Abstract
Cell-derived vesicles represent a recently discovered mechanism for intercellular communication. We investigated their potential role in interaction of microbes with host organisms. We provide evidence that different stimuli induced isolated neutrophilic granulocytes to release microvesicles with different biologic properties. Only opsonized particles initiated the formation of microvesicles that were able to impair bacterial growth. The antibacterial effect of neutrophil-derived microvesicles was independent of production of toxic oxygen metabolites and opsonization or engulfment of the microbes, but depended on β2 integrin function, continuous actin remodeling, and on the glucose supply. Neutrophil-derived microvesicles were detected in the serum of healthy donors, and their number was significantly increased in the serum of bacteremic patients. We propose a new extracellular mechanism to restrict bacterial growth and dissemination.
Key Points
Neutrophilic granulocytes stimulated with opsonized particles produce microvesicles (MVs) that are able to impair bacterial growth.
Antibacterial effect correlates with number and size of aggregates between bacteria and MVs and depends on cytoskeletal reorganization of MVs.
Introduction
Cell-derived vesicles (such as exosomes, ectosomes, microvesicles, shedding microvesicles, and microparticles) represent a recently discovered mechanism for cell-cell communication.1–3 Exosomes are small (50-100 nm) vesicles released from multivesicular bodies.4 They are involved in antigen presentation5–7 and cell-to-cell transfer of receptors8 or RNA,9,10 thereby influencing or reprogramming neighboring cells and often promoting tumorigenesis.8,11 Exosomes also play a role in host defense against microorganisms: tracheobronchial epithelial cells produce exosome-like vesicles with antiviral activity,12 and macrophage-derived exosomes are able to transfer pathogen-associated molecular patterns of opportunistic intracellular pathogens to uninfected cells.13 Larger vesicles, called microvesicles (MVs) or microparticles express tissue factor on their surface that is capable of initiating coagulation.14 Both exosomes and MVs of different cellular origin were detected in various body fluids and selective enrichment was related to specific diseases.15–19
Neutrophilic granulocytes (PMNs) play a critical role in innate immune mechanisms by engulfing, killing, and degrading various microorganisms. PMNs produce larger vesicles (named by the authors alternatively as ectosomes, microparticles, or MVs) after incubation with various stimuli.19–22 Microparticles obtained from PMNs stimulated by chemotactic agents or phorbol esters activated cytokine (IL-6) secretion from endothelial cells23 and platelets,24 thereby contributing to the procoagulant effect of leukocyte-derived microparticles.25 Chemotactic peptide-induced PMN-ectosomes increase the secretion of the anti-inflammatory cytokine transforming growth factor β26 and interfere with the maturation of monocyte-derived dendritic cells.27,28 Auto-antibody–induced microparticles were suggested to be involved in the pathogenesis of vasculitis.19 However, potential contribution of PMN-derived MVs to antimicrobial defense has not been investigated.
This study shows that stimulation of peripheral blood PMNs by different agents resulted in the release of MVs of different composition and different functional properties. Importantly, specific MVs were able to reduce the growth of nonopsonized bacteria more efficiently than intact PMNs. The antibacterial effect of PMN-derived MVs was associated with their ability to aggregate bacteria on their surface. PMN-derived MVs were detectable in significantly higher numbers in the serum of bacteremic patients than in healthy donors. MVs from bacteremic patients were able to aggregate bacteria ex vivo. We propose a new extracellular mechanism by which neutrophils contribute to the battle against invading microbes.
Methods
Materials
Saponin was from Merck; cytochrome c, lucigenin, phorbolmyristate acetate (PMA), paraformaldehyde, formyl-methionyl-leucyl-phenylalanin (fMLF), cytochalasin B (CB), jasplakinolide, latrunculin A, wortmannin, and PKH2GL-1KT cell membrane stain from Sigma-Aldrich, CXCL12 from PeproTech, sterile endotoxin-free HBSS from ThermoScientific, Ficoll from Phadia, the blocking anti-CD18 antibody (clone TS1/18) from BioLegend. All other reagents were of research grade. H-medium contained 145mM NaCl, 5mM KCl, 1mM MgCl2, 0.8mM CaCl2, 5mM glucose, 10mM HEPES, pH = 7.4. GFP-expressing S aureus29 was a kind gift of Professor William Nauseef (University of Iowa).
Preparation of MVs from PMNs and human serum
Venous blood was drawn from healthy adult volunteers according to procedures approved by the Institutional Review Board of the Semmelweis University. Neutrophils were obtained by dextran sedimentation followed by Ficoll-Paque gradient centrifugation as previously described.30 The preparation contained more than 95% PMN and less than 0.5% eosinophils. PMNs (typically 4.5 × 106cell in 450 μL HBSS) were incubated with or without activating agent (added in 50 μL) for 20 minutes at 37°C on a linear shaker (80 rpm/min), unless indicated otherwise. After incubation, PMNs were sedimented (500g, 5 minutes, 4°C) and the supernatant was filtered through a 5-μm pore sterile filter (Sterile Millex Filter Unit, Millipore). The filtered fraction was sedimented again (15 700g, 10 minutes, 4°C). The sediment was resuspended in HBSS at the original incubation volume unless indicated otherwise. Protein concentration was determined by the Bradford protein assay using BSA as standard. Serum of healthy donors and bacteremic patients was filtered and sedimented as described. This study was conducted in accordance with the Declaration of Helsinki.
Opsonization of bacteria
Bacteria (9 × 108 in 900 μL HBSS) were opsonized with 100 μL pooled normal human serum for 15 minutes at 37°C. After opsonization, bacteria were centrifuged (5 minutes, 4°C, 5000g), and washed by HBSS. The concentration of bacteria was set to OD600 = 1.0.
Measurement of bacterial growth and superoxide production
Bacterial growth was followed in a plate reader (Labsystems iEMS Reader MF, Thermos Scientific) on the basis of changes in optical density at 650 nm, as described.30 Production of superoxide (O·−) was determined with lucigenin-based chemiluminescence as previously described.30
Microvesicle detection by flow cytometry
CD11b-labeled (see next section) PMNs were activated, and MVs were separated as described. A FACSCalibur analytic flow cytometer (Becton Dickinson) was used. An initial microvesicle-size gate was set with the help of calibrating GFP-expressing bacteria (S aureus, diameter 0.8 μm) and 3.8 μm SPHERO rainbow alignment particles (Spherotech). The absolute count of MVs was measured for 15 s. We defined PMN-derived MVs as particles less than 1.5 μm in diameter and expressing surface marker CD11b.
Fixation and staining of MVs and PMNs
MVs or PMNs were layered on cover slips and fixed with fixation buffer (4% paraformaldehide, 0.1% saponin in HBSS) for 5 minutes at room temperature. When indicated, samples were permeabilized with 0.5% Triton X-100 for 3 minutes at room temperature after fixation. After washing with HBSS, MVs were visualized by R-PE conjugated anti-CD11b monoclonal Ab (1 μg/mL, 30 minutes; Dako), or with FITC-conjugated anti-CD18 monoclonal Ab (1 μg/mL, 30 minutes; Dako). Myeloperoxidase (MPO) was detected with anti-MPO monoclonal Ab (4 μg/mL, 30 minutes; Abcam), and FITC-conjugated anti–mouse goat Fab (4 μg/mL, 30 minutes; Invitrogen) as secondary antibody. F-actin was labeled with Alexa 488–Phalloidin (0.1 wt/vol %, 30 minutes; Invitrogen), phosphatidylserine was stained with FITC-conjugated annexin-V (used as indicated by the manufacturer; Becton Dickinson Bioscience). As nucleic acid marker propidium iodide was used (1 μg/mL, 10 minutes; Invitrogen). On each sample, autofluorescence and isotype antibodies were measured as controls.
Transmission, confocal, and video microscopy
For confocal and video microscopy experiments, Zeiss LSM510 confocal laser scanning microscope equipped with 40 ×/1.3 and with 63 ×/1.3 oil immersion objective (Plan-Neofluar, Zeiss) was used. In video microscopic experiments stage and objective were heated to 37°C and slips were coated with BSA (10 wt/vol %) for 1 hour at room temperature. Images were analyzed with LSM Image Browser software (Zeiss). For transmission microscopy experiments, a Leica DMI6000B inverted microscope was used (Leica Microsystems; objective: 63 ×/1.3 phase contrast, camera: Leica DFC480 CCD). Images were analyzed with ImageJ 1.42 software (NIH). In transmission and confocal microscopy, 9 regions of interest per sample were investigated.
Electron microscopy of MVs
Pelleted MVs were fixed at room temperature for 60 minutes with 4% paraformaldehyde in PBS. The preparations were postfixed in 1% OsO4 (Taab) for 30 minutes. After rinsing with distilled water, the pellets were dehydrated by ethanol, followed by block staining with 1% uranyl-acetate in 50% ethanol for 30 minutes, and embedded in Taab 812 (Taab). After overnight polymerization at 60°C and sectioning for EM, the ultrathin sections were analyzed with a Hitachi 7100 electron microscope equipped by Veleta, a 2k × 2k MegaPixel side-mounted TEM CCD camera (Olympus). Electron micrographs were edited by Photoshop CS3 (Adobe).
Dynamic light scattering
Dynamic light scattering (DLS) measurements were performed at room temperature, using an ALV goniometer with a MellesGriot diode-pumped solid-state laser (CVI MellesGriot) at 457.5-nm wavelength (type 58 BLD 301). The radius of the particles was calculated using sphere approximation.17
Mass spectrometry analysis
Proteins were extracted from MVs using a previously described methanol extraction protocol.31,32 Extracted proteins were digested in solution with trypsin (Promega), tryptic peptides lyophilized, and resuspended in 0.5% acetic acid, as previously described.32 MVs extracts were analyzed with a Thermo LTQ linear ion trap using a previously described 2D-LC-MS/MS approach.32,33 For database searching of tandem mass spectra charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using SequestSorcerer (Sage-N), and a FASTA-formatted human protein database (Human RefSeq, 2007) was searched. Searches were performed with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 1.2 Da. Iodoacetamide derivative of cysteine was specified as a fixed modification. Oxidation of methionine was specified as a variable modification.
Scaffold Version 3.0.00 (Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm.34 Protein identifications were accepted if they could be established at greater than 95.0% probability assigned by the Protein Prophet algorithm.32 Quantitative assessment and comparison of MV protein composition with different stimuli was based on a previously described spectral counting-based approach.33,35
Immunoblotting
MVs were lysed in 4 × Laemmli sample buffer, boiled, run on 10% (wt/vol) polyacrylamide gels and transferred to nitrocellulose membranes. After blocking for 1 hour in PBS containing 5% albumin and 0.1% (wt/vol) Tween 20, blots were incubated with anti–lactoferrin polyclonal antibody in 1:1000 dilution or anti–β-actin mAb (both from Sigma-Aldrich) in 1:10 000 or anti-MPO antibody (Cell Signaling, Danvers) in 1:500 dilution in PBS containing 5% ovalbumin. Bound antibody was detected with enhanced chemiluminescence using horseradish peroxidase–conjugated anti–rabbit-Ig (from donkey) or anti–mouse-Ig (from sheep) secondary antibodies (GE Healthcare) used in 1:5000 dilution.
Results
Effect of various stimulants on MV formation
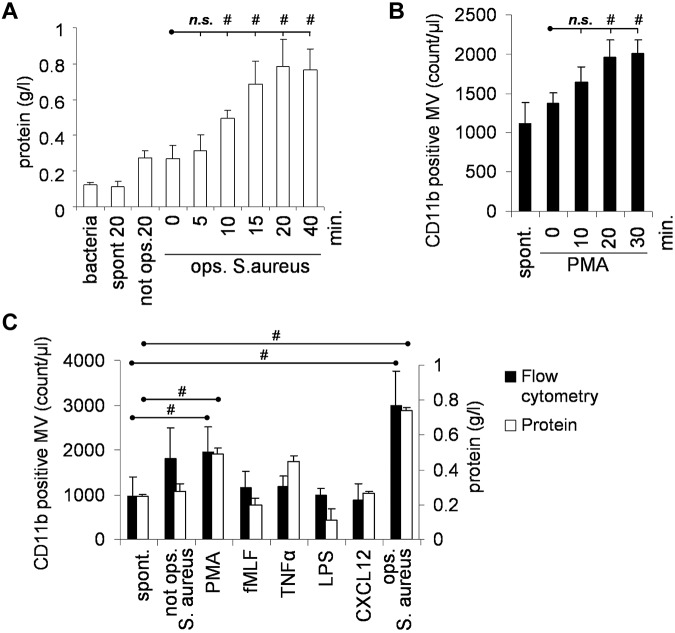
Release of MVs by isolated peripheral blood PMNs was followed by 2 independent methods: on the basis of protein content and by flow cytometry after staining the cells with antibodies against the PMN-marker CD11b. Using 2 different stimuli, we observed by both methods similar kinetics, and maximum MV formation was reached by 20 minutes (Figure 1A-B). In all subsequent experiments, 20 minutes stimulation time was used. We observed spontaneous MV formation in resting cells (referred to as s-MVs). Chemotactic agents (such as the formyl peptide fMLF or the chemokine CXCL12 or lipopolysaccharide [LPS]) did not influence either the number or the protein content of the formed MVs (Figure 1C). The protein kinase C activating pharmacologic agent phorbolmyristate acetate (PMA) induced an approximately 2-fold increase in MV formation (p-MVs). The highest amount of MV formation was induced by incubation of PMNs with S aureus, opsonized with pooled normal human serum (b-MVs). Nonopsonized bacteria did not significantly increase MV formation. Zymosan opsonized with normal serum also enhanced release of MVs from PMNs (data not shown). Formation of b-MVs could be prevented by inhibition of actin polymerization with cytochalasin B, but it was not affected by diphenylene-iodonium (DPI), an inhibitor of the NADPH oxidase, or in the absence of glucose or in the presence of deoxyribonuclease (DNase; not shown).
Figure 1.
Effect of various stimulants on MV formation. Amount of formed MVs was followed on the basis of protein concentration (A-C) or flow cytometry after staining PMNs with anti-CD11b antibody (bacteria were not stained at all). (B-C) Stimulation of 9 × 106/mL PMNs was carried out with 108/mL opsonized S aureus (A-C) or 100nM PMA (B-C) or 1μM fMLF (C) for 20 minutes or 20 ng/mL TNFα for 30 minutes, 100 ng/mL LPS for 120 minutes or 100 ng/mL CXCL-12 for 5 minutes or 108/mL S aureus with or without opsonization (C). In panel A the protein content of bacteria cosedimented with MVs is shown. In panel C and all later figures, this value has been subtracted. Bars show mean ± SEM, n = 4; #P < .05.
The cell count, uptake of vital dyes, morphology by light microscopy, superoxide production, and elimination of bacteria did not differ significantly between PMNs sedimented after b-MV formation and those without any stimulus (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In fact, repeated incubation with opsonized bacteria resulted in repeated formation of b-MVs.
Characterization of PMN-derived MVs
By fluorescence microscopy we observed vesicular structures that could be stained with the fluorescent lipid intercalating dye PKH2GL-1KT (not shown) and with antibodies against CD11b (Figure 2A), CD18 (not shown), the 2 chains of the major neutrophil integrin Mac1 and CD177 (not shown). On treatment of the MVs with detergents or distilled water both lipid and protein staining disappeared, and MVs were no longer detected by flow cytometry (data not shown). These results support the vesicular nature and outside-out orientation of the MV preparation.17 MVs could also be stained with annexin V (Figure 2B), indicating that phosphatidylserine was present in the outer leaflet of PMN-derived MVs, as described for other exosome and microparticle preparations.3
Figure 2.
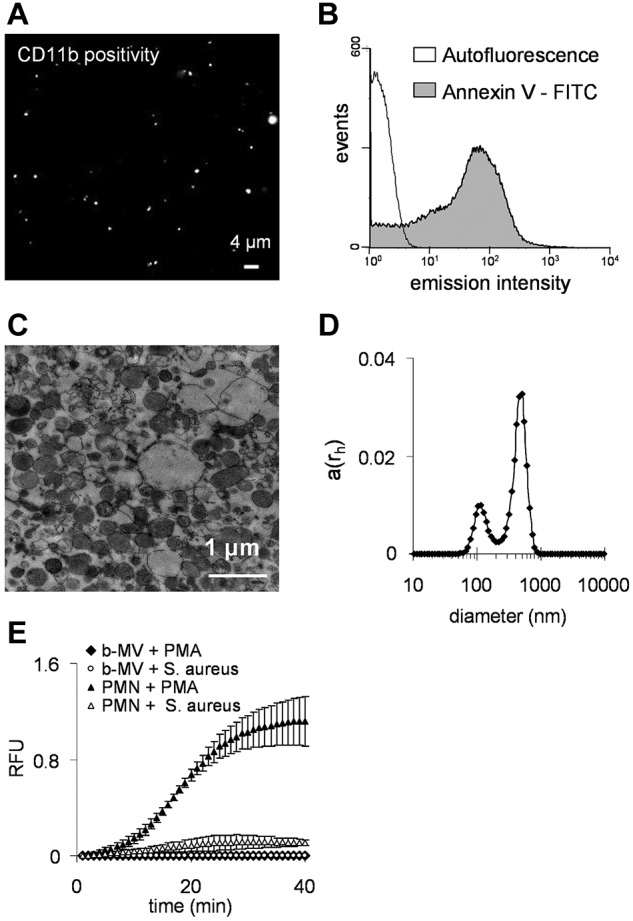
Characterization of PMN-derived MVs. (A) Fluorescence microscopy image of separated MVs. CD11b was marked with anti-CD11b R-PE conjugated monoclonal antibody. Original magnification is 630×. (B) Flow cytometry analysis of binding of FITC-conjugated annexin to phosphatidylserine. (C) Representative electron microscopic image of MVs. Original magnification is 10 000×. (D) Analysis of isolated MVs by dynamic light scattering. The x-axis is set to logarithmic scale, a(rh) denotes the coefficient of autocorrelation function of the scattered electric field. (E) Superoxide production of 106 PMNs or MVs derived from 107 PMNs stimulated by 100nM PMA (dots show mean of measured RFU ± SEM, n = 4).
The size of the MVs was estimated by dynamic light scattering where we identified 2 vesicle populations: a smaller fraction with a diameter around 100 nm, and a major fraction with diameter of approximately 500 nm (range 200-800 nm; Figure 2D). These data were confirmed by electron micrographs (Figure 2C). Thus, our MVs are larger than ectosomes produced by chemotactic stimuli in PMNs,20,21 but correspond in size to microparticles.23,24
In contrast to intact neutrophils, PMN-derived MVs did not produce superoxide on stimulation with PMA or opsonized bacteria (Figure 2E). It is important to note that all these properties of PMN-derived MVs (size, surface properties, and inability to produce superoxide) were independent of the conditions used to stimulate MV formation.
Effect of PMN-derived MVs on bacterial growth
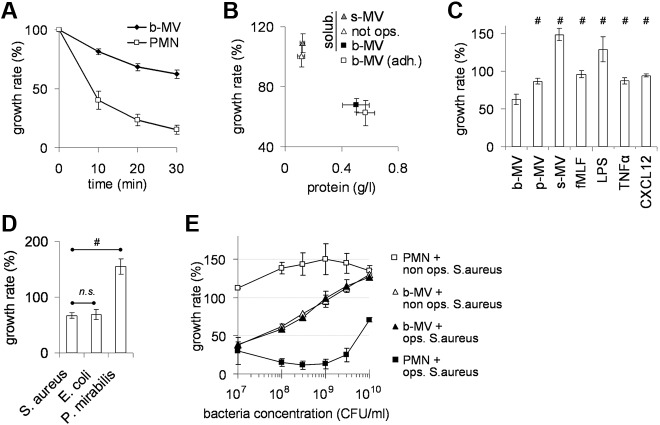
When PMNs were incubated with a tenfold excess of opsonized bacteria, the proportion of proliferating bacteria gradually decreased so that by 30 minutes only approximately 20% of the original number remained (Figure 3A). When the same experiment was carried out with b-MVs formed from an identical number of PMNs, approximately 60% of the original number of bacteria was present after 30 minutes of incubation (Figure 3A). Antibacterial effect of MVs depended on opsonization of the initiating particles but it was independent of the suspended or adherent state of the producing PMNs (Figure 3B). Zymosan opsonized with normal serum also initiated the formation of antibacterial MVs but serum alone was not effective (not shown). Presence of normal serum had no effect on either type of MVs.
Figure 3.
Effect of PMNs derived MVs on bacterial growth. (A) Effect of PMNs (9 × 106/mL) and of b-MVs (derived from the same amount of PMNs) in time on growth of fresh, opsonized S aureus (9 × 107/mL). Points indicate mean ± SEM, n = 47. (B) MVs were collected from equal number of suspended PMNs incubated with opsonized or nonopsonized S aureus or from adherent PMNs incubated with opsonized S aureus. (C) MVs were collected after incubation of PMNs with the indicated agents (concentration and times are described in Figure 1C) and applied at equal protein concentration. (D) Effect of the same amount of b-MVs on the indicated, opsonized bacteria. (E) Antibacterial capacity of 9 × 106/mL PMNs (■) and of b-MVs (▴; derived from the same amount of PMNs) on opsonized (full symbols) and nonopsonized (empty symbols) S aureus. In panels B through E bacterial growth after 30 minutes incubation is shown; 100% represents the initial bacterial count (9 × 107/mL) that did not change in the absence of nutrients, but increased during the 30 minutes incubation period in the presence of nutrients. In panel B points indicate mean of protein concentration and of bacterial growth rate, ± SEM, n = 4; in panels C through E bars indicate mean ± SEM, n = 4; #P < .05.
Next, we investigated bacterial growth in the presence of MVs induced by different stimuli. MVs were applied at equal protein concentration. Figure 3C shows that MVs formed spontaneously (s-MVs) or induced by fMLF, LPS, TNFα, or CXCL12 failed to impair bacterial growth. The incubation time of PMNs with certain stimuli (LPS, CXCL12, or TNFα) was extended up to 1 or 2 hours, but the resulting MVs had neither higher protein content nor developed any antibacterial effect. P-MVs impaired bacterial growth slightly, but there was a statistically significant difference between the antibacterial effect of b-MVs and p-MVs. The antibacterial effect of b-MVs was not limited to S aureus, the bacteria used for initiation of MV formation, as b-MVs impaired the growth of E coli to a similar extent. However, b-MVs were ineffective against Proteus mirabilis (Figure 3D). Thus, PMNs are able to form MVs both spontaneously and on stimulation with different agents. However, only particles (bacteria or zymosan) opsonized with normal serum stimulated the release of MVs that significantly interfere with the growth of selected types of bacteria.
Characterization of the antibacterial effect of b-MVs
We compared the antibacterial capacity of b-MVs and intact PMN at different bacterial loads (Figure 3E). Intact PMNs were able to efficiently eliminate opsonized S aureus up to a 300-fold excess. At a 1:1 ratio of b-MVs (expressed in PMN-equivalent) to bacteria, b-MVs were as effective as PMNs, but their antibacterial capacity gradually decreased as the bacterial load increased. The antibacterial effect of b-MVs did not differ between opsonized and nonopsonized bacteria. This finding is in sharp contrast to the critical role of opsonization in induction of b-MV formation (Figures 1C and 3B). When intact PMNs were incubated with nonopsonized S aureus, bacterial growth was not inhibited. Thus, the efficacy of b-MVs against nonopsonized bacteria exceeded that of intact PMNs at most concentrations of bacteria.
We also tested the effect of various pharmacologic inhibitors and incubation conditions on the antibacterial effect of b-MVs. Treatment of b-MVs with agents interfering with actin polymerization and cytoskeletal structure, such as cytochalasin B, latrunculin A, and jasplakinolide, prevented the antibacterial effect of b-MVs (Figure 4A). Incubation of b-MVs and bacteria in the absence of glucose or in the presence of wortmannin, an inhibitor of phosphatidylinositol-3-kinase also impaired antibacterial activity. Inhibition of β2 integrin function with blocking antibodies against CD18 or in the presence of EDTA significantly reduced the antibacterial effect. On the other hand, inhibition of NADPH oxidase by DPI did not alter the antibacterial activity of b-MVs. On disruption of the vesicular structure by distilled water or detergents, the antibacterial effect of b-MVs completely disappeared (Figure 4A). Filtration through 450-nm pore size filters also resulted in significant diminution of the antibacterial effect of b-MVs (not shown). We interpret these data to indicate that inhibition of bacterial growth by b-MVs depends on intact vesicular and cytoskeletal structure and requires metabolic activity by the MVs.
Figure 4.
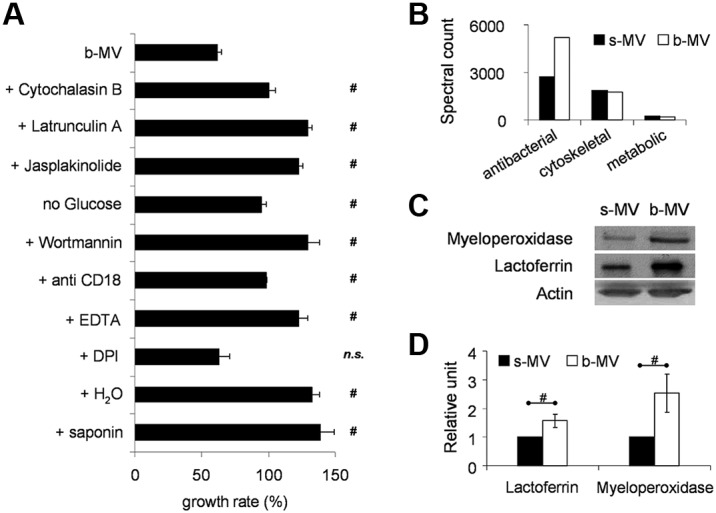
Characterization of the antibacterial effect of b-MVs. (A) Effect of various inhibitors (cytochalasin B 10μM; latrunculinA 10μM; jasplakinolide 1μM; wortmannin 300nM; all added 5 minutes before incubation), absence of glucose or presence of blocking CD18-antibodies (clone TS1/18), or 1mM EDTA or 1μM DPI or pretreatment with distilled water or with saponin (1 mg/mL) on antibacterial effect of b-MVs. Change in S aureus growth was measured after 30 minutes incubation with the indicated sample. 100% represents the initial bacterial count that did not change in the absence of nutrients, but increased during the 30 minutes incubation period when inactive or destroyed vesicles were present. It was controlled that inhibitors in the applied concentration and time as well as absence of glucose by themselves did not affect bacterial growth. Bars and points indicate mean, ± SEM, n = 4; #P < .05. (B) Proteomic analysis of the contents of PMN-derived MVs. Equal amount of total protein was analyzed in the different samples. Proteins were ranked by spectral count.32 (C) Western blot of lactoferrin and myeloperoxidase of s-MVs and b-MVs produced by 2 × 107 PMNs. Actin is shown as loading control. (D) Densitometric analysis of lactoferrin and myeloperoxidase signal related to total protein content of MVs (± SEM; n = 5); #P < .05. See also supplemental Tables 1 and 2.
Protein composition of MVs
The protein composition of MVs formed spontaneously (s-MVs), induced by PMA (p-MVs), or by opsonized S aureus (b-MVs) was determined by proteomic analysis. We identified 282 proteins with 95% confidence from the 3 types of MVs (supplemental Table 1). These proteins were subjected to Ingenuity Pathways Knowledge Base analysis (Ingenuity Systems), as previously described.32 The major biologic processes consisted of cytoskeletal regulation (42 proteins), adhesion (26 proteins), host defense and inflammation (55 proteins), and metabolism, including enzymes of the glycolytic pathway (39 proteins). The cellular localization of the majority of proteins was the cytosol (107 proteins) or plasma membrane (62 proteins), although 28 proteins were primarily associated with granules. Plasma membrane components identified consisted of the 2 chains of the complement receptor CR3 (the integrin Mac-1 composed of CD11b + CD18), FcγRIII, and the 2 membrane-bound chains (gp91phox and p22phox) of the phagocytic NADPH oxidase. Interestingly, the cytosolic components of the oxidase were only detectable in p-MVs but not in b-MVs. Eosinophil peroxidase was either adherent to the anionic surface of MVs or indicates MVs-formation by contaminating eosinophils. Several proteins that were previously described as typical constituents of exosomes, such as tetraspanins (CD63, CD81, CD82, CD9), alix, and TSG101,4 were not identified by this analysis.
The relative amounts of each protein were compared between b-MVs and s-MVs, using spectral counts.33,35 The amount of antibacterial proteins was almost doubled in b-MVs, whereas no difference was observed in the amount of cytoskeletal or metabolic proteins (Figure 4B). Analyzing at the level of individual proteins, 29 proteins were identified from the 100 most highly expressed proteins that were increased by at least 40% in b-MVs, including 11 of the 20 most highly expressed proteins (supplemental Table 2). Analysis of those 29 proteins showed that 15 proteins possessed antibacterial activity and 26 proteins were previously identified in neutrophil granules.36,37 The difference in the amount of lactoferrin and myeloperoxidase (MPO) was verified by Western blotting (Figure 4C-D).
Localization of certain proteins within b-MVs was assessed by comparison of staining of b-MVs before and after permeabilization by Triton X-100. Actin, MPO, and lactoferrin could be visualized on CD11b-positive vesicles only in permeabilized samples, whereas staining of CD11b was not altered after permeabilization. No staining with propidium iodide was detected in nonpermeabilized or in permeabilized MVs (data not shown).
Bacterial aggregation is required for the antibacterial effect of b-MVs
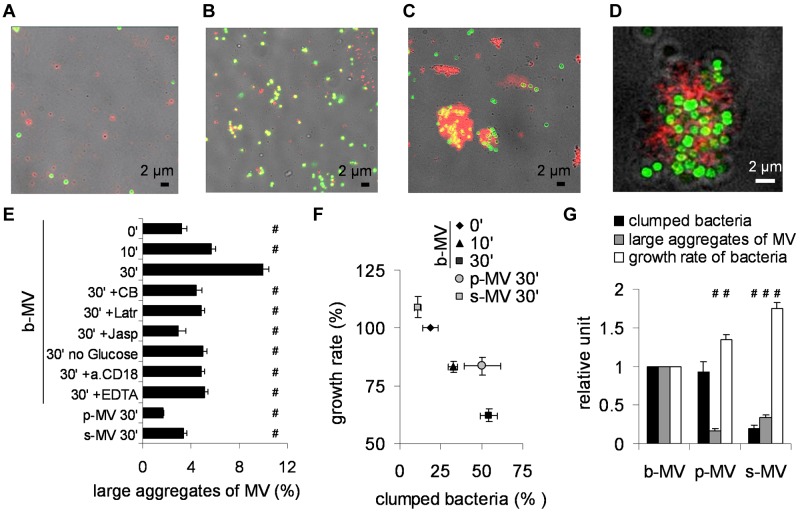
The interaction of b-MVs with bacteria was followed by fluorescence microscopy. Isolated b-MVs were labeled with anti CD11b antibody (red) and S aureus expressed green fluorescent protein. Typical pictures are shown in Figure 5. At the beginning of the incubation period, red b-MVs and green bacteria are largely separated (Figure 5A). After 10 minutes incubation, colocalization of isolated b-MVs and S aureus occurs frequently (Figure 5B), whereas after 30 minutes incubation, large aggregates of b-MVs and bacteria were observed (Figure 5C). In thin confocal microscopic sections of aggregates, the separation of the 2 colors is clearly visible (Figure 5D), indicating that b-MVs did not ingest bacteria. No aggregation could be detected when either b-MVs or bacteria were incubated separately for 30 minutes or b-MVs were pretreated with detergent (not shown). The ratio of large (more than 1.5 μm) aggregates of CD11b-positive b-MVs and bacteria to all CD11b-stained b-MVs increased from 2% at the beginning of incubation to 10% at 30 minutes (Figure 5E). Incubation of s-MVs or p-MVs with bacteria for 30 minutes did not result in any increase of the ratio of large aggregates (Figure 5E). Similar to the effect on bacterial growth (Figure 4A), b-MVs incubated with bacteria in the presence of agents that inhibit cytoskeletal reorganization or β2 integrin function or in the absence of glucose failed to form large aggregates (Figure 5E). A clear negative relationship could be observed between bacterial growth and proportion of large, aggregated b-MVs (supplemental Figure 2A). In addition to S aureus, b-MVs also formed large aggregates with E coli but not with P mirabilis (data not shown).
Figure 5.
Potential mechanism of the antibacterial effect of b-MVs. (A-C) Representative transmission fluorescence microscopy images (of 35 similar ones from 4 experiments) about coincubation of b-MVs with opsonized S aureus at time points 0 (A), 10 minutes (B), and 30 minutes (C). Red marks indicate CD11b-positivity and green marks show GFP-expressing S aureus. (D) Confocal microscopy image of a representative clump (35 areas of interest were investigated from 4 experiments). Z-axis was 1 μm. Red and green marks are the same as described. (E) Statistical analysis of images taken from 4 independent experiments. Means were made on each sample by 3 individual investigators, and averaged. On the x-axis the ratio of bacteria and CD11b-positive, at least 1.5-μm wide aggregates to all CD11b-positive events is presented. Concentration of inhibitors is the same as detailed in Figure 4. Error bars show SEM, n = 4; #P < .05. (F) Relation of growth rate to the ratio of clumped bacteria (bacteria within CD11b-positive environment compared with all bacteria). Error bars represent SEM, n = 4. (G) Graphic representation of the proportion of clumped bacteria, large aggregates, and growth rate of bacteria in the presence of 3 different MV preparations. Error bars represent SEM, n = 4.
To further investigate the relationship of these aggregates to bacterial survival, the percentage of bacteria present in the aggregates was analyzed and related to bacterial growth (Figure 5F, supplemental Figure 2B). Incubation of b-MVs with S aureus for 30 minutes resulted in aggregation of more than 55% of all bacteria present, and decreased the growth rate to 60%. In all other experimental conditions, the ratio of aggregated bacteria was lower and the growth rate higher showing an inverse relation between percent aggregated bacteria and growth rate (Figure 5F). Although p-MVs induced aggregation of almost 50% of bacteria (Figure 5F), the size of those aggregates was much smaller (Figure 5E-G), and the antibacterial effect of p-MVs was significantly less than that of b-MVs (Figure 5G). Apparently, the inhibition of bacterial growth is dependent on both the number and size of the bacterial aggregates. Taken together, our data suggest that formation of large aggregates between bacteria and MVs may play a critical role in the antibacterial effect of b-MVs. This suggestion was also supported by video microscopy: in the course of 45 minutes, freely diffusing bacteria underwent visible division cycles, but the size of bacterial clumps aggregated with b-MVs did not grow (see the supplemental Video).
To determine whether bacteria survived in the large aggregates with b-MVs, classic microbiologic dilution assays were performed after mechanical disintegration of aggregates. No difference in the number of wells where bacterial growth occurred was observed in the presence or absence of b-MVs, although there was a significant difference in the rate of growth. These experiments suggest that impaired bacterial growth by b-MVs was because of a bacteriostatic, rather than bactericidal, effect. A bacteriostatic effect is in agreement with the finding that GFP did not diffuse out of bacteria clumped with b-MVs (Figure 5D, supplemental Video).
PMN-derived MVs in healthy individuals and bacteremic patients
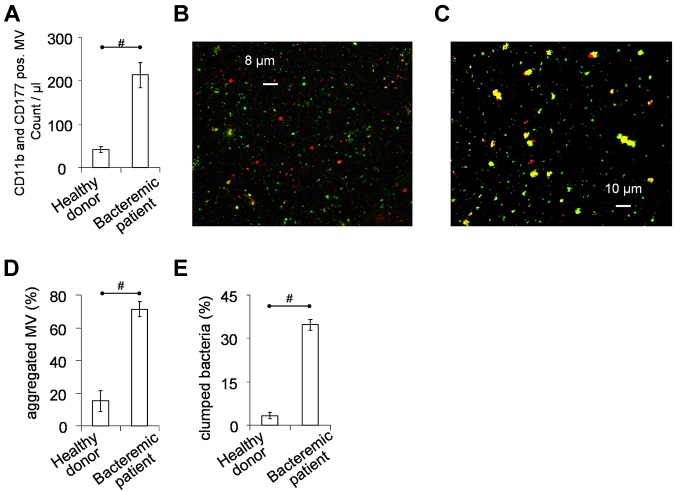
Neutrophil-derived MVs identified by double-staining with antibodies against CD11b and CD177 were routinely detected in the serum of healthy donors. The number of such MVs was 5- to 6-fold higher in serum from patients with documented S aureus bacteremia and fever within 24 hours before drawing the blood sample (Figure 6A; clinical data of the patients are summarized in supplemental Table 3).
Figure 6.
PMN derived MVs in vivo and ex vivo. MVs were separated from serum, using the same conditions as applied for isolated PMNs. (A) Concentration of PMN derived MVs in serum of healthy donors (n = 6) or patients with clinically verified S aureus bacteremia (n = 12), measured by flow cytometry after staining with anti-CD11b and anti-CD177 antibodies. (B-E) MVs separated from serum were incubated with not opsonized S aureus for 30 minutes at 37°C in HBSS with gentle shaking. Interaction was followed by fluorescent microscopy at the end of incubation. (B) Representative image of MVs from healthy donors (of 35 similar images from 4 independent experiments), whereas panel C is representative of MVs from bacteremic patients (53 similar images from 6 independent experiments). Red shows CD11b positivity, green represents S aureus. Statistical analysis (with same criteria as in Figure 5) of microscopic images shows amount of aggregated MVs to all MVs (D), and amount of clumped bacteria to all bacteria (E). Bars show mean ± SEM; #P < .05.
Next we investigated the interaction of PMN-derived MVs isolated from blood serum with GFP-expressing S aureus under ex vivo conditions. We observed formation of large and CD11b positive aggregates between bacteria and MVs from bacteremic patients (Figure 6C), whereas clump formation was only rarely detected with MVs of healthy individuals (Figure 6B). Statistical analysis revealed an approx. Four-fold higher ratio of large aggregates in case of MVs from bacteremic patients than from healthy individuals (Figure 6D). The ratio of clumped bacteria was 7- to 8-fold higher with bacteremic MVs than with MVs from healthy sera (Figure 6E). These data indicate that PMN-derived MV production is increased during bacteremia and bacteremic MVs initiate the formation of similar aggregates ex vivo as observed with b-MVs produced in vitro.
Discussion
Various stimuli were shown earlier to initiate the formation of PMN-derived extracellular vesicles that affect endothelial cells,23,25 platelets,24 or monocytes and macrophages.26–28 However, this is the first report about the direct effect of PMN-derived MVs on bacteria.
Our results showed that MVs released from peripheral blood PMNs by various stimuli have similar size and orientation, but differ in protein composition and functional properties. Only stimulation of PMNs with opsonized particles resulted in production of MVs that were capable of significant reduction of bacterial growth. The antibacterial effect of b-MVs was associated with vesicles that are larger than either the exosomes or the neutrophil granules. In addition, the presence of plasma membrane markers on the vesicle surface suggests that they are not intracellular granules released from neutrophils.
The mechanism(s) by which b-MVs inhibited bacterial growth differed from those of intact PMNs. Although b-MVs expressed the membrane components of the phagocytic NADPH oxidase, gp91phox, and gp22phox, none of the cytosolic subunits of the oxidase were identified and superoxide generation by b-MVs could not be detected. The antibacterial effect of PMNs required bacterial opsonization, whereas the bacteriostatic effect of b-MVs was totally independent of opsonization (Figure 3E). PMNs typically engulf microorganisms and carry out intracellular killing within phagosomes. In contrast, our confocal micrographs indicate that b-MVs do not engulf bacteria; rather they concentrate the microorganism on their surface. Finally, b-MVs are bacteriostatic, whereas PMNs are bactericidal.
The antibacterial effect of b-MVs also differs in significant aspects from the properties of the recently described neutrophil extracellular traps (NETs).38 Formation of NETs typically requires 2 to 4 hours,39 whereas maximum b-MVs-formation was reached in 20 minutes. NETs are destroyed by DNase38 but b-MV release and activity were not affected by DNase treatment. NET formation depends on superoxide production by Nox2,40 whereas both the formation and effect of b-MVs were independent of respiratory burst activity. LPS and PMA are the most effective stimuli of NET-formation, whereas antibacterial MVs were only induced by opsonized particles. No filamentous structures were visible in b-MVs, and permeabilized MVs were not stained by propidium iodide. In contrast, the antibacterial effect of b-MVs depended on an intact vesicular structure, an intact cytoskeleton, and on an adequate supply of glucose.
The mechanism of impaired bacterial growth by b-MVs probably depends on multiple factors. In the presented experiments, 2 specific properties of b-MVs have been revealed. First, the formation of large aggregates with bacteria was associated with their antibacterial activity (Figure 5F-G, supplemental Figure 2, and supplemental Video) and only occurred in the presence of bacteria and b-MVs (as opposed to s-MVs or p-MVs). Aggregation was prevented by inhibition of cytoskeletal reorganization or β2 integrin function and depended on glucose metabolism and phosphatidylinositol-3-kinase activity of b-MVs. In a recent study, firm binding of particles to the surface of macrophages was shown to require active involvement of the actin cytoskeleton.41 We suggest that a similar mechanism may be involved in the integrin-dependent interaction of b-MVs with bacteria that requires continuous actin remodeling. The second specialty of b-MVs is the enrichment of PMN granule proteins, many of which are known to be antibacterial. The detected difference in protein composition of b-MVs, compared with s-MVs, suggests that incorporation of granule constituents into MVs is important to the development of antibacterial activity. It is possible that in the large extracellular aggregates bacteria are exposed to granule proteins concentrated in MVs, thereby limiting bacterial growth.
PMN-derived MVs were detected in the serum of healthy donors, and their number was markedly increased in the serum of patients with an active S aureus infection (Figure 6A). Similar observations were made earlier in meningococcal septic patients.18 These data suggest that bacteria induce formation of MVs under in vivo conditions. In contrast to healthy donors, PMN-derived MVs isolated from bacteremic sera showed definitive aggregation ability ex vivo (Figure 6B-E). Taken into account that sequestration and immobilization of bacteria by PMN-derived MVs was completely independent of opsonization, this mechanism could contribute to limitation of microbial growth in the early stages of infection.
Supplementary Material
Acknowledgments
The authors are indebted to Drs D. Veress for help with DLS measurements, E. Ostorházi for valueable advices, G. Szanda for help with confocal microscopy, and A. Mócsai and T. Németh for antibodies and advices. The devoted technical help of E. Fedina, E. Horváth-Seres, K. Marosvári, and F. Kolonics is greatly appreciated.
This work was supported by grants from the Hungarian National Research Fund (OTKA K81277 and K75084), the Health Research Council (ETT), and TÁMOP (grants 4.2.1/B-09/1/KMR-2010-0001 and 4.2.2/B10/1-2010-0013).
Footnotes
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.T. and A.L. designed and performed the majority of experiments and prepared the figures; R.C.-K., and A.V.N. worked out methods for quantitation of MVs; G.N. and E.B. called our attention to microvesicles and continuously exchanged data; Z.I. selected and characterized the patients; Á.K. performed electron microscopy; D.P. and K.M.-L. performed the proteomic analysis; and E.L. supervised, coordinated and financed the experimental work, and had major role in writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Erzsébet Ligeti, Dept of Physiology, Semmelweis University, Tűzoltó u 37-47, 1094, Budapest, Hungary; e-mail: ligeti@puskin.sote.hu.
References
- 1.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78(9):838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 3.Gyorgy B, Szabo TG, Pasztoi M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons M, Raposo G. Exosomes: vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21(4):575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thery C, Regnault A, Garin J, et al. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147(3):599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 8.Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 9.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20(11):1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- 12.Kesimer M, Scull M, Brighton B, et al. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23(6):1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110(9):3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller I, Klocke A, Alex M, et al. Intravascular tissue factor initiates coagulation via circulating microvesicles and platelets. FASEB J. 2003;17(3):476–478. doi: 10.1096/fj.02-0574fje. [DOI] [PubMed] [Google Scholar]
- 15.VanWijk MJ, VanBavel E, Sturk A, Nieuwland R. Microparticles in cardiovascular diseases. Cardiovasc Res. 2003;59(2):277–287. doi: 10.1016/s0008-6363(03)00367-5. [DOI] [PubMed] [Google Scholar]
- 16.Nomura S, Ozaki Y, Ikeda Y. Function and role of microparticles in various clinical settings. Thromb Res. 2008;123(1):8–23. doi: 10.1016/j.thromres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Gyorgy B, Modos K, Pallinger E, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117(4):e39–48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- 18.Nieuwland R, Berckmans RJ, McGregor S, et al. Cellular origin and procoagulant properties of microparticles in meningococcal sepsis. Blood. 2000;95(3):930–935. [PubMed] [Google Scholar]
- 19.Hong Y, Eleftheriou D, Hussain AA, et al. Anti-neutrophil cytoplasmic antibodies stimulate release of neutrophil microparticles. J Am Soc Nephrol. 2012;23(1):49–62. doi: 10.1681/ASN.2011030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess C, Sadallah S, Hefti A, Landmann R, Schifferli JA. Ectosomes released by human neutrophils are specialized functional units. J Immunol. 1999;163(8):4564–4573. [PubMed] [Google Scholar]
- 21.Gasser O, Hess C, Miot S, Deon C, Sanchez JC, Schifferli JA. Characterisation and properties of ectosomes released by human polymorphonuclear neutrophils. Exp Cell Res. 2003;285(2):243–257. doi: 10.1016/s0014-4827(03)00055-7. [DOI] [PubMed] [Google Scholar]
- 22.Duarte TA, Noronha-Dutra AA, Nery JS, et al. Mycobacterium tuberculosis-induced neutrophil ectosomes decrease macrophage activation. Tuberculosis (Edinb) 2012;92(3):218–225. doi: 10.1016/j.tube.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Mesri M, Altieri DC. Endothelial cell activation by leukocyte microparticles. J Immunol. 1998;161(8):4382–4387. [PubMed] [Google Scholar]
- 24.Pluskota E, Woody NM, Szpak D, et al. Expression, activation, and function of integrin alphaMbeta2 (Mac-1) on neutrophil-derived microparticles. Blood. 2008;112(6):2327–2335. doi: 10.1182/blood-2007-12-127183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesri M, Altieri DC. Leukocyte microparticles stimulate endothelial cell cytokine release and tissue factor induction in a JNK1 signaling pathway. J Biol Chem. 1999;274(33):23111–23118. doi: 10.1074/jbc.274.33.23111. [DOI] [PubMed] [Google Scholar]
- 26.Gasser O, Schifferli JA. Activated polymorphonuclear neutrophils disseminate anti-inflammatory microparticles by ectocytosis. Blood. 2004;104(8):2543–2548. doi: 10.1182/blood-2004-01-0361. [DOI] [PubMed] [Google Scholar]
- 27.Eken C, Gasser O, Zenhaeusern G, Oehri I, Hess C, Schifferli JA. Polymorphonuclear neutrophil-derived ectosomes interfere with the maturation of monocyte-derived dendritic cells. J Immunol. 2008;180(2):817–824. doi: 10.4049/jimmunol.180.2.817. [DOI] [PubMed] [Google Scholar]
- 28.Eken C, Martin PJ, Sadallah S, Treves S, Schaller M, Schifferli JA. Ectosomes released by polymorphonuclear neutrophils induce a MerTK-dependent anti-inflammatory pathway in macrophages. J Biol Chem. 2010;285(51):39914–39921. doi: 10.1074/jbc.M110.126748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz J, Leidal KG, Femling JK, Weiss JP, Nauseef WM. Neutrophil bleaching of GFP-expressing staphylococci: probing the intraphagosomal fate of individual bacteria. J Immunol. 2009;183(4):2632–2641. doi: 10.4049/jimmunol.0804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rada BK, Geiszt M, Kaldi K, Timar C, Ligeti E. Dual role of phagocytic NADPH oxidase in bacterial killing. Blood. 2004;104(9):2947–2953. doi: 10.1182/blood-2004-03-1005. [DOI] [PubMed] [Google Scholar]
- 31.Dean WL, Lee MJ, Cummins TD, Schultz DJ, Powell DW. Proteomic and functional characterisation of platelet microparticle size classes. Thromb Haemost. 2009;102(4):711–718. doi: 10.1160/TH09-04-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uriarte SM, Powell DW, Luerman GC, et al. Comparison of proteins expressed on secretory vesicle membranes and plasma membranes of human neutrophils. J Immunol. 2008;180(8):5575–5581. doi: 10.4049/jimmunol.180.8.5575. [DOI] [PubMed] [Google Scholar]
- 33.Cummins TD, Barati MT, Coventry SC, Salyer SA, Klein JB, Powell DW. Quantitative mass spectrometry of diabetic kidney tubules identifies GRAP as a novel regulator of TGF-beta signaling. Biochim Biophys Acta. 2010;1804(4):653–661. doi: 10.1016/j.bbapap.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 35.Powell DW, Weaver CM, Jennings JL, et al. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24(16):7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1(5):440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg HF. RNase A ribonucleases and host defense: an evolving story. J Leukoc Biol. 2008;83(5):1079–1087. doi: 10.1189/jlb.1107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 39.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191(3):677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi M, Hakkim A, Brinkmann V, et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 2009;114(13):2619–2622. doi: 10.1182/blood-2009-05-221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flannagan RS, Harrison RE, Yip CM, Jaqaman K, Grinstein S. Dynamic macrophage “probing” is required for the efficient capture of phagocytic targets. J Cell Biol. 2010;191(6):1205–1218. doi: 10.1083/jcb.201007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.