-WT. Next, we studied the role of MK2-mediated PIAS1 S522 phosphorylation in KLF2 promoter activity. As shown in Fig. 3C, transduction of Ad-PIAS1-S522A mutant but not WT significantly inhibited MK2-mediated KLF2 promoter activity. We also found that transduction of Ad-PIAS1-S522A inhibited eNOS expression induced by MK2 overexpression (Fig. 3D). Taken together, these data support a critical role of PIAS1 S522 phosphorylation and subsequent p53-SUMOylation in MK2-mediated KLF2 and eNOS expression.
Figure 3. MK2-mediated PIAS1 phosphorylation regulates p53-SUMOylation and subsequently enhances KLF2 promoter activity and eNOS mRNA expression.
(A, B) Effects of p53 SUMOylation on KLF2 promoter activity (A) and eNOS mRNA expression (B) in HUVECs transduced with Ad-LacZ, Ad-MK2, or Ad-PIAS1 were evaluated using dual-luciferase reporter assays (A) and real-time quantitative PCR (B). HUVECs were transduced with Ad-LacZ, -MK2, -PIAS1, or -p53-KR for 18 hrs and further transfected with the KLF2 promoter activity reporter construct for 18 hrs, then assayed for firefly and Renilla luciferase activities (A). After 18 hrs of the adenovirus transduction, HUVECs were assayed for eNOS mRNA levels (B). (C) HUVECs were transduced with Ad-LacZ, -PIAS1-WT, -PIAS1-S522A, or -MK2 for 18 hrs, and KLF2 promoter activity (C) or eNOS mRNA levels (D) were detected as described for A–B. *P < 0.05, **P < 0.01 (n=3 mice; mean ± S.D.). (E) A signaling scheme describing the relationship between the inhibitory effect of p53 on KLF2-eNOS expression and MK2-induced PIAS1-S522 phosphorylation which leads to p53 SUMOylation.
MK2-mediated PIAS1 phosphorylation inhibits inflammatory genes expression under TNF stimulation
We found that PIAS1-S522 phosphorylation by MK2 suppressed NF-κB transactivation (Figs. 1E and H). To test if MK2-mediated PIAS1 phosphorylation and its transrepression of NF-κB were involved in EC inflammation, we transduced ECs with Ad-PIAS1-WT and -PIAS1-S522A to assay for the expression of proinflammatory genes regulated by NF-κB (Figs. 4A–D). We found that overexpression of Ad-PIAS1-WT significantly inhibited the TNF-induced E-selectin, ICAM-1, VCAM-1, and MCP-1 mRNA expression in ECs, but interestingly this inhibitory effect was lost for the Ad-PIAS1-S522A mutant (Figs. 4A–D). We confirmed that the protein expression levels of Ad-PIAS1-WT and Ad-PIAS1-S522A were similar in ECs (Fig. 4E). These data suggest a crucial role of PIAS1 S522 phosphorylation in TNF-mediated negative feedback mechanism against endothelial inflammation (Fig. 4F).
Figure 4. PIAS1 phosphorylation mutant decreases EC inflammation.
HUVECs were transduced with Ad-LacZ, Ad-PIAS1-WT, or Ad-PIAS1-S522A for 18 hrs followed by treatment with TNF. Cells were evaluated for mRNA expression for E-selectin (A), ICAM-1 (B), VCAM-1(C), and MCP-1 (D) by real-time quantitative PCR. Values are mean ± S.D. (n=3); **P < 0.01. (E) Protein expression of PIAS1-WT and PIAS1-S522A was evaluated by Western blotting using anti-PIAS1. (F) A scheme describing the effects of TNF and subsequent PIAS1-S522 phosphorylation-mediated negative feedback on inflammatory gene expression in ECs.
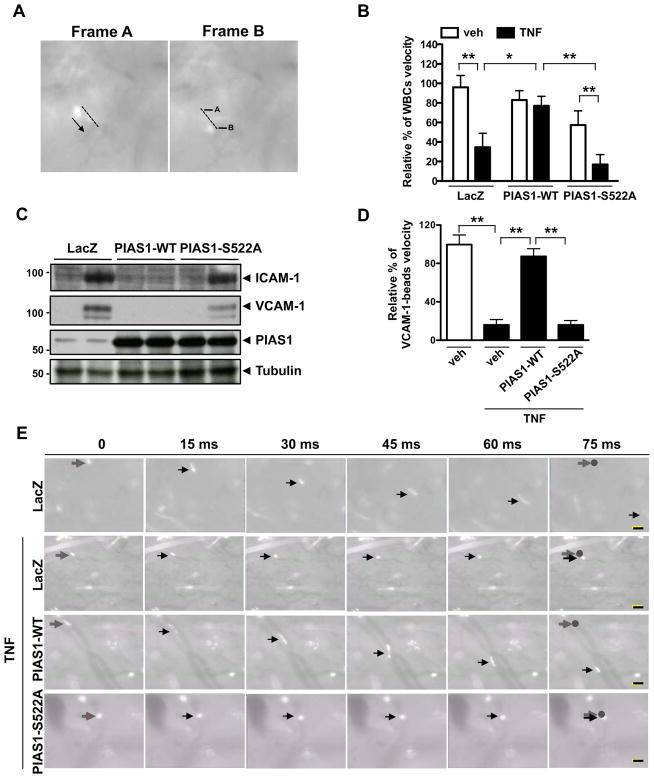
PIAS1 Ser522 phosphorylation inhibits TNF-mediated endothelial inflammation and leukocyte rolling in vivo
Velocity of leukocyte rolling is a good indicator for evaluating the inflammatory state of the endothelium. To study the role of PIAS1 Ser522 phosphorylation in vivo, we examined leukocyte rolling in microvessels using a mouse skin graft model. To avoid transplant rejection, we used an autologous skin transplant in which a piece of skin from an ear of a C57BL/6 mouse was grafted onto the flank of the same animal. It has previously been demonstrated that grafted skin is vascularized and that leukocytes can be imaged in microvessels within the graft13. First we transduced ear skin preparations with Ad-LacZ, Ad-PIAS1, or Ad-PIAS1-S522A for 1 hr and then transplanted them to the graft bed. Seven days after transplantation, we examined the transduction efficiency of the adenovirus vector by β-gal staining (Supplemental Fig. VIA). We detected β-gal staining in Ad-LacZ transduced skin grafts but not in non-transduced grafts. In addition, PIAS1 expression was ~2 fold increased by transduction of Ad-PIAS1 and -PIAS1-S522A compared to Ad-LacZ (Supplemental Fig. VIB). Using intravital microscopy, we next studied leukocyte rolling in microvessels 3 hrs after peritoneal injection of saline or TNF16. Leukocyte rolling velocity was determined by measuring the distance that each leukocyte traveled along the vessel wall between two successive frames (Fig. 5A). The baseline leukocyte rolling velocity showed no significant differences among Ad-LacZ, -PIAS1-WT, and -PIAS1-S522A-transduced skin grafts (Fig. 5B and Supplemental videos I–III). However, the average rolling velocity was decreased by ~2 times in the graft of TNF-injected animals compared to saline injected mice (Fig. 5B and Supplemental videos I and IV). Interestingly, we found that TNF-mediate leukocyte rolling was significantly inhibited by transduction of Ad-PIAS1-WT but not of Ad-PIAS1-S522A mutant (Fig. 5B and Supplemental video V and VI).
Figure 5. PIAS1 Ser522 phosphorylation inhibits TNF-mediated leukocyte rolling in the skin transplants.
After 7 days of transplantation of skin grafts that were transduced with Ad-LacZ, Ad-PIAS1-WT, or Ad-PIAS1-S522A, the rolling and velocity of leukocytes (A–B) or fluorescent beads coated with anti-VCAM-1 (D–E) in the grafts were determined by intravital microscopy with and without 3 hrs of TNF injection. (A) Representative images with indications of distance traveled by leukocytes between two adjacent frames. (B) Quantification of leukocyte rolling and velocity in the skin graft’s micro-vessels. A total of ≈100 leukocytes was counted from two different fields in each skin graft. *P < 0.05 and **P < 0.01 (n=4 mice; mean ± S.D.). (C) Protein expression of VCAM-1, ICAM-1, PIAS1, and tubulin was determined by immunoblotting using whole tissue lysates from skin grafts as indicated. The figure is representative of two independent experiments. (D) Quantification of the roling velocity of anti-VCAM-1 antibody-coated fluorescent beads in the skin graft’s micro-vessels. A total of ≈ 60–100 beads were counted from two different microscope fields in each skin graft. **P < 0.01 (n=3; mean ± S.D.). (E) Captured images at each 15 ms show movement of anti-VCAM-1 antibody-coated fluorescent beads in the skin graft’s micro-vessel. Grey and black arrows indicate an anti-VCAM-1 antibody-coated fluorescent bead in the vessel. Grey arrow and circle: the position of the bead at 0 ms. Bars; 20 μm.
To investigate the role of PIAS1-S522 phosphorylation in the endothelial inflammation in this model, first we determined VCAM-1 and ICAM-1 protein expression in the skin graft. Consistent with our in vitro experiments showing mRNA expression of inflammatory genes (Fig. 4), overexpression of PIAS1-WT significantly inhibited VCAM-1 and ICAM-1 protein expression, but compared with PIAS1-WT we could not detect significant inhibitory effects of PIAS1-S522A mutant on the TNF-mediated VCAM-1 and ICAM-1 induction (Fig. 5C). Next to examine the role of PIAS1 S522 phosphorylation in regulating endothelial VCAM-1 expression in vivo, we injected fluorescent microspheres coated with anti-VCAM-1 into C57BL/6 mice that received transplantation of skin grafts transduced with Ad-LacZ, Ad-PIAS1-WT, or Ad-PIAS1-S522A 7 days ago as described above. After 3 hrs of TNF treatment we found that the velocity of anti-VCAM-1-coated fluorescent beads was significantly slowed down in Ad-LacZ transduced control. Note that the beads were better-visualized than rolling leukocytes in blood vessels. Overexpression of PIAS1-WT improved bead velocity, but the mutant of PIAS1-S522 failed to do so, suggesting a crucial role of PIAS1 S522 phosphorylation in regulating TNF-mediated endothelial VCAM-1 expression (Figs. 5D, E, and Supplemental videos VII–X). These data again suggest a critical role of PIAS1-S522 phosphorylation in endothelial inflammation.
Discussion
In this study we have uncovered a novel mechanism in ECs through which an inflammatory kinase MK2 can promote transrepression activity of PIAS1 through its S522 phosphorylation. We found that expression of PIAS1 had a transrepressive effect on NF-κB while knockdown of PIAS1 reversed this phenotype, a novel finding in ECs. When we examined the relationship between MK2 and PIAS1 in regulating NF-κB transactivation, we found that MK2 phosphorylated PIAS1 S522 (Figs. 1C–E) and this modification enhanced PIAS1’s transrepressive function on NF-κB (Figs. 1F–H). Furthermore, in contrast to IKKα-mediated S90 PIAS1 phosphorylation, MK2-mediated S522 PIAS1 phoshorylation increased its SUMO E3 ligase activity, then leading to p53-SUMOylation (Fig. 2). We found this MK2-mediated p53-SUMOylation increased KLF2 promoter activity as well as subsequent eNOS expression (Fig. 3). We also found an anti-inflammatory effect of PIAS1 S522 phosphorylation in vivo (Fig. 5). Collectively, our results have revealed a novel negative feedback pathway through which a pro-inflammatory kinase MK2 plays a unique anti-inflammatory role by regulating both NF-κB transrepression and SUMO E3 ligase activity of PIAS1 via phosphorylation of PIAS1 S522 as shown in Fig. 6A.
Figure 6. A schematic drawing showing how stimulation by TNF activates MK2-mediated NF-κB transactivation independent of the canonical IKKα-IκBα-NF-κB pathway in endothelial cells.
(A) TNF-mediated activation of IKKα not only induces NF-κB transactivation through the canonical TNF-IKKα-NF-κB signaling pathway but also negatively regulates this event by phosphorylating PIAS1 at the S90 residue8. Similar to IKKα, TNF-mediated activation of MK2 leads to NF-κB transactivation and also activates a negative feedback mechanism through which MK2 phosphorylates PIAS1 at the S522 residue to enhance its transrepression of NF-κB and p53 SUMOylation, p53 nuclear export, and subsequent KLF2-mediated eNOS expression. (B) The domafin structure of PIAS1. SAP domain: scaffold attachment factor A and B. PINIT: Pro-Ile-Asn-Ile-Thr motif. RLD: RING-finger-like zinc binding domain, protein-protein interactions, interacts with the SUMO conjugase Ubc9, sumoylation. AD: highly acidic domain. S/T: serine/threonine rich region. PIAS1 S90 site lie in the NF-κB interacting region, whereas PIAS1 S522 site does not lie in any of PIAS1’s functional domains involved in NF-κB binding or its SUMO E3 ligase activity (RLD) domain18.
PIAS1 plays an important role as a transrepressor of NF-κB to regulate inflammation. It has previously been shown that PIAS1 interferes with p65’s DNA-binding ability by selectively binding to the C-terminus region of p65, leading to the inhibition of NF-κB target genes like IκBα in response to TNF7, 17. Furthermore TNF-mediated activation of IKKα not only induces NF-κB transactivation through the canonical TNF-IKKα-NF-κB signaling pathway but also negatively regulates this event by phosphorylating PIAS1 at S908. This concept of restrictive inflammatory gene activation through a negative feedback loop is similar to the mechanism we have identified in this study. Similar to IKKα, TNF-mediated activation of MK2 leads to phosphorylation of PIAS1 S522 and inhibits NF-κB transactivation (Fig. 1D). Also interesting to note is that the S522 site does not lie in any of the PIAS1’s functional domains that are involved in its binding to p65 or its SUMO E3 ligase activity (a RING-finger-like zinc binding domain, RLD, Fig. 6B)18. Possible interplay between S90 and S522 phosphorylation on PIAS1 activity requires further investigation.
Although MK2 did not affect the upstream events of the canonical NF-κB signaling pathway, it enhanced NF-κB transactivation (Supplemental Fig. I and Fig. 1). Previously, Gorska et al. have reported that MK2 enhances NF-κB activity by blocking nuclear retention of p38 to prevent excessive phosphorylation of mitogen- and stress-activated protein kinase-1 (MSK1). In turn, by reducing MSK1 activity, MK2 prevents p65 export from the nucleus, leading to sustained NF-κB activation19. We assume that TNF can simultaneously trigger MK2 to sequester p38 and phosphorylate PIAS1, thereby activating the proinflammatory p38/MK2-MSK1 pathway and the newly revealed anti-inflammatory MK2-PIAS1 pathway to form a coordinated regulatory negative feedback loop. This loop is expected to provide a balanced inflammatory response in the physiological state that may become dys-regulated in vascular pathologies.
Post-translational modifications are key regulators of signal transduction due to their ability to transiently alter numerous physiological and pathological pathways. This is especially true in complicated processes like inflammation in which a careful regulation of signaling is the difference between the physiological and the pathological states. Our study here expands the role of MK2 in endothelial inflammation, by activating a novel negative feedback loop through PIAS1 phosphorylation that can curb pathological inflammation. A disruption to this delicate balance between MK2’s pro-inflammatory and anti-inflammatory roles could tip the scale towards endothelial inflammation and dysfunction.
Supplementary Material
Acknowledgments
This work is supported by grants from the National Institute of Health (NIH) to Dr. Abe (HL-088637, HL-064839, HL-077789, and HL-102746), Dr. Fujiwara (HL-064839, HL-102746), and Dr. Morrell (HL094547 and HL093179). Dr. Abe is a recipient of Established Investigator Awards of the American Heart Association (AHA) (0740013N).
Footnotes
Disclosures
None.
References
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 3.Deguchi JO, Aikawa M, Tung CH, Aikawa E, Kim DE, Ntziachristos V, Weissleder R, Libby P. Inflammation in atherosclerosis: visualizing matrix metalloproteinase action in macrophages in vivo. Circulation. 2006;114:55–62. doi: 10.1161/CIRCULATIONAHA.106.619056. [DOI] [PubMed] [Google Scholar]
- 4.Collins T, Cybulsky MI. NF-kappaB: pivotal mediator or innocent bystander in atherogenesis? J Clin Invest. 2001;107:255–264. doi: 10.1172/JCI10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukundan L, Odegaard JI, Morel CR, Heredia JE, Mwangi JW, Ricardo-Gonzalez RR, Goh YP, Eagle AR, Dunn SE, Awakuni JU, Nguyen KD, Steinman L, Michie SA, Chawla A. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat Med. 2009;15:1266–1272. doi: 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKellar GE, McCarey DW, Sattar N, McInnes IB. Role for TNF in atherosclerosis? Lessons from autoimmune disease. Nat Rev Cardiol. 2009;6:410–417. doi: 10.1038/nrcardio.2009.57. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, Cheng G, Wu H, Shuai K. Negative regulation of NF-kappaB signaling by PIAS1. Mol Cell Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, Yang Y, Chernishof V, Loo RR, Jang H, Tahk S, Yang R, Mink S, Shultz D, Bellone CJ, Loo JA, Shuai K. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 9.Kumar A, Kim CS, Hoffman TA, Naqvi A, Dericco J, Jung SB, Lin Z, Jain MK, Irani K. p53 impairs endothelial function by transcriptionally repressing Kruppel-Like Factor 2. Arterioscler Thromb Vasc Biol. 2011;31:133–141. doi: 10.1161/ATVBAHA.110.215061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heo KS, Lee H, Nigro P, Thomas T, Le NT, Chang E, McClain C, Reinhart-King CA, King MR, Berk BC, Fujiwara K, Woo CH, Abe J. PKCzeta mediates disturbed flow-induced endothelial apoptosis via p53 SUMOylation. The Journal of cell biology. 2011;193:867–884. doi: 10.1083/jcb.201010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M, Berk BC. Mitogen-activated protein kinase (ERK1/2) activation by shear stress and adhesion in endothelial cells. Essential role for a herbimycin-sensitive kinase. J Clin Invest. 1996;98:2623–2631. doi: 10.1172/JCI119083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 13.Morrell CN, Murata K, Swaim AM, Mason E, Martin TV, Thompson LE, Ballard M, Fox-Talbot K, Wasowska B, Baldwin WM., 3rd In vivo platelet-endothelial cell interactions in response to major histocompatibility complex alloantibody. Circ Res. 2008;102:777–785. doi: 10.1161/CIRCRESAHA.107.170332. [DOI] [PubMed] [Google Scholar]
- 14.Gaestel M. MAPKAP kinases - MKs - two’s company, three’s a crowd. Nat Rev Mol Cell Biol. 2006;7:120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Kim CS, Hoffman TA, Naqvi A, Dericco J, Jung SB, Lin Z, Jain MK, Irani K. p53 impairs endothelial function by transcriptionally repressing Kruppel-Like Factor 2. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:133–141. doi: 10.1161/ATVBAHA.110.215061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozer K, Adanali G, Siemionow M. Late effects of TNF-alpha-induced inflammation on the microcirculation of cremaster muscle flaps under intravital microscopy. J Reconstr Microsurg. 2002;18:37–45. doi: 10.1055/s-2002-19708. [DOI] [PubMed] [Google Scholar]
- 17.Jang HD, Yoon K, Shin YJ, Kim J, Lee SY. PIAS3 suppresses NF-kappaB-mediated transcription by interacting with the p65/RelA subunit. J Biol Chem. 2004;279:24873–24880. doi: 10.1074/jbc.M313018200. [DOI] [PubMed] [Google Scholar]
- 18.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 19.Gorska MM, Liang Q, Stafford SJ, Goplen N, Dharajiya N, Guo L, Sur S, Gaestel M, Alam R. MK2 controls the level of negative feedback in the NF-kappaB pathway and is essential for vascular permeability and airway inflammation. J Exp Med. 2007;204:1637–1652. doi: 10.1084/jem.20062621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo CH, Shishido T, McClain C, Lim JH, Li JD, Yang J, Yan C, Abe J. Extracellular signal-regulated kinase 5 SUMOylation antagonizes shear stress-induced antiinflammatory response and endothelial nitric oxide synthase expression in endothelial cells. Circ Res. 2008;102:538–545. doi: 10.1161/CIRCRESAHA.107.156877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.