Abstract
Retrotransposon and retroviral RNA delivery to particle assembly sites is essential for their replication. mRNA and Gag from the Ty1 retrotransposon colocalize in cytoplasmic foci, which are required for transposition and may be sites for virus-like particle (VLP) assembly. To determine which Ty1 components are required to form mRNA/Gag foci, localization studies were performed in a Ty1-less strain expressing galactose-inducible Ty1 plasmids (pGTy1) containing mutations in GAG or POL. Ty1 mRNA/Gag foci remained unaltered in mutants defective in Ty1 protease or deleted for POL. However, Ty1 mRNA containing a frameshift mutation (Ty1fs) that prevents the synthesis of all proteins accumulated in the nucleus. Ty1fs RNA showed a decrease in stability that was mediated by the cytoplasmic exosome, nonsense mediated decay, and the processing-body. Localization of Ty1fs RNA remained unchanged in an nmd2Δ mutant. When Gag and Ty1fs mRNA were expressed independently, Gag provided in trans increased Ty1fs RNA level and restored localization of Ty1fs RNA in cytoplasmic foci. Endogenously expressed Gag also localized to the nuclear periphery independent of RNA export. These results suggest that Gag is required for Ty1 mRNA stability, efficient nuclear export, and localization into cytoplasmic foci.
Keywords: retrotransposon, Gag, mRNA, nuclear export, Ty1, Saccharomyces
The Saccharomyces Ty1 element belongs to a widely distributed group of transposable elements called long terminal repeat (LTR) retrotransposons or Pseudoviruses, which resemble retroviruses in genome organization and replication (1, 2) (see Fig. 1A for a summary of Ty1 replication). Ty1 is transcribed by RNA polymerase II to yield an abundant 5.7 kb genomic RNA (gRNA) that can be translated or packaged into virus-like particles (VLPs). The translation products are Gag-p49 and Gag-Pol-p199, the latter being less abundant (~3%) because it is the product of a +1 ribosomal frameshift (3). The Gag-p49 and Gag-Pol-p199 precursor proteins form immature VLPs that undergo proteolytic processing by Ty1 protease (PR) to form mature Gag and Pol proteins and functional VLPs. Gag-p49 is cleaved by PR near its C-terminus to form Gag-p45, which is the major capsid protein. Gag-p45 contains a functional nucleic acid chaperone domain that is analogous to retroviral nucleocapsid proteins (4). The Gag-Pol-p199 precursor is also cleaved to form Gag-p45, in addition to the enzymes required for retrotransposition: PR, integrase (IN), and reverse transcriptase (RT). After processing of precursor proteins within VLPs, the encapsidated Ty1 gRNA is reverse transcribed into cDNA. To complete the replication cycle, Ty1 cDNA and IN are imported into the nucleus via a bipartite nuclear localization signal present on IN (5, 6), and the cDNA is integrated into the host genome.
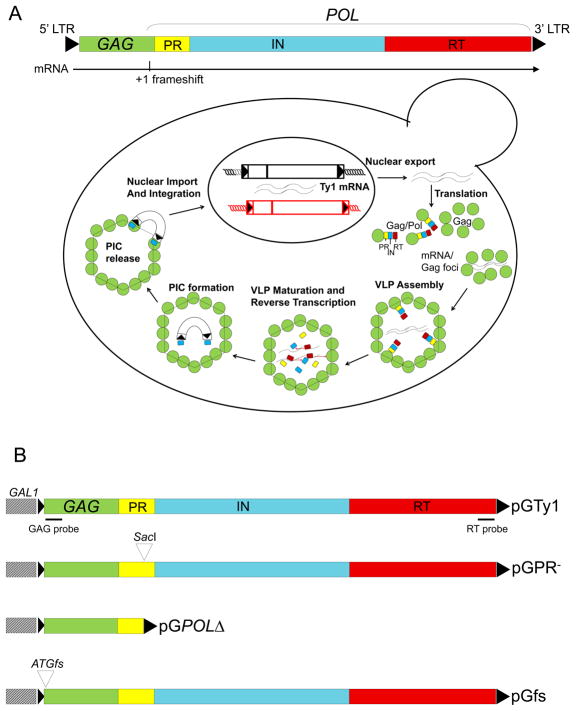
Fig. 1.
Ty1 retrotransposition. (A) The functional organization of Ty1 showing the Ty1 mRNA and the site of the +1 ribosomal frameshift is presented at the top. The Ty1 life cycle is depicted below. Ty1 is transcribed by RNA pol II to form a 5.7 kb transcript, which functions as either gRNA for packaging into VLPs or mRNA for translation into Gag-p49 or Gag-Pol-p199. Gag accumulates in the cytoplasm to become the capsid protein of Ty1 VLPs. POL encodes the enzymes required for Ty1 transposition: PR, IN, and RT. Translation of POL requires a +1 ribosomal frameshift at the 3′ end of GAG yielding Gag-Pol-p199. Prior to VLP formation, Gag and presumably Gag-Pol form mRNA/Gag foci, which are sites where VLPs cluster and may be required for assembly. Ty1 mRNA is specifically packaged as a dimer into VLPs. Ty1 PR cleaves Gag-p49 and Gag-Pol-p199 precursors to form mature proteins within VLPs, such as Gag-p45 and IN-p71. An IN/RT heteromer reverse transcribes Ty1 mRNA into cDNA. A pre-integration complex (PIC) containing Ty1 cDNA and IN is imported into the nucleus via a nuclear localization signal present on IN. Retrotransposition is completed by IN-mediate insertion into a new location in the host genome. (B) pGTy1 plasmids. Overexpression plasmids contain Ty1 fused to the GAL1 promoter in a 2μ-based multicopy shuttle plasmid. The position of the hybridization probes for FISH-IF is shown. The following pGTy1 mutants were used in this study: pGPR−, which renders PR enzymatically inactive, pGPOLΔ, which encodes an inactive PR and lacks RT and IN, and pGfs, which does not yield Ty1 proteins.
Ty1 gRNA shares two key trafficking steps with its retroviral relatives (7). Both Ty1 and retroviral gRNAs are exported from the nucleus, translated, and packaged into VLPs or viral particles. Since retroviral gRNA can be spliced to form subgenomic mRNAs, a variety of mechanisms have evolved to allow nuclear export and cytoplasmic accumulation of unspliced gRNA, which is required for assembly of infectious particles. For example, HIV-1 encodes a master regulator called Rev, which inhibits splicing of mRNAs containing a Rev response element and promotes nuclear export of partially spliced forms of viral mRNAs and unspliced gRNA (8). HIV-1 Gag then binds cytoplasmic gRNA, and this RNP complex is recruited to the plasma membrane for virus assembly, although it is not known if there is a specific cellular environment where Gag first encounters the gRNA (9–11). In contrast, cis-acting sequences flanking the src gene of Rous sarcoma virus (RSV) interact with host export proteins TAP and Dpb5, which modulate the accumulation of unspliced gRNA in the cytoplasm (12–14). Interestingly, RSV Gag enters the nucleus transiently, where it binds gRNA and promotes nuclear export (15–19). It has been suggested that this discrete population of RNA is selectively packaged. Unlike retroviruses, Ty1 mRNA is not spliced; therefore, it is also the gRNA (1). To date, it is not clear whether this pool of gRNA is differentiated into packaged RNA versus translated mRNA.
The trafficking and utilization of Ty1 mRNA during retrotransposition is unique in several respects. Given that Ty1 mRNA is a relatively stable transcript and accumulates to 5–10% of total mRNA in haploid cells, Ty1 proteins and VLPs are present in low amounts (20–25). Transposition events also occur at a low rate. However, overexpression of Ty1 from a GAL1 promoter in a multicopy plasmid (pGTy1) results in visualization of VLPs by electron microscopy and more than a 10,000-fold increase in retrotransposition events. Interestingly, this occurs with only a 5–15 fold increase in Ty1 mRNA. Therefore, it can be inferred that the use of Ty1 gRNA as a template for translation and reverse transcription increases dramatically during galactose-induced expression of pGTy1.
Recent studies have begun to clarify this paradox. After export from the nucleus via the Mex67p pathway, Ty1 mRNA colocalizes with Gag in cytoplasmic granules, referred to as T-bodies, or mRNA/Gag foci, in about 30% of mitotic cells (26, 27). So far, two general classes of genes that affect the formation or appearance of mRNA/Gag foci have been identified. The first class is involved in transcription by RNA polymerase II (pol II) (26). For example, mutations in the RNA pol II subunit Rpb1p, Mediator subunits Srb2p and Srb5p, or the Ty1 transcriptional regulator Spt21p do not markedly change Ty1 mRNA levels or transposition frequency yet enhance the formation of mRNA/Gag foci. The second class is genes required for RNA degradation or storage in processing (P) bodies (27, 28). Deletion of P-body components can decrease the number and alter the appearance of Ty1 mRNA/Gag foci, in addition to decreasing the levels of retrotransposition-competent VLPs, mature Ty1 proteins, and packaged gRNA. There is conflicting evidence that Ty1 mRNA/Gag foci occur in P-bodies (26–28). However, P-body components are important cofactors for Ty1 (27–29) and Ty3 retrotransposition (30) and Ty3 VLP assembly (30).
Here, we investigated the Ty1 components required to form mRNA/Gag foci. We provide evidence that Ty1 transcripts defective or deleted for PR, IN, and RT coding sequence still form foci with Ty1 Gag. These results show that either unprocessed (p49) or mature (p45) Gag is sufficient for forming mRNA/Gag foci. When Gag is absent, Ty1 RNA accumulates in the nucleus and the RNA level and stability decreases. When Gag is provided in trans, Ty1 RNA colocalizes with Gag in the cytoplasm and the RNA level returns to normal. Endogenously expressed Gag can also be detected in association with the nuclear envelope in wild type cells. Therefore, Gag enhances Ty1 mRNA stability, nuclear export and colocalization into cytoplasmic foci.
Results and Discussion
POL products are not necessary for Ty1 mRNA/Gag localization into cytoplasmic foci
To determine the Ty1 components required to form Ty1 mRNA/Gag foci, we analyzed Ty1 mutants containing deletion or inactivation of POL-encoded products (Fig. 1B). Galactose induction of pGTy1 mutants was performed in a S. paradoxus strain that lacks Ty1 elements to minimize any contribution from endogenous Ty1 expression (31, 32). Localization of Ty1 mRNA and Gag (p49 and p45) was analyzed by fluorescence in situ hybridization and immunofluorescence (FISH-IF) microscopy using a GAG-DIG probe (Fig. 1B) and anti-VLP antiserum, respectively. As shown previously (27), multiple Ty1 mRNA and Gag foci appeared in about one third of cells overexpressing pGTy1 and colocalized in 96% of these cells (Fig. 2A). Inactivating Ty1 PR (pGPR−) (33, 34) or deleting most of PR, and all of the IN and RT coding sequence (pGPOLΔ), but leaving the 3′ LTR intact did not alter the colocalization of mRNA/Gag foci. As expected, Ty1 mRNA and Gag signals in the Ty1-less control strain (DG1768) were indistinguishable from background levels (data not shown) (27). Since POL products are not required for localization of Ty1 mRNA into cytoplasmic foci, Gag-p49 may be the only Ty1 protein required. Our results also support and extend recent work suggesting that formation of mRNA/Gag foci precedes maturation of VLPs (35).
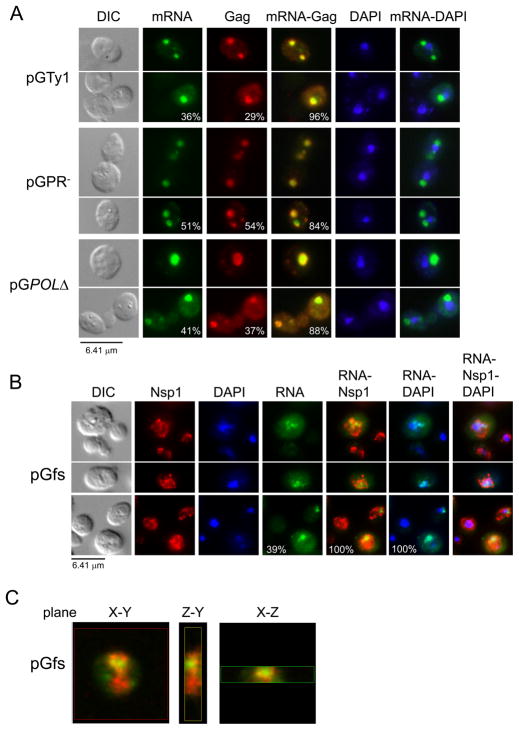
Fig. 2.
Localization of Ty1 mRNA into discrete cytoplasmic foci requires Gag. Cells lacking chromosomal Ty1 elements but containing wild type or mutant pGTy1 plasmids were induced with galactose for 16 hr at 20°C and visualized by differential interference microscopy (DIC). Ty1 mRNA and Gag were detected by FISH-IF, and DNA was stained with DAPI. Ty1 mRNA was detected using a GAG-DIG probe and Gag was detected with anti-Ty1 VLP antiserum. (A) Ty1 mRNA and Gag colocalized in discrete cytoplasmic foci in cells expressing pGTy1 (wild type), and mutants pGPR- and pGPOLΔ. The merge of Ty1 mRNA/Gag and Ty1 mRNA/DAPI stains is shown. Percentages of cells from the same population containing cytoplasmic Ty1 mRNA and Gag foci, and the propensity of these to colocalize are indicated. (B) Nuclear retention of Ty1fs RNA. Anti-Nsp1p antibody was used to delineate the nuclear envelope. The merges of Ty1 RNA/Nsp1p/DAPI are on the right. Percentages of cells containing Ty1 RNA foci, Ty1 RNA foci within the boundaries of Nsp1p staining, and the percentages of Ty1 RNA/DAPI overlap are shown. Only Nsp1p positive cells were counted. (C) Nuclear localization of Tyfs RNA observed by confocal microscopy. The X-Y, Y-Z, and X-Z planes for one focal point are shown. Ty1fs RNA is shown in green and the nuclear envelope is in red.
Ty1fs RNA remains in the nucleus in the absence of Gag
To examine if Gag is required for the localization of Ty1 mRNA into cytoplasmic foci, a single base was deleted adjacent to the GAG start codon in pGTy1 to create pGfs. This mutant produces full-length transcript (Ty1fs RNA) upon galactose-induction but does not synthesize detectable levels of Gag or Pol proteins (J. Wang and D.J. Garfinkel, unpublished results), and contains a premature termination codon (31). A Ty1-less strain containing pGfs was induced with galactose for 16 hr and subjected to FISH-IF analysis using a GAG-DIG probe, and antisera against VLPs and the nuclear pore component Nsp1p (36), which outlines the nuclear envelope (Fig. 2B). Thirty-nine percent of the cells contained a Ty1fs gRNA focus, which localized within the boundaries of the Nsp1p stain and colocalized with the nuclear stain DAPI. The Ty1fs RNA signal appeared fainter compared to Ty1 mRNA, which is consistent with previous work showing that the level of Ty1fs RNA decreases when expressed from its native promoter (31). As expected, there was only a background signal detected with the anti-VLP antiserum (data not shown). Confocal microscopy of these samples showed that Ty1fs mRNA localized within the nucleus (Fig. 2C, Sup. Movie 1). Interestingly, FIV and HIV-1 gRNA also localize at the nuclear rim independent of Gag or a packaging signal (37).
Degradation and nuclear retention of Ty1fs RNA
To determine the level of Ty1fs RNA expressed from pGfs and identify the pathways used to degrade this abnormal transcript, total RNA from isogenic wild type and mutant strains defective in RNA degradation were subjected to Northern analysis (Fig. 3, Sup. Fig. 1, Sup. Table 1). The cells were induced for pGTy or pGfs expression for 2 and 16 hr to recapitulate the level of Ty1 gRNA observed in S. cerevisiae (21), and monitor changes in RNA level that may be dependent on the length of induction. Signals were normalized to ACT1 (Actin) unspliced and spliced transcripts. As expected for a transcript containing a premature termination codon, the level of Ty1fs RNA decreased 5–10 fold in wild type cells induced for 2, 4, 8 or 16 hr (Sup. Fig. 1). To quantify the difference in Ty1fs RNA stability, the level of the Ty1 mRNA and Ty1fs transcripts were determined after culturing cells in 2% glucose for 1 hr following induction for 16 hr (Sup. Fig. 2), a treatment that quickly represses GAL1 transcription The Ty1 and Tyfs transcripts decreased 1.25- and 12.5-fold, respectively, relative to the level of ACT1 mRNA after glucose shut-off. Together, these results show that Ty1fs RNA is unstable when compared with the long-lived wild type transcript (25), and suggests that Gag may enhance the stability of Ty1 mRNA.
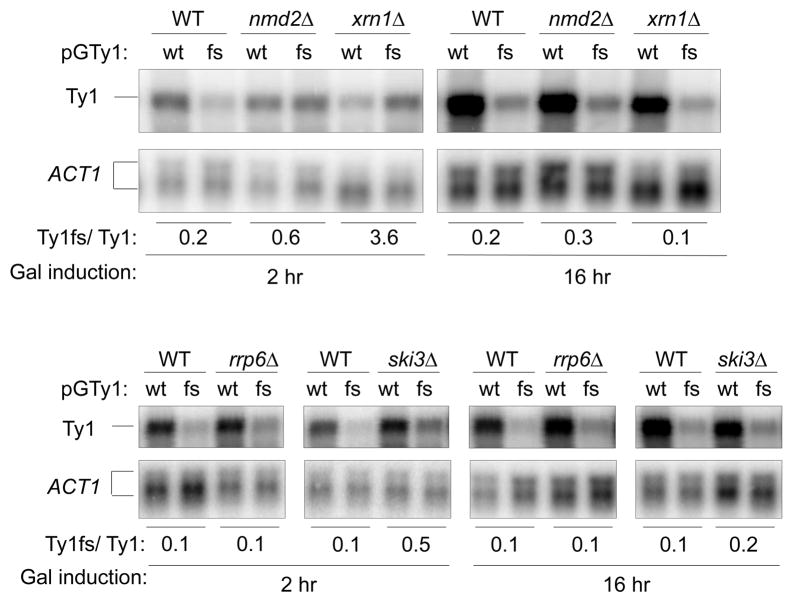
Fig 3.
Level of wild type Ty1 (WT) and Ty1fs (fs) RNAs in nmd2Δ, xrn1Δ, rrp6Δ, and ski3Δ mutants. Total RNA was extracted from wild type (+) and mutant (Δ) strains after galactose induction for 2 and 16 hr. RNA was analyzed by Northern blot using 32P-labeled Ty1 and ACT1 riboprobes. Both the unspliced and spliced ACT1 transcripts were quantified. The Ty1/ACT1 ratios were normalized to the wild type control (Sup. Table 1), which is considered to be 1. The relative amount of Ty1fs RNA/Ty1 mRNA is shown at the bottom.
We examined mutants defective in nonsense-mediated decay (NMD; nmd2Δ), the cytoplasmic (ski3Δ) or nuclear exosome (rrp6Δ), and the P-body (xrn1Δ) for rescue of Ty1fs RNA when compared with Ty1 mRNA. Rescue was observed at 2 hr of galactose induction in the nmd2Δ, xrn1Δ, and ski3Δ mutants, but not in rrp6Δ(Fig. 3, Sup. Fig. 1A). However, the relative amount of Ty1fs RNA decreased with the length of galactose induction in the nmd2Δ, ski3Δ, and xrn1Δ mutants. For example, Ty1fs RNA accumulation reached a maximum by 2 hr in the nmd2Δ mutant, and then decayed to the level present in the wild type NMD2 strain (data not shown, Sup. Fig. 1B). The time-dependent difference in RNA decay suggests the possibility that each pathway degrades Ty1fs RNA at a different rate, a property that has been observed for other transcripts (38). In agreement with previous work (39, 40), exported Ty1fs RNA is recognized as an aberrant transcript containing a premature termination codon by NMD and then is targeted to the P-body for destruction. Also, Ty1fs RNA apparently is degraded by the cytoplasmic exosome (Fig. 3). The lack of rescue observed in the rrp6Δ mutant suggests that Ty1fs RNA is not recognized as an aberrant transcript by the nuclear exosome. The level of wild type Ty1 mRNA remained about the same in all the mutants except the P-body exonuclease mutant xrn1Δ(Fig. 3, Sup. Table 1) Surprisingly, the level of wild type Ty1 mRNA decreased 10-fold while Ty1fs RNA increased 3-fold after 2 hr of galactose-induction in the absence of XRN1, which resulted in an apparent overestimate of the rescue. One possibility is that the increased amount of a Ty1 or possibly a GAL1 noncoding transcript observed in an xrn1Δ mutant represses GAL1-promoted Ty1 transcription via modifications in chromatin (41–43). However, this view fails to explain the increased amount of Ty1fs RNA in the xrn1Δ mutant, which probably affects Ty1fs RNA stability more than its synthesis. Together, these results also raise the possibility that inefficient nuclear export of Ty1fs RNA occurs throughout galactose induction.
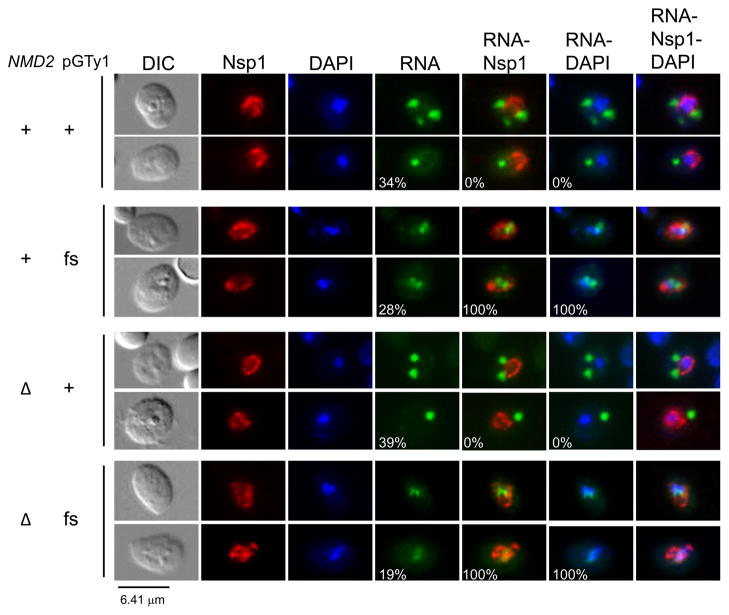
Since Ty1fs RNA is recognized by NMD, which occurs in the cytoplasm (40), we examined the localization of Ty1 RNA in NMD2 wild type and nmd2Δ mutant strains after 16 hr of induction (Fig. 4). In the NMD2 strain, similar percentages of cells with cytoplasmic Ty1 mRNA (34%) or nuclear Ty1fs RNA (28%) foci were observed. Wild type Ty1 mRNA localized in foci in 39% of nmd2Δ mutant cells, and the foci were cytoplasmic. Ty1fs RNA foci were detected in 19% of nmd2Δ cells. However, these foci remained nuclear, since they colocalized with DAPI-stained material and were within the boundaries of the nuclear pore marker Nsp1p. In addition, comparable percentages of cells with nuclear Ty1fs RNA foci were observed in the presence (28%) or absence (19%) of NMD. Therefore, the failure to detect cytoplasmic Ty1fs RNA foci in an nmd2Δ mutant is not due to RNA turnover, but rather is associated with the absence of Gag.
Fig. 4.
Nonsense Mediated Decay does not affect nuclear retention of Ty1fs RNA. Ty1 RNA and Gag localization in an nmd2Δ mutant strain containing either wild type pGTy1 or pGfs were analyzed by FISH-IF. DNA was stained with DAPI, Ty1 RNA was detected using a GAG-DIG probe, and Nsp1p was detected with anti-Nsp1p antiserum. Representative images illustrating the percentages of cells containing Ty1 RNA foci (cytoplasmic or nuclear) and overlaps between Ty1 RNA-Nsp1p and Ty1 RNA-DAPI, are shown.
Gag restores Ty1fs RNA level and nuclear export
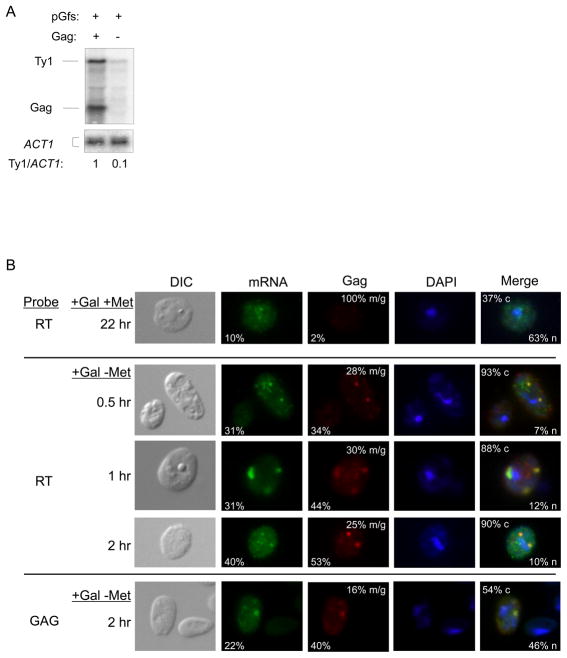
We developed a system to distinguish Ty1fs RNA from mRNA encoding Gag-p49 (Fig. 5). This system consists of expressing Ty1fs RNA from a URA3-based pGTy1 plasmid and Gag from the POLΔ mutant using the MET25 promoter on a TRP1-based vector (pMPOLΔ). Thus, Ty1fs RNA expression is induced in the presence of galactose, whereas Gag mRNA expression from pMPOLΔ is repressed when methionine is present. These plasmids were introduced into a Ty1-less strain and expressed either concomitantly or sequentially. To determine if the expression of Gag in trans influences the level of full-length Ty1fs RNA (Fig. 5A), Northern analysis was performed with total RNA from cells expressing both pGfs and pMPOLΔ or pGfs and an empty vector for 48 hr, and a probe that detected both transcripts. The level of Ty1fs RNA increased 10-fold in the presence of Gag and was similar to the level of GAG transcript. Given that Ty1fs RNA is unstable (Sup. Fig. 2), it is very likely that Gag restores Ty1fs RNA level by preventing its degradation, although early work suggested the possibility that Gag overexpression might stimulate endogenous Ty1 transcription (44).
Fig. 5.
Gag provided in trans restores Ty1fs mRNA level, nuclear export, and formation of cytoplasmic mRNA/Gag foci. (A) Northern analysis of Ty1fs RNA in the presence or absence of Gag expressed from pMPOLΔ. Total RNA was extracted from strains expressing pGfs (expresses Ty1fs RNA) and Gag (pMPOLΔ: expresses a transcript encoding Gag) or pGfs and an empty vector (−) induced with galactose for 48 hr. RNA was hybridized with 32P-labeled riboprobes corresponding to Ty1 GAG and ACT1. Note that the Ty1 GAG probe will detect both the Ty1fs RNA (5.7 kb) and GAG mRNA (2 kb) from pMPOLΔ. Ty1/ACT1 ratios were estimated as described in Fig 4. (B) Ty1fs RNA dynamics when Gag is provided in trans. Cells containing pMPOLΔ and pGfs were grown in media containing galactose (+Gal) and methionine (+Met) to induce pGfs only, or lacking methionine (+Gal − Met) to induce both plasmids, for indicated time lengths prior to FISH-IF analysis. Ty1 mRNA was detected with either GAG-DIG or RT-DIG probes as indicated. Note that the GAG-DIG probe will detect both Ty1fs mRNA and truncated POLΔ mRNA, while the RT-DIG probe will only detect Ty1fs RNA (Fig. 1B). Gag was detected with an anti-Ty1 VLP antiserum. Representative images shown illustrate the percentages of cells containing Ty1 mRNA or Gag foci, propensities for mRNA/Gag (m/g) overlap, and percentages of cytoplasmic (c) or nuclear (n) localization of Ty1 mRNA.
Does coexpression of Gag in trans also restore nuclear export and formation of mRNA/Gag foci (Fig. 5B)? We can distinguish which transcript is being expressed based on the probe used for FISH. The GAG-DIG probe detects both full-length Ty1fs and truncated POLΔ transcripts, whereas the RT-DIG probe only detects the Ty1fs mRNA (see Fig. 1B). Cells were first grown in medium containing galactose and methionine for 22 hr to induce only the transcript from pGfs and then analyzed by FISH-IF. Ty1fs RNA was detected using an RT-DIG probe. Under these conditions, induction of pGfs led to 10% of cells containing Ty1fs RNA, out of which 67% contained RNA with nuclear retention. Only 2% of the cells contained Gag foci, which always colocalized with cytoplasmic RNA foci. These Gag foci probably result from leaky expression of the MET25 promoter in pMPOLΔ even in the presence of methionine. This indicates that repression of pMPOLΔ by methionine was not complete but was sufficient to minimize nuclear export of Ty1fs RNA by Gag in most cells.
To allow expression of both plasmids, cells were washed and grown in medium containing galactose but lacking methionine for a period of 30 min to 2 hr (Fig. 5B). At designated time points, cells were analyzed by FISH-IF using either an RT-DIG or GAG-DIG probe and anti-VLP antiserum. Between 30 min and 2 hr in the absence of methionine, Gag was detected in 34–53% of the cells. FISH-IF analysis with the RT-DIG probe detected Ty1fs RNA in 31–40% of cells. In more than 88% of these cells Ty1fs RNA now localized into cytoplasmic foci. Colocalization of Ty1fs RNA with Gag was observed 25–30% of the time. Ty1fs RNA remained associated with the nucleus in only 7–12% of cells containing Ty1 RNA. These results show that Gag-p49 can act in trans to facilitate Ty1 RNA nuclear export. Failure to completely export Ty1fs RNA may be caused by lower expression from the MET25 promoter relative to expression from GAL1, or possible defects in the export function of the Gag-p49 precursor relative to Gag-p45. When FISH-IF was performed using the GAG-DIG probe at 2 hr, Ty1 RNA from both pGfs and pMPOLΔ was present in foci in 22% of cells. Gag was detected in 40% of cells with 16% mRNA/Gag colocalization. At 2 hr, we observed 90% cytoplasmic localization of Ty1fs RNA when detected with the RT-DIG probe. However, when RNA was detected with the GAG-DIG probe, we observed only 54% cytoplasmic localization of Ty1 RNA in cells containing detectable transcripts. This suggests that Gag may prefer to export the full-length Ty1fs RNA rather than the truncated POLΔ transcript, which could be due to a sequence within POL or the RNA structure formed when POL is present. Taken together, these results suggest that Gag-p49 alone is necessary and sufficient to enhance nuclear export of Ty1 mRNA and form cytoplasmic mRNA/Gag foci.
Nuclear association of endogenous Gag in a conditional mex67 mutant and wild type cells
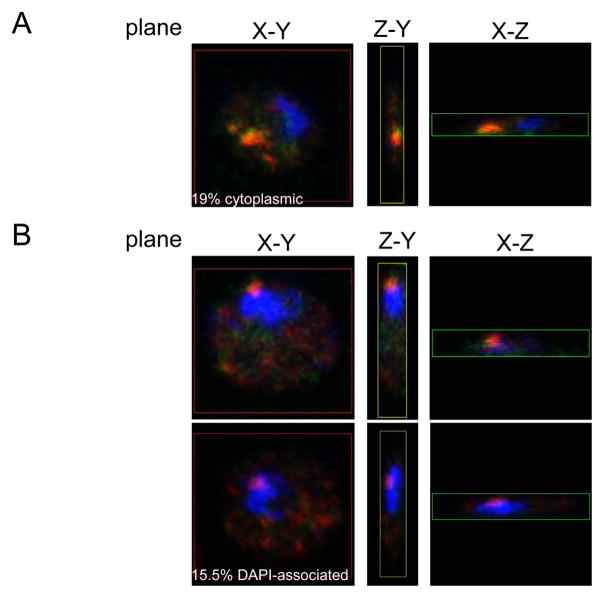
Our results suggest that either Gag itself or some other Gag-dependent RNA binding protein may shuttle between the nucleus and the cytoplasm to promote Ty1 mRNA export, yet Gag is not present in the nucleus at steady state (26–28, 35). To address the possibility that the nuclear localization of Ty1 Gag is only transient, we analyzed endogenous Ty1 mRNA and Gag localization in S. cerevisiae cells containing a mex67-5 temperature-sensitive allele, since MEX67 is required for Ty1 mRNA nuclear export (26). Two experiments were performed in an attempt to trap Gag in the nucleus. Both experiments required a growth temperature that minimized RNA export while maintaining mRNA/Gag foci, since mex67-5 cells become increasingly deficient in export at temperatures above 30°C, while mRNA/Gag foci begin to disperse into smaller puncta at 30°C (27). In the first experiment, cells were grown for 20 hr at 20°C, and then incubated for 1–2 hr at 30°C. Fixed cells were analyzed by FISH-IF and confocal microscopy (Fig. 6). Fifty-eight cells were observed to have three different localizations for Gag: 1) cytoplasmic foci (Fig. 6A, 19%: 11/58 cells), 2) scattered cytoplasmic puncta or no signal (data not shown) (65.5%: 38/58 cells), and 3) Close proximity to DAPI stained material (Fig. 6B, 15.5%: 9/58 cells). Ty1 Gag, mRNA and DAPI stains were also visualized at multiple focal planes (0.2 or 0.25 μm intervals) in selected cells to generate a three-dimensional reconstruction. Analysis of individual planes of each Gag focus showed Gag associated with the periphery or possibly within the DAPI stained material (Fig. 6B). Gag was usually observed associated with Ty1 mRNA regardless of its localization. However, a similar staining pattern was observed in an isogenic MEX67 wild type strain (data not shown).
Fig. 6.
Mex67-5 cells exhibit perinuclear mRNA-Gag foci. Temperature-sensitive mex67-5 cells were grown for 20 hr at 20°C and then shifted to 30°C for 1–2 hr. Cells were processed for FISH-IF and images were taken with a confocal microscope. Ty1 mRNA was detected with an RT-DIG probe (shown in green) and Gag was detected with an anti-Ty1 VLP antiserum (shown in red). DNA was stained with DAPI (shown in blue). Representative images of the X-Y, Y-Z, and X-Z planes for one focal point of each cell with a type of Gag localization: (A) cytoplasmic, and (B) association with DAPI stained material.
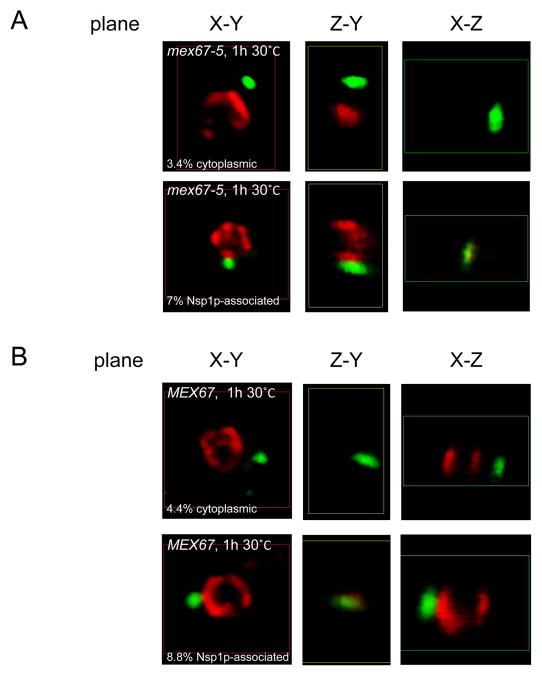
To confirm and extend the results obtained with DAPI staining to delineate the nucleus, cells were grown and analyzed under comparable conditions using Nsp1p and VLP antisera (Fig. 7, Sup. Movie 2). To distinguish background signal from Gag foci, imaging was performed alongside a Ty1-less strain (DG1768), which likely resulted in an underestimate of the number of cells containing Gag foci. Ty1 Gag and Nsp1p were visualized at multiple focal planes (0.25 μm intervals), to generate a three-dimensional reconstruction. Three Gag localization patterns were observed: 1) cytoplasmic foci, which were distinctly separate from the Nsp1p stain (Fig. 7A, top panels, 3.4%: 22/655 cells), 2) scattered cytoplasmic puncta or no signal (data not shown, 89.6%: 587/655 cells), and 3) Nsp1p-associated foci, which were adjacent or overlapping with Nsp1p (Fig. 7A, bottom panels, 7%: 46/655 cells). Although Ty1 Gag foci were present in close association with the nuclear envelope, we were unable to detect Gag within the nuclear envelope. When an isogenic MEX67 wild type strain was analyzed under identical conditions, we observed a similar percentage of cells with cytoplasmic (4.4%: 16/363 cells) and Nsp1p-associated Gag foci (8.8%: 32/363 cells; Fig. 7B), which reinforces the analysis described above using DAPI staining. In addition, wild type cells continuously grown at the permissive temperature displayed the same pattern as those shifted to 30°C (data not shown). Therefore, Ty1 Gag appears to collect near the nuclear envelope in 8% of mitotic cells, whether or not Ty1 mRNA export is inhibited. Although an association between Ty1 Gag and the nuclear envelope was not detected in previous studies (26–28), it is probably due to use of a DNA stain as a nuclear marker, rather than a marker for the nuclear envelope or nuclear pores. Similar localization has been reported for Ty3 Gag overexpressed from the GAL10 promoter, but these were shown to consist of assembled VLPs at the nuclear pore (45). This is unlikely to be the case here, because VLPs are rarely observed by electron microscopy when chromosomal Ty1 elements are expressed from their endogenous promoters (24, 27). Our work supports the idea that Gag, prior to assembling into VLPs, can associate with the nuclear envelope and stabilize Ty1 mRNA as it exits the nucleus.
Fig. 7.
Mex67-5 and wild type cells exhibit Gag foci in close association with the nuclear envelope. Temperature-sensitive mex67-5 cells (A) and isogenic MEX67 cells (B) were grown for 20 hr at 20°C and then shifted to 30°C for 1 hr. Cells were processed for IF and images were taken with a confocal microscope. Ty1 Gag was detected with an anti-Ty1 VLP antiserum (shown in green) and Nsp1p was detected with anti-Nsp1p antiserum (shown in red). Representative images of the X-Y, Y-Z, and X-Z planes for one focal point of each cell were assembled in MIPAV. Examples of Gag localization in the cytoplasm or associated with Nsp1p are shown for each strain.
The nuclear export of specific unspliced mRNAs using a virally encoded nucleocytoplasmic shuttle protein has been described for several retroviruses (37, 46–50). With this mechanism, the shuttle protein contains a nuclear localization signal (NLS), a nuclear export signal (NES), an RNA-binding domain, and domains that bind karyopherins (e.g. importins and exportins), which are cellular factors involved in nucleocytoplasmic translocation. For example, RSV Gag possesses NLSs in its nucleocapsid and matrix (MA) domains, which can mediate nuclear entry in association with importin family members Imp-α and Imp-11 (16, 19, 49, 51). Once inside the nucleus, RSV Gag or MA binds unspliced gRNA (16, 52), which may promote Gag dimerization. The combination of RNA-binding and Gag-multimerization unmasks an NES present on the p10 domain of Gag (18, 53), resulting in export of mRNA/Gag complexes. In addition, binding of gRNA with Gag inhibits interactions with importins, and the viral RNA packaging site stimulates Gag-Crm1p interaction, which is necessary for nuclear export (16). Thus, the gRNA may regulate the ordered association of host import and export functions that mediate gRNA/Gag trafficking and viral assembly.
In budding yeast retrotransposons, mutations in Ty3 nucleocapsid (NC) that disrupt binding of Gag3 to Ty3 RNA lead to the accumulation of Gag3 in the nucleus (54). Furthermore, the Schizosaccharomyces pombe Tf1 Gag protein contains an NLS that is required for both transposition and an interaction with the nuclear pore protein Nup124p (55). Previous work indicates that Ty1 Gag is localized to the cytoplasm as mRNA/Gag foci or VLPs (26, 27, 35). At present, an NLS has not been identified in Ty1 Gag. Although Gag contains a putative NES (L-TF-L-YNT-F-Q-I between residues 217 and 226) that conforms to the Class 1D consensus for yeast (56), alanine scanning mutagenesis of this region compromised Ty1 transposition but failed to block nuclear export of Ty1 mRNA, Gag, or formation of mRNA/Gag foci (data not shown). Nuclear export of a model protein containing a NLS fused to GFP (56) also failed when the candidate Gag NES was added to the protein.
Since nuclear association of Ty1 Gag was not observed using the two-plasmid expression system, we hypothesized that Gag may have a transient nuclear localization. Our results suggest that Gag collects at the nuclear periphery, and enhances Ty1 mRNA export using host machinery. Given that Mex67p is required for Ty1 mRNA export (26), the Mex67p pathway may mediate export of mRNA/Gag complexes.
Although we could not show that Ty1 Gag enters the nucleus or contains a NES, other proteins involved in the nuclear export of RNA also exhibit a predominantly cytoplasmic localization. For example, Dbp5p, a DEAD-box ATPase in yeast localizes mainly to the cytoplasm and the cytoplasmic face of the nuclear pore (57), is involved in remodeling of RNPs prior to release from the nuclear pore and is required for normal export of poly(A) RNA (58–60). Although steady-state localization of Dpb5p is cytoplasmic, defects in the Ran or Crm1p pathways result in visual nuclear localization of Dbp5p (61). A human DEAD-box protein, DDX3, which is involved in Rev-mediated nuclear export of HIV-1 gRNA, localizes to the cytoplasm and the nuclear periphery, because it is constitutively exported by CRM-1 (62, 63). Hence, DDX3 is only detected in the nucleus when CRM-1 is inhibited by leptomycin B. Similar results are also observed for RSV (19) and FIV Gag (50), with FIV Gag reported to localize to the nuclear periphery prior to the leptomycin B studies (37). Although FIV Gag nuclear shuttling is CRM1-sensitive, it seems to function independently of a classical NES (50). Thus, Ty1 Gag may also traffic into the nucleus without a classical NES, but we have yet to visualize nuclear-localized Gag. In addition, further examination by confocal microscopy of a possible overlap of Gag foci with the nucleus in xrn1Δ and lsm1Δ P-body mutants (27), reveals that Gag is present at the nuclear periphery (Checkley and Garfinkel, unpublished data).
What role does the nuclear interaction between Ty1 gRNA and Gag play during the process of retrotransposition? Like RSV and FIV gRNA, Ty1 gRNA is likely exported from the nucleus independently of Gag and undergoes multiple rounds of translation. As the level of Gag increases, which occurs during induction of pGTy1, it promotes export of Ty1 gRNA and may direct the transcript to sites of VLP assembly. In addition, Gag likely stabilizes Ty1 mRNA since Ty1fs RNA is unstable and coexpression of Gag restores Ty1fs RNA level. Nonetheless, early interactions between Ty1 gRNA and Gag immediately after nuclear export may facilitate the accumulation of cytoplasmic mRNA/Gag foci, which are sites where VLPs cluster and may be involved in VLP assembly (27). Although it remains to be determined where HIV-1 gRNA first interacts with Gag in the cytoplasm (17), an emerging theme from Ty1, RSV, foamy virus, and FIV interactions raise the possibility that HIV-1 Gag may interact with its gRNA soon after export.
In this study, we have shown that Ty1 Gag enhances Ty1 mRNA accumulation, nuclear export, and localization of this RNA into cytoplasmic foci. The development of a two-plasmid expression system in the Ty1-less strain allowed us to separate the expression and detection of a full-length Ty1 mRNA from that of Gag in the same cell. This dual-plasmid system will be useful to help define the domains in Ty1 mRNA and Gag that interact and whether these sequences affect additional steps in the Ty1 replication cycle. This system also could be used to identify host cofactors involved in Gag-mediated Ty1 mRNA export and further our understanding of similar processes that occur with retroviral gRNAs.
Materials and Methods
Yeast strains, plasmids, and growth conditions
Standard yeast genetic and microbiological procedures were used (64, 65). The Ty1-less S. paradoxus strains DG1768 (MATα his3-Δ200hisG ura3 gal3 Spo−) (32), and DG2204 (MATα his3-Δ200hisG ura3 trp1) were used in this work. Strain DG1768 was transformed to uracil prototrophy with plasmids pGTy1, pGPOLΔ, pGPR−, and pGfs. DG2204 was transformed with both pBDG1130 (pGfs) and pMACB36 (pMPOLΔ). pGTy1 (pGTy1H3Cla) contains an active Ty1 element fused at its transcriptional initiation site to the GAL1 promoter (66). pGPOLΔ(pBDG1130, pGTy1pol-Δ1702-5562) contains a deletion between BglII restriction enzyme (RE) sites, which are in PR and adjacent to the stop codon in POL, that removes most of POL (deletion of 1702-5562) (31). pGPR− (pJEF1266, pGTy1pr-1702SacI) contains an in-frame SacI linker (codons S-E-L-G) inserted into the PR domain, which renders PR enzymatically inactive (33, 34). pGfs (pBDG1112, pGTy1gag-ATGfs) had a single A nucleotide deleted adjacent to the start codon in GAG, which creates a -1 frameshift mutation and a BamHI RE site (31). With the exception of pMACB36, all plasmids are present on 2μ-based multicopy shuttle vectors carrying the URA3 gene. The plasmid pMACB36 has the MET25 promoter in place of GAL1 and is present in a TRP1-based vector. To create pMACB36, the GAL1 promoter was replaced by the MET25 promoter using standard cloning procedures, and then the URA3 marker was replaced by the TRP1 marker by homologous recombination in yeast (67). Briefly, the GAL1 promoter from pBDG1130 was removed by EcoRI and XhoI RE digest. The MET25 promoter was amplified from S. cerevisiae strain BY4742 using the following primers pMET25-F616-EcoRI and pMET25-R1000-XhoI (Sup. Table 2). This PCR fragment was then ligated into the cut BDG1130 vector to make plasmid pMACB33. To replace the URA3 marker for the TRP1 marker, pMACB33 was cut with StuI, which cuts only once in URA3. Homologous recombination between linearized pMACB33 and a PCR fragment containing the TRP1 marker with URA3 flanking sequences was carried out after transformation into strain DG1251 (MATα his3-Δ200 ura3-167 trp1-hisG spt3-101) and Trp+ Ura3− recombinants were obtained. pMACB36 was recovered from a Trp+ Ura3− recombinant by transformation of total yeast DNA into E. coli DH5α. S. paradoxus strain DG1768 deleted for NMD2, SKI3, RRP6, and XRN1 were constructed as previously described (67, 68), using flanking or terminal gene specific primers conserved between S. cerevisiae and S. paradoxus isolates (69) (Sup. Table 2), and tools provided on the Welcome Trust Sanger Institute (http://www.sanger.ac.uk/research/projects/genomeinformatics/sgrp.html) and Saccharomyces Genome databases (http://www.yeastgenome.org/). Correct deletants containing KanMX were verified by PCR and then transformed with pGTy1 or pGfs. DG2204 was transformed with pGfs and pRS424 (TRP1/2μ-vector) (70) or pMPOLΔ. Strains were grown in synthetic complete (SC) media lacking uracil (SC-Ura) containing 2% raffinose (pH 6.5) at 30°C until log phase, then placed in SC-Ura media containing 2% galactose and grown at 20°C for 14–16 hr for microscopic analyses, or 2–16 hr for Northern analyses. For shutting off GAL1 expression, glucose (2% final concentration) was added to a 16 hr galactose culture, followed by incubation at 20°C for 1 hr. For Northern analysis of cells coexpressing pGfs and pMPOLΔ or pGfs and an empty vector, cells were grown in SC-Ura plus 2% raffinose, diluted 25-fold into SC-Ura-Trp-Met plus 2% galactose, and grown for 48 hr at 20°C. For sequential expression of pGTy1fs and pMPOLΔ, cells were first grown in synthetic minimal media (SD) containing histidine (H), leucine (L), lysine (K), 2% raffinose, and 5 mM methionine at 20°C overnight. To first induce pGfs, cells were resuspended in SD+H+L+K containing 2% galactose and 5mM methionine and grown at 20°C for 22 hr. To express pMPOLΔ, cells were washed and resuspended in SD+H+K+L medium containing 2% galactose but lacking methionine, and then grown at 20°C for 2 hr. Isogenic S. cerevisiae strains containing the temperature-sensitive allele mex67-5 (THJY2232; MATa his3-Δ1 leu2-Δ0 trp1-ΔhisG ura3-Δ0 mex67-5) or wild type MEX67 (THJY2210) were kindly provided by F. Malagon and T. Jensen. Cells were grown in SC-Ura containing 2% glucose for 20 hr at 20°C, and shifted to 30°C for 1–2 hr. Cells were fixed at 30°C and analyzed by FISH-IF and confocal microscopy.
Northern analysis
Total RNA was isolated using the MasterPure yeast purification kit (Epicentre Biotechnologies (Madison, WI) (71). Northern blot hybridization and phosphorimage analysis were performed as previously described (31, 72). 32P-labeled riboprobes were synthesized from Ty1 GAG and ACT1 coding sequence using the MAXscript in vitro transcription kit (Applied Biosystems/Ambion, Woodland, TX) and [α-32P]UTP (3000 Ci/mmol) (PerkinElmer, Waltham, MA).
Oligonucleotide probes used for fluorescent in situ hybridization (FISH)
Ty1 mRNA was detected with either a GAG or an RT oligonucleotide labeled at the 3′ end with digoxigenin (DIG)-dUTP using the DIG Oligonucleotide Tailing Kit, 2nd Generation (Roche Applied Science, Indianapolis, IN). The GAG and RT oligonucleotides antisense TyA-322 or antisense RT-4219 (Sup. Table 2), respectively, were synthesized and HPLC-purified by Invitrogen (Carlsbad, CA).
Antibodies used for immunofluorescence microscopy (IF)
Gag was detected using a rabbit anti Ty1-VLP antibody (kindly provided by Alan Kingsman) at dilutions of 1:1000–2000 for IF. Either AF594 (for colabeling with Ty1 RNA) or AF488 (for colabeling with Nsp1) conjugated anti-rabbit (Invitrogen) was used at 1:100–200. DIG-labeled probes were detected using Fluorescein (FITC) conjugated sheep anti-DIG Fab fragment (Roche Applied Sciences) at 1 ng/μl. Nsp1p was detected using a mouse monoclonal antibody from Abcam (Cambridge, MA) at a dilution of 1:250–500. AF555 conjugated anti-mouse (Invitrogen) was used at 1:100–200.
FISH and IF microscopy
The FISH-IF procedure has been described previously (27). For most experiments, 80–200 cells per strain were scored to obtain quantitative data for localization of Ty1 RNA and Gag signal. For confocal analyses, the number of cells scored is mentioned in the text. Signals below background were not considered for quantification. Percentages of Ty1 mRNA and Gag foci were calculated from cells that were stained with Nsp1p antibody and DAPI. Percentage of Ty1 mRNA/Gag colocalization was calculated from cells containing mRNA/Gag overlap over the total number of cells containing Gag foci. Percentage of cytoplasmic localization of Ty1 mRNA was calculated from cells containing at least one visible foci of mRNA in the cytoplasm over the total number of cells containing Ty1 mRNA. Likewise, percentage of nuclear retention of Ty1 RNA was calculated from cells only containing Ty1 mRNA/Nsp1p/DAPI stain overlap over the total number of cells containing Ty1 mRNA. Fluorescence imaging was performed using a Nikon Eclipse E-1000 microscope equipped with a Nikon C-CU Universal condenser, an oil immersion Nikon Plan Fluor DIC 100x objective and a Hamamatsu Orca-ER c4742-95 cooled charge-coupled device (CCD) camera (Hamamatsu, Japan). The Openlab 5.0 software (Improvision, PerkinElmer, Waltham, MA) was used to acquire images and merge fluorescent images to give the composite images shown. For all experiments involving the Nsp1p antibody, image acquisition was carried out using a Zeiss Axio Observer microscope equipped with an AxioCam HSm Camera and analyzed with AxioVision v4.6 software (Carl Zeiss Microscopy, LLC, North America). Exposure times used to capture fluorescent and DAPI images were kept consistent throughout each experiment. Figures were constructed with Adobe Photoshop software (Adobe Systems, San Jose, CA).
Confocal images involving DAPI as a nuclear marker were performed using an Olympus FV1000 confocal microscope (Olympus America Inc., Center Valley, PA) using a 60X, 1.42 NA (numerical aperture), oil objective lens or a Zeiss LSM 510 confocal microscope (Carl Zeiss Microscopy, LLC, North America) using a 63X, 1.42 NA oil objective lens. The x, and y resolutions were 0.33, and z resolution was either 0.2 or 0.25 μm, respectively. The DAPI channel was acquired using a 405 nm laser and collecting emitted light from 420–480 nm. The green channel was acquired with a 488 nm laser and collecting light from 505–550 nm. The red channel was acquired with a 561 nm laser and collecting light above 570 nm. Confocal images involving Nsp1p as a nuclear marker were performed using a Zeiss LSM 510 confocal microscope (Carl Zeiss Microscopy, LLC, North America) using a 63X, 1.42 NA oil objective lens and a z resolution of 0.25 μm. The green channel was acquired with a 488 nm laser and collecting light from 503–530 nm. The red channel was acquired with a 543 nm laser and collecting light above 560 nm. Images from both experiments were processed with MIPAV (Medical Image Processing, Analysis, and Visualization) software (http://mipav.cit.nih.gov). Noise was reduced in the green and red channels in the following way: each 2D slice image was filtered with a median filter using a 3×3 square kernel to reduce salt and pepper noise, each 2D slice image was filtered with a Gaussian filter using a standard deviation of 1 pixel to further reduce noise. Specific cells were cropped from the 3D color image in order to ease visual interpretation of these images. The contrast of the red, green, and blue channels was separately enhanced to clearly show Nsp1p, Ty1 Gag and RNA signals in final images. Intensity profiles along straight lines through Ty1 Gag and RNA signals were displayed in order to determine whether Gag and RNA signals actually colocalized with each other, and were not apparently overlapping because of blurring caused by the spatial resolution of the optical microscope. Gag foci were determined to be DAPI- or Nsp1-associated if Ty1 Gag signal was adjacent to, overlapping, or surrounded by the DAPI or Nsp1p signal.
Supplementary Material
Sup. Fig. 1. Induction of wild type Ty1 (+) and Ty1fs (fs) mRNA in an nmd2Δ mutant. (A) Total RNA was extracted from wild type (+) and nmd2Δ(Δ) strains after galactose induction for 2 (lanes 1–4), 4 (lanes 5–8), 8 (lanes 9–12) and 16 hr (lanes 13–16). RNA was analyzed as described in the Fig. 3 legend. (B) The fold-expression of pGfs RNA normalized to wild type pGTy1 mRNA for NMD2 (diamond) and nmd2Δ(square) strains is plotted against the length of galactose induction in hours.
Sup. Fig. 2. Stability of wild type Ty1 (+) and Ty1fs (fs) transcripts after glucose (Glc) shut off. Total RNA was extracted from wild type strains after galactose induction for 16 hr (lanes 1 and 2), followed by incubation for 1 hr in 2% glucose (lanes 3 and 4). RNA was subjected to Northern analysis as described in the Fig. 3 legend.
Sup. Table 1. Quantification of Northern hybridization signals from wild type, nmd2Δ, xrn1Δ, rrp6Δ, and ski3Δ strains
Sup.Table 2. Oligonucleotide sequences
Three-dimensional reconstruction of Ty1fs RNA (green) and Nsp1p (nuclear envelope marker, red). Ty1fs RNA is found within the boundaries of the nuclear envelope.
Three-dimensional reconstruction of Ty1 Gag (green) and Nsp1p (red).
Acknowledgments
We thank Francisco Malagon and Agniva Saha for helpful discussions, Jeffrey Strathern for sharing laboratory resources, Benjamin Luttge for critical review of the manuscript, and Kimberley Peifley, De Chen and the UGA Coverdell Microscopy Center for technical assistance with the confocal microscopes. This work is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, the University of Georgia Research Foundation, and NIH grant GM095622 awarded to D.J.G. This work is also funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN2612008000001E. J.A.M. is supported by the National Science Foundation Graduate Research Fellowship 1011RH25213. The contents of this publication do not necessarily reflect the views or policies of the Department of Health and Human Services, and mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
References
- 1.Voytas DF, Boeke JD. Ty1 and Ty5 of Sacharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 614–630. [Google Scholar]
- 2.Peterson-Burch BD, Voytas DF. Genes of the Pseudoviridae (Ty1/copia retrotransposons) Mol Biol Evol. 2002;19(11):1832–1845. doi: 10.1093/oxfordjournals.molbev.a004008. [DOI] [PubMed] [Google Scholar]
- 3.Kawakami K, Pande S, Faiola B, Moore DP, Boeke JD, Farabaugh PJ, Strathern JN, Nakamura Y, Garfinkel DJ. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135(2):309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem. 2000;275(25):19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 5.Kenna MA, Brachmann CB, Devine SE, Boeke JD. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18(2):1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore SP, Rinckel LA, Garfinkel DJ. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18(2):1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson C, Malim M. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic. 2006;7(11):1440–1450. doi: 10.1111/j.1600-0854.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- 8.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 9.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106(45):19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6(11):e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5(10):e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeBlanc JJ, Uddowla S, Abraham B, Clatterbuck S, Beemon KL. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virology. 2007;363(2):376–386. doi: 10.1016/j.virol.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogert RA, Lee LH, Beemon KL. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70(6):3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paca RE, Ogert RA, Hibbert CS, Izaurralde E, Beemon KL. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J Virol. 2000;74(20):9507–9514. doi: 10.1128/jvi.74.20.9507-9514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbitt-Hirst R, Kenney S, Parent L. Genetic evidence for a connection between Rous Sarcoma Virus Gag nuclear trafficking and genomic RNA packaging. Journal of Virology. 2009;83(13):6790–6797. doi: 10.1128/JVI.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc Natl Acad Sci U S A. 2010;107(20):9358–9363. doi: 10.1073/pnas.1000304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parent LJ. New insights into the nuclear localization of retroviral Gag proteins. Nucleus. 2011;2(2):92–97. doi: 10.4161/nucl.2.2.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheifele L, Kenney S, Cairns T, Craven R, Parent L. Overlapping roles of the Rous Sarcoma Virus Gag p10 domain in nuclear export and virion core morphology. Journal of Virology. 2007;81(19):10718–10728. doi: 10.1128/JVI.01061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous Sarcoma Virus Gag polyprotein. Proc Natl Acad Sci U S A. 2002;99(6):3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 21.Curcio MJ, Garfinkel DJ. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol. 1992;12(6):2813–2825. doi: 10.1128/mcb.12.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curcio MJ, Hedge AM, Boeke JD, Garfinkel DJ. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1990;220(2):213–221. doi: 10.1007/BF00260484. [DOI] [PubMed] [Google Scholar]
- 23.Elder RT, St John TP, Stinchcomb DT, Davis RW, Scherer S, Davis RW. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):581–591. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- 24.Garfinkel DJ, Boeke JD, Fink GR. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42(2):507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 25.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malagon F, Jensen TH. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28(19):6022–6032. doi: 10.1128/MCB.00684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Checkley MA, Nagashima K, Lockett S, Nyswaner KM, Garfinkel DJ. P-bodies are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol. 2010;30(2):382–398. doi: 10.1128/MCB.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5′ to 3′ mRNA decay factors colocalize with Ty1 Gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol. 2010;84(10):5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Devine SE. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics. 2003;164(3):867–879. doi: 10.1093/genetics/164.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beliakova-Bethell N, Beckham C, Giddings TH, Jr, Winey M, Parker R, Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12(1):94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garfinkel DJ, Nyswaner K, Wang J, Cho JY. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics. 2003;165(1):83–99. doi: 10.1093/genetics/165.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore SP, Liti G, Stefanisko KM, Nyswaner KM, Chang C, Louis EJ, Garfinkel DJ. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast. 2004;21(8):649–660. doi: 10.1002/yea.1129. [DOI] [PubMed] [Google Scholar]
- 33.Monokian GM, Braiterman LT, Boeke JD. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene. 1994;139(1):9–18. doi: 10.1016/0378-1119(94)90517-7. [DOI] [PubMed] [Google Scholar]
- 34.Youngren SD, Boeke JD, Sanders NJ, Garfinkel DJ. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988;8(4):1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malagon F, Jensen TH. T-body formation precedes virus-like particle maturation in S. cerevisiae RNA biology. 2011;8(2):1–6. doi: 10.4161/rna.8.2.14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61(6):979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- 37.Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J Virol. 2010;84(13):6352–6366. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker R. RNA Degradation in Saccharomyces cerevisae. Genetics. 2012;191(3):671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125(6):1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuperwasser N, Brogna S, Dower K, Rosbash M. Nonsense-mediated decay does not occur within the yeast nucleus. RNA. 2004;10(12):1907–1915. doi: 10.1261/rna.7132504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22(5):615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32(5):685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, Nicolas A, Thermes C, Morillon A. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475(7354):114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 44.Roth J, Kingsman S, Kingsman A, Martin-Rendon E. Possible regulatory function of the Saccharomyces cerevisiae Ty1 retrotransposon core protein. Yeast. 2000;16:921–932. doi: 10.1002/1097-0061(200007)16:10<921::AID-YEA588>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 45.Beliakova-Bethell N, Terry LJ, Bilanchone V, DaSilva R, Nagashima K, Wente SR, Sandmeyer S. Ty3 nuclear entry is initiated by viruslike particle docking on GLFG nucleoporins. J Virol. 2009;83(22):11914–11925. doi: 10.1128/JVI.01192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renault N, Tobaly-Tapiero J, Paris J, Giron ML, Coiffic A, Roingeard P, Saib A. A nuclear export signal within the structural Gag protein is required for prototype foamy virus replication. Retrovirology. 2011;8(1):6. doi: 10.1186/1742-4690-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hope TJ. Viral RNA export. Chem Biol. 1997;4(5):335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 48.Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28(8):419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 49.Parent LJ, Gudleski N. Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation. J Mol Biol. 2011;410(4):553–564. doi: 10.1016/j.jmb.2011.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemler I, Saenz D, Poeschla E. Feline Immunodeficiency Virus Gag is a nuclear shuttling rotein. J Virol. 2012 doi: 10.1128/JVI.00692-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J Virol. 2006;80(4):1798–1806. doi: 10.1128/JVI.80.4.1798-1806.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kenney S, Lochmann T, Schmid C, Parent L. Intermolecular interactions between retroviral Gag proteins in the nucleus. Journal of Virology. 2008;82(2):683–691. doi: 10.1128/JVI.02049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ako-Adjei D, Johnson MC, Vogt VM. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J Virol. 2005;79(21):13463–13472. doi: 10.1128/JVI.79.21.13463-13472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larsen LS, Beliakova-Bethell N, Bilanchone V, Zhang M, Lamsa A, Dasilva R, Hatfield GW, Nagashima K, Sandmeyer S. Ty3 nucleocapsid controls localization of particle assembly. J Virol. 2008;82(5):2501–2514. doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dang VD, Levin HL. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol Cell Biol. 2000;20(20):7798–7812. doi: 10.1128/mcb.20.20.7798-7812.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic. 2008;9(12):2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x. [DOI] [PubMed] [Google Scholar]
- 57.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18(15):4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17(9):2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17(9):2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28(5):850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 61.Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999;18(20):5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 63.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119(3):381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 64.Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology. 1991:194. [PubMed] [Google Scholar]
- 65.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1986. [Google Scholar]
- 66.Garfinkel DJ, Mastrangelo MF, Sanders NJ, Shafer BK, Strathern JN. Transposon tagging using Ty elements in yeast. Genetics. 1988;120(2851484):95–108. doi: 10.1093/genetics/120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manivasakam P, Weber SC, McElver J, Schiestl RH. Microhomology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Research. 1995;23:2799–2800. doi: 10.1093/nar/23.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 69.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJ, van Oudenaarden A, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A. 2009;106(37):15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008;178(1):197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup. Fig. 1. Induction of wild type Ty1 (+) and Ty1fs (fs) mRNA in an nmd2Δ mutant. (A) Total RNA was extracted from wild type (+) and nmd2Δ(Δ) strains after galactose induction for 2 (lanes 1–4), 4 (lanes 5–8), 8 (lanes 9–12) and 16 hr (lanes 13–16). RNA was analyzed as described in the Fig. 3 legend. (B) The fold-expression of pGfs RNA normalized to wild type pGTy1 mRNA for NMD2 (diamond) and nmd2Δ(square) strains is plotted against the length of galactose induction in hours.
Sup. Fig. 2. Stability of wild type Ty1 (+) and Ty1fs (fs) transcripts after glucose (Glc) shut off. Total RNA was extracted from wild type strains after galactose induction for 16 hr (lanes 1 and 2), followed by incubation for 1 hr in 2% glucose (lanes 3 and 4). RNA was subjected to Northern analysis as described in the Fig. 3 legend.
Sup. Table 1. Quantification of Northern hybridization signals from wild type, nmd2Δ, xrn1Δ, rrp6Δ, and ski3Δ strains
Sup.Table 2. Oligonucleotide sequences
Three-dimensional reconstruction of Ty1fs RNA (green) and Nsp1p (nuclear envelope marker, red). Ty1fs RNA is found within the boundaries of the nuclear envelope.
Three-dimensional reconstruction of Ty1 Gag (green) and Nsp1p (red).