Abstract
Charcot-Marie-Tooth (CMT) disease is the most common hereditary neuropathy resulting from mutations in >30 genes expressed in either the Schwann cells or the axon of peripheral nerves. The disease is classified into demyelinating (CMT1), axonal (CMT2) or intermediate (CMTI) based on electrophysiological and pathological findings. Our study focused on the identification of a novel disease mutation in a large Sardinian family with CMT2 of autosomal dominant (AD) inheritance. All available family members were clinically evaluated and samples were collected from consenting individuals. Initially, we excluded known CMT2 genes/loci in this family. We then conducted a genome-wide linkage analysis and mapped the gene to chromosome 9q33–q34. Refined linkage and haplotype analyses defined an 11.6-Mb candidate region with a maximum LOD score of 8.06. Following exclusion of several candidate genes from the region, we targeted the LRSAM1 (leucine-rich repeat and sterile alpha motif-containing 1) gene, very recently found to be associated with autosomal recessive CMT2 in one family. For a more efficient investigation of this large gene, already available proband RNA (cDNA) was initially analyzed. Targeted DNA analysis then confirmed a novel LRSAM1 splice-site (c.2047-1G>A) mutation, causing a frameshift that introduces a stop codon three amino acids further down the new reading frame (p.Ala683ProfsX3). This mutation is located in the C-terminal RING finger motif of the encoded protein and leads to premature truncation of the protein. In the course of our work, a second LRSAM1 mutation dominantly transmitted was identified by another group. Our data further confirms that LRSAM1 mutations are associated with CMT2 of AD inheritance.
Keywords: CMT2, axonal neuropathy, novel LRSAM1 splicing mutation
Introduction
Charcot-Marie-Tooth (CMT) disease or hereditary motor and sensory neuropathy affects 1 in 2500 people and is the most common inherited neurological disorder.1 The clinical presentation is typically of a slowly progressive distal muscle weakness and atrophy with distal sensory loss, high steppage gait, foot deformities, and decreased or absent tendon reflexes.2 Age of onset is variable. CMT is grouped into demyelinating, axonal and intermediate forms based on electrophysiological and pathological findings. The demyelinating types are characterized by reduced motor nerve conduction velocities (MNCVs<38 m/s) and mainly myelin abnormalities on nerve biopsy. The axonal types are characterized by normal or slightly reduced MNCV (>38 m/s) and primarily axonal degeneration on nerve biopsy.3, 4 Intermediate types of CMT are characterized by median MNCVs in the range of 25–45 m/s.5 Inheritance of CMT can be autosomal dominant (AD) that is the most common,6 X-linked,7 or autosomal recessive (AR).8 This diversity results from a large number of mutations in multiple causative genes that are expressed either by myelinating Schwann cells, axons or both. More than 40 loci and about 30 causative CMT genes have thus far been identified (http://www.molgen.ua.ac.be/CMTMutations). Genes in which mutations have been associated with the AD axonal form of the disease (CMT2) include MPZ,9 NEFL,10 GARS,11 RAB7,12 MFN2,13 HSPB1,14 GDAP1,15 HSPB816 and DNM2.17
Recently, two novel LRSAM1 mutations were associated with axonal CMT neuropathies.18, 19 The p.Glu638AlafsX7 mutation has been reported in a family with AR axonal CMT and the p.Leu708ArgfsX28 mutation has been identified in a family with AD axonal CMT (CMT2). The leucine-rich repeat and sterile alpha motif-containing 1 (LRSAM1) protein is a RING finger protein with multiple functions that has a role in receptor endocytosis.
We hereby report a novel splice-site mutation in LRSAM1 in a large Italian CMT2 family. The mutation leads to aberrant acceptor site usage and a concomitant protein frameshift thus confirming the association of LRSAM1 gene mutations with the CMT2 phenotype.
Materials and methods
Subjects and samples
In all, 18 patients and 22 unaffected family members were evaluated neurologically in detail by at least two of the co-author neurologists. Family history was obtained indicating segregation of the disease in, at least, four generations. Standard motor and sensory nerve conduction studies were performed for all available family members.
Blood was collected from consenting individuals and genomic DNA was isolated using the Qiagen Gentra Puregene Blood Kit (Qiagen, Dusseldorf, Germany). Total RNA from a lymphoblastoid cell line of the proband and a control was extracted using the Qiagen RNeasy kit (Qiagen). Ethical approval was granted by the corresponding Institutional Ethics Committees.
Linkage studies
A total 22 available family members were genotyped at 169 markers (Supplementary Table S1) with an average spacing of 24.2 cM, using the genome-wide screening set 6a (Research Genetics, Inc., Huntsville, AL, USA) and following our previously described methodology.20 Linkage analysis was performed using the LINKAGE package of programs.21 Lod scores (Z) were calculated under an AD inheritance model and a disease allele frequency of 1 in 10 000. Two-point lod scores between the disease in the family and each genotyped marker locus were calculated using the MLINK program (The Rockefeller University, New York, NY, USA). Multipoint lod scores were calculated using the LINKMAP program (The Rockefeller University).
Fine mapping of the candidate region was performed with 40 family members and 28 marker loci spanning a 34.3-cM region from D9S934 to D9S158.
Haplotype analysis
Haplotypes of all individuals were constructed using the Cyrillic program (Cyrillic Software, Oxfordshire, UK) and further manual editing.
Mutation analysis
Genomic DNA sequencing of the candidate genes was performed using the proband DNA. PCR primers were designed using the Primer3 program (Whitehead Institute for Biomedical Research, Cambridge, MA, USA) and are available upon request. PCR amplification of candidate genes was performed using standard methods and PCR products were purified with the QIAquick PCR purification kit (Qiagen) or ExoSAP-IT for PCR product clean-up (Affymetrix, Santa Clara, CA, USA). All coding exons of the relevant genes and 15 bp on both flanking sites were covered both by forward and reverse strand sequencing using the CEQ dye terminator (Beckman Coulter, Nyon, Switzerland) or BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA) and the Beckman Coulter CEQ analyzer (Beckman Coulter) or ABI 3130xl Genetic Analyzer (Applied Biosystems) according to the manufacturer's protocol. Sequence traces were automatically compared with the normal gene sequences as listed in the GenBank database, using the CEQ8000 investigator software (Beckman Coulter) or ABI SeqScape software (Applied Biosystems).
All available family members were analyzed for the identified mutation to obtain direct evidence of the mutation event and its co-segregation with the disease in the family. Specific primers for amplification of the LRSAM1 exons 24 and 25 and the relevant PCR conditions are available upon request.
RNA analysis
Reverse transcription PCR of isolated RNA was performed using the Protoscript First strand cDNA synthesis kit (New England Biolabs, Ipswich, MA, USA) with oligo-dT primers, according to the kit's manual. Whole-cDNA PCR amplification of the LRSAM1 gene was performed using long PCR (Crimson LongAmp Taq DNA Polymerase, New England Biolabs). PCR products were analyzed on a 1.5% agarose gel with ethidium bromide staining, then purified and sequenced as described above, using internal primers (available upon request). Sequence traces were automatically compared with the normal cDNA LRSAM1 gene sequence.
Cloning
Patient cDNA was amplified with primers 5′-AAGAGAAGCAGCAGCGAGAG-3′ and 5′-GCCCAGATGATCTTCCTCAA-3′, flanking the identified LRSAM1 mutation. PCR products were cloned into SmaI-digested pcDNA3-EGFP vectors and transfected into Escherichia coli. Plasmid DNA was isolated from individual cloned colonies and then sequenced.
Western blot analysis
Proteins were isolated from lymphoblastoid cell lines and their concentration was determined by the Bradford method. A total of 20 μg of protein extracts were separated by gel electrophoresis, membrane blotted and incubated with the anti-LRSAM1 mouse antibody (1:300, Abcam, Cambridge, UK), followed by incubation with a goat anti-mouse secondary antibody (1:2000, Santa Cruz Biotechnology, Inc., Heidelberg, Germany) conjugated to horseradish peroxidase (HRP). HRP was visualized using blotting luminal reagents and exposure to X-ray films. As a control, 4 μg of protein extracts were processed in the same methodology, using the anti-GAPDH mouse antibody (1:2000, Santa Cruz Biotechnology, Inc.).
Results
Clinical features
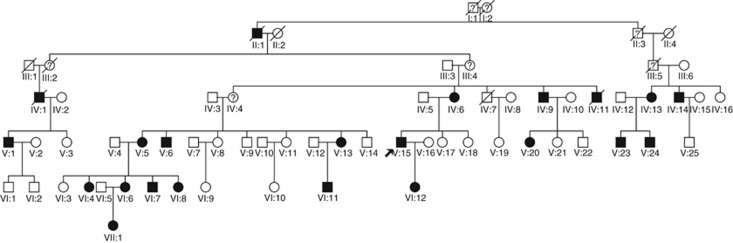
The proband (V.15) is a member of the large Sardinian family (family number 234) shown in Figure 1. His initial symptoms were of distal weakness mainly in the left lower limb with ipsilateral foot drop, at about 34 years of age. On first examination, at the age of 43, there was moderate foot drop on the left; he could not stand on his heels and had difficulties standing on his toes, bilaterally. There was bilateral weakness of ankle dorsiflexion more on the left (MRC scale 3 on the left and 4 on the right). There was distal atrophy in the lower legs and feet, more on the left. Vibration sense was lost at both ankles and reduced at knees; tactile and heat-pain sensitivity were reduced at legs and feet, more on the left. There was no evidence of weakness, atrophy or of any sensory abnormalities in the upper limbs. He reported occasional cramps in the calf muscles and erectile dysfunction.
Figure 1.
Pedigree of the Sardinian CMT2 family 234.
The MNCV of the left deep peroneal nerve was slightly reduced at 31.5 m/s, with a prolonged distal latency at 11.5 ms and a reduced motor action potential (MAP) amplitude (0.8 mV). The left median nerve MNCV was 50.1 m/s, with a distal latency of 3.6 ms and MAP amplitude of 5.4 mV. No sensory potentials were detectable from both sural nerves.
The clinical phenotype in the other affected members of the family had a similar pattern of initial symptoms clinical presentation, being more prominent in the lower limbs, in the majority of cases bilaterally. Table 1 summarizes the clinical findings. The age of onset varied from 15 (a female, VI.4) to 50 (her mother, V.5). The disease course was slowly progressive in all affected individuals, characterized by distal weakness, atrophy and multimodal hypoesthesia in the lower limbs, diffusing to the upper limbs in later years. Patients remain ambulant throughout their lifespan, with one exception (patient VI.4, wheelchair bound since the age of 31 years old). Symptoms of episodic numbness, paresthesias and cramps were reported by all patients. Autonomic disturbances include erectile dysfunction and, in a single case (IV.9), trophic disorder of the foot with mutilating arthropathy.
Table 1. Summary of clinical presentation.
| PID | AAE/Sex | SSO | ULDA | LLDA | Pes cavus | TRP | TRA | VS | MN | DPN |
|---|---|---|---|---|---|---|---|---|---|---|
| V.5 | 55/F | 50 | No | Yes | Yes | ± | − | Lost | 49.1 | 39.3 |
| VI.4 | 31/F | 15 | ± (Thenar) | Yes | Yes | + | − | Lost | 54.2 | 39.1 |
| VI.7 | 29/M | Asymptomatic | No | No | Slight | + | − | Lost | 52.7 | 42.1 |
| V.13 | 34/F | Asymptomatic | No | No | No | + | − | Lost | 56.2 | 37.8 |
| VI.11 | 20/M | Asymptomatic | No | No | No | − | − | Lost | 59.2 | 45.1 |
| V.15 | 43/M | 34 | No | Yes | Yes | + | − | Lost | 50.1 | 31.5 |
| VI.12 | 22/F | Asymptomatic | No | No | Slight | + | − | Pres | 57.8 | 52.9 |
| IV.13 | 55/F | 39 | No | Slight | Slight | + | − | Lost | 54.8 | 38.9 |
| V.23 | 15/M | Asymptomatic | No | No | Yes | ++ | − | Reduced | 52.1 | 40.5 |
| V.24 | 27/M | Asymptomatic | No | No | No | ++ | − | Reduced | 55.9 | 45.3 |
Abbreviations: AAE, age at examination; DPN, deep peroneal nerve motor conduction velocity (m/s); F, female; LLDA, lower limbs distal atrophy; M, male; MN, median nerve motor conduction velocity (m/s); PID, patient identification; SSO, subjective symptoms onset (age in years); TRA, tendon reflex Achilles; TRP, tendon reflex patella; ULDA, upper limbs distal atrophy; VS, vibration sense.
Linkage and haplotype analyses
Genome-wide screening and two-point linkage analysis of the family (Supplementary Table S1), mapped the disease in the family to chromosome 9q33–q34. The candidate region was further confirmed with fine mapping and a maximum two-point lod score of 8.06 at locus D9S63. Two-point linkage analysis data of the fine mapped region are provided in Table 2. Haplotype analysis (Supplementary Figure S2) revealed a common disease haplotype in all affected individuals that further supports the linkage data. The candidate region is defined as the 11.6-Mb interval between marker loci D9S1872 and D9S1861 and contains 196 genes.
Table 2. Two-point linkage analysis results of the candidate region.
| Recombination fraction | ||||||
|---|---|---|---|---|---|---|
| Marker locus | Distance between marker loci in Mb | 0.0 | 0.01 | 0.05 | 0.1 | 0.2 |
| D9S934 | 0 | −infinity | 3.61 | 3.90 | 3.67 | 2.86 |
| D9S2145 | 0.321 | −infinity | 3.66 | 4.08 | 3.92 | 3.11 |
| D9S1872 | 0.373 | −infinity | 5.08 | 5.44 | 5.21 | 4.21 |
| D9S1850 | 1.661 | 5.55 | 5.44 | 5.00 | 4.43 | 3.23 |
| D9S116 | 0.170 | 4.20 | 4.13 | 3.80 | 3.39 | 2.53 |
| D9S1823 | 0.080 | 4.48 | 4.40 | 4.08 | 3.65 | 2.74 |
| D9S282 | 0.217 | 3.20 | 3.13 | 2.84 | 2.47 | 1.71 |
| D9S1881 | 3.108 | 6.09 | 5.98 | 5.50 | 4.89 | 3.61 |
| D9S778 | 0.217 | 6.53 | 6.41 | 5.92 | 5.29 | 3.97 |
| D9S290 | 4.331 | 7.77 | 7.63 | 7.05 | 6.30 | 4.73 |
| D9S61 | 0.043 | 5.53 | 5.42 | 5.00 | 4.45 | 3.28 |
| D9S121 | 0.231 | 5.60 | 5.49 | 5.04 | 4.47 | 3.28 |
| D9S752 | 0.149 | 6.59 | 6.45 | 5.91 | 5.21 | 3.72 |
| D9S115 | 0.297 | 7.13 | 7.01 | 6.50 | 5.82 | 4.37 |
| D9S1831 | 0.174 | 7.14 | 7.01 | 6.48 | 5.80 | 4.35 |
| D9S63 | 0.233 | 8.06 | 7.91 | 7.34 | 6.59 | 5.01 |
| D9S1861 | 0.666 | −infinity | −1.01 | 0.76 | 1.22 | 1.19 |
| D9S118 | 0.048 | −infinity | 4.83 | 5.19 | 4.96 | 4.00 |
| D9S64 | 0.961 | −infinity | 3.24 | 3.83 | 3.80 | 3.12 |
| D9S1847 | 0.345 | −infinity | 1.99 | 2.73 | 2.86 | 2.48 |
| D9S1830 | 0.279 | −infinity | −0.07 | 1.39 | 1.83 | 1.73 |
| D9S1199 | 0.115 | −infinity | 1.90 | 2.63 | 2.75 | 2.37 |
| D9S164 | 0.425 | −infinity | −1.52 | 0.38 | 0.95 | 1.05 |
| D9S66 | 0.412 | −infinity | −2.79 | −1.10 | −0.38 | 0.14 |
| D9S1826 | 1.931 | −infinity | −2.83 | −1.01 | −0.19 | 0.40 |
| D9S158 | 0.650 | −infinity | −6.92 | −3.53 | −1.95 | −0.58 |
Highest two-point lod score value is indicated in bold.
Mutation analysis
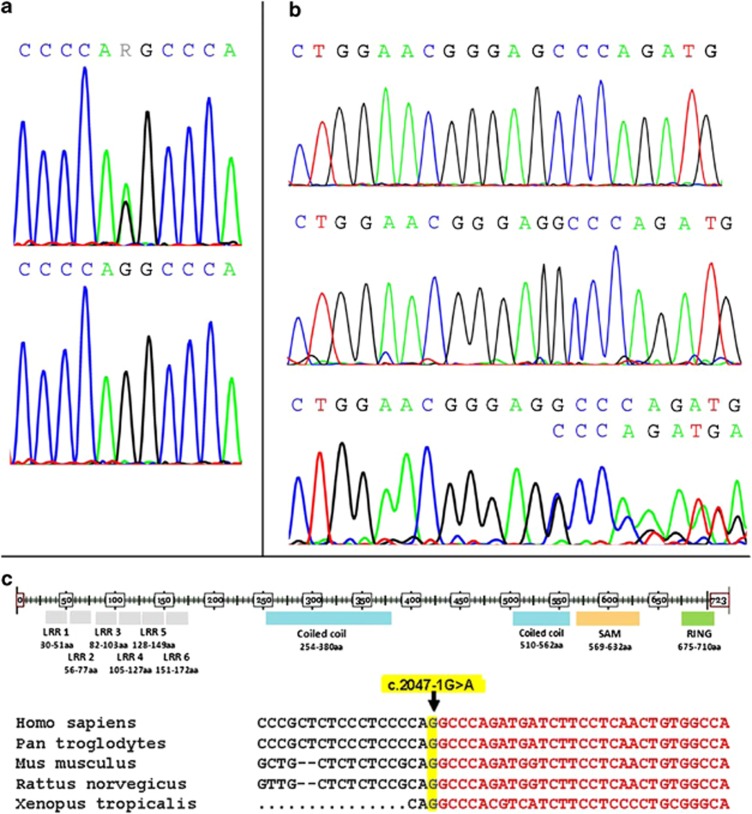
We initially selected seven candidate genes from this region, all expressed in the nervous system, encoding syntaxin-binding protein 1, dynamin 1 protein, neuronal calcium-binding proteins, heat shock protein 5, cerebral endothelial cell adhesion molecule protein, mitogen-activated protein kinase-associated protein 1 and Rab9 effector protein with kelch motifs. We sequenced all their exons and intron–exon boundaries and did not find any disease-associated mutation in any of these genes. After exclusion of mutations in the seven selected candidate genes, sequence analysis of the LRSAM1 gene was performed. As total RNA of the proband was already available from our previous investigations, and to cover this very large gene in a more efficient way, we initially looked for alterations at the RNA level (cDNA). Deletion of a G corresponding to either c.2046 or c.2047 has been detected (Figure 2b), which was further investigated at the DNA level by analysis of both exons 24 and 25 and revealed a splice-site c.2047-1G>A (IVS25-1G>A) mutation (Figure 2a). This mutation is altering the splicing recognition site AG at the intron 24/exon 25 boundary to AA. The splicing mechanism seems to recognize instead the next AG, one base downstream as exon 25 starts with a G. This results in the deletion of the first DNA base of exon 25 at the RNA level (Figure 2b), causing a frame shift. At the protein level, amino acid 683 is changed from alanine to proline and a stop codon is introduced after the next amino acid. Western blot analysis further proved the prediction that a truncated protein is produced missing >30 amino acids, by the presence of a second 79-kDa protein product in the patient sample as compared with the 84-kDa protein that is present in both the unaffected and patient samples (Figure 3).
Figure 2.
Sequences showing the mutation in genomic DNA of the proband and a normal control individual (a) and the resulting sequences of mutant cloned cDNA, normal cDNA and mixed trace from proband cDNA (b). LRSAM1 protein domains and DNA sequence of the LRSAM1 gene region encompassing the identified mutation region, in various organisms (c).
Figure 3.
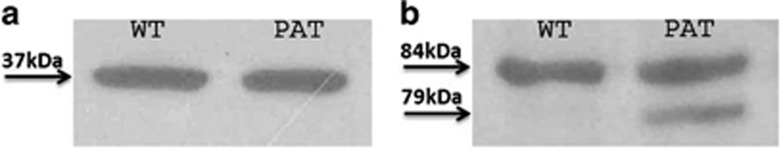
Western blot analysis of the GAPDH (a) and LRSAM1 (b) proteins from a normal control individual (WT) and the proband (PAT). A second smaller LRSAM1 protein product of 79 kDa is observed in the patient sample, proving the predicted truncation of the protein.
Co-segregation of the mutation with the disease in the family was confirmed by sequence analysis of exons 24 and 25 in the proband and further sequence analysis of exon 25 in all available members of the family.
Discussion
We identified a novel splice-site LRSAM1 gene mutation (c.2047-1G>A (IVS25-1G>A), p.Ala683ProfsX3) in a large Sardinian family that is associated with AD axonal neuropathy (CMT2). The alteration is located within the C-terminal RING finger motif region of the LRSAM1 protein and it affects a highly conserved region in different species, including Pan troglodytes, Mus musculus, Rattus norvegicus and Xenopus tropicalis (Figure 2c). Clinical and electrophysiological features of patients in this family are typical of CMT2 including distal weakness and atrophy beginning in the lower limbs, with sensory disturbances and pes cavus, normal MNCVs or slightly reduced only in the deep peroneal nerve and frequent absence of the sural nerve action potential. In this family the disorder usually has a late onset, with several affected individuals being asymptomatic until adult age, and the course is very slow, with very late functional impairment.
The reported mutation results in a 79-kDa protein that lacks a functional RING finger motif domain. As the activity of most E3 ubiquitin ligases has been reported to be specified by the RING domain,22 this mutation most likely disturbs LRSAM1 E3 ligase activity. The other two axonal CMT recently reported LRSAM1 mutations similarly cause disturbance or absence of the RING domain. The p.Glu638AlafsX7 mutation that has been reported in AR axonal CMT,18 results in truncation of the protein at amino acid 645, totally abolishing the RING finger domain. The p.Leu708ArgfsX28 mutation that has been reported in AD axonal CMT,19 results in the exchange of leucine at the C-terminal end of the RING motif to arginine and the production of an altered and elongated protein, thus also altering the RING finger domain of LRSAM1. However, the mechanisms by which each of these distinct LRSAM1 gene mutations contribute to the development of the sensori-motor axonal neuropathy still remains to be elucidated.
Interestingly, it has recently been suggested that LRSAM1 may have a neuroprotective role against mutant huntingtin in Huntington's disease.23 Through microarray analysis and real-time PCR confirmation, six genes (LRSAM1, NASP, RAB9B, ERP29, TAP1 and PFDN5) were found to be upregulated distinctly in R6/2 mice with 300 CAG repeats as compared with R6/2 mice with 150 CAG repeats.23 Further study of LRSAM1 and ERP29 expression in a primary striatal culture model of Huntington's disease, has lead to the hypothesis that increased expression, in particular of LRSAM1, results in a delayed disease onset, latent disease progression and longer lifespan.23
Ubiquitination has an important role in selective protein degradation and membrane protein trafficking,24 and it is involved in a number of cellular processes that include the cell cycle, cell transformation and signal transduction.24, 25 LRSAM1, also known as Tsg101-associated ligase or RING finger E3 ubiquitin ligase, has been reported to regulate a Tsg101-associated complex responsible for the sorting of cargo into cytoplasm-containing vesicles that bud at the multivescicular body and at the plasma membrane.26 LRSAM1 polyubiquitinates lysine residues in the C-terminus of uncomplexed Tsg101 resulting in proteasomal degradation.27 Further elucidation of the precise functions of LRSAM1 and of other proteins interacting with LRSAM1 and other E3-ubiquitin ligases may result in the identification of novel therapeutic targets for disease modification in inherited CMT neuropathies and other neurodegenerative diseases.
Acknowledgments
This study has been supported by the Muscular Dystrophy Association of the USA (grants to Lefkos Middleton (1994–1999) and to Kyproula Christodoulou (2002–2003)) and the Association Française contre les Myopathies of France (grants to Lefkos Middleton (1995–1996) and to Kyproula Christodoulou (2009–2012)).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Barisic N, Claeys KG, Sirotkovic-Skerlev M, et al. Charcot-Marie-Tooth disease: a clinico-genetic confrontation. Ann Hum Genet. 2008;72:416–441. doi: 10.1111/j.1469-1809.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- Harding AE, Thomas PK. Genetic aspects of hereditary motor and sensory neuropathy (types I and II) J Med Genet. 1980;17:329–336. doi: 10.1136/jmg.17.5.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TD. Hereditary motor-sensory neuropathies. Charcot-Marie-Tooth syndrome. Neurol Clin. 1989;7:9–23. [PubMed] [Google Scholar]
- Berger P, Young P, Suter U. Molecular cell biology of Charcot-Marie-Tooth disease. Neurogenetics. 2002;4:1–15. doi: 10.1007/s10048-002-0130-z. [DOI] [PubMed] [Google Scholar]
- Mastaglia FL, Nowak KJ, Stell R, et al. Novel mutation in the myelin protein zero gene in a family with intermediate hereditary motor and sensory neuropathy. J Neurol Neurosurg Psychiatry. 1999;67:174–179. doi: 10.1136/jnnp.67.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. I. Neurologic, genetic, and electrophysiologic findings in hereditary polyneuropathies. Arch Neurol. 1968;18:603–618. doi: 10.1001/archneur.1968.00470360025002. [DOI] [PubMed] [Google Scholar]
- Fischbeck KH, ar-Rushdi N, Pericak-Vance M, et al. X-linked neuropathy: gene localization with DNA probes. Ann Neurol. 1986;20:527–532. doi: 10.1002/ana.410200414. [DOI] [PubMed] [Google Scholar]
- Harding AE, Thomas PK. Autosomal recessive forms of hereditary motor and sensory neuropathy. J Neurol Neurosurg Psychiatry. 1980;43:669–678. doi: 10.1136/jnnp.43.8.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrosu MG, Vaccargiu S, Marrosu G, et al. Charcot-Marie-Tooth disease type 2 associated with mutation of the myelin protein zero gene. Neurology. 1997;50:1397–1401. doi: 10.1212/wnl.50.5.1397. [DOI] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, et al. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, De Jonghe P, Coen K, et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:22–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova IV, Irobi J, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Claramunt R, Pedrola L, Sevilla T, et al. Genetics of Charcot-Marie-Tooth disease type 4A: mutations, inheritance, phenotypic variability, and founder effect. J Med Genet. 2005;42:358–365. doi: 10.1136/jmg.2004.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BS, Zhao GH, Luo W, et al. Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Hum Genet. 2005;116:222–224. doi: 10.1007/s00439-004-1218-3. [DOI] [PubMed] [Google Scholar]
- Fabrizi GM, Ferrarini M, Cavallaro T, et al. Two novel mutations in dynamin-2 cause axonal Charcot-Marie-Tooth disease. Neurology. 2007;69:291–295. doi: 10.1212/01.wnl.0000265820.51075.61. [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Bedard K, et al. Mutation in the gene encoding ubiquitin ligase LRSAM1 in patients with Charcot-Marie-Tooth disease PLoS Genet 201061–7.pii: e1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterman MA, Sorrentino V, Kasher PR, et al. A frameshift mutation in LRSAM1 is responsible for a dominant hereditary polyneuropathy. Hum Mol Genet. 2011;21:358–370. doi: 10.1093/hmg/ddr471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou K, Zamba E, Tsingis M, et al. A novel form of distal hereditary motor neuronopathy maps to chromosome 9p21.1-p12. Ann Neurol. 2000;48:877–884. [PubMed] [Google Scholar]
- Lathrop GM, Lalouel J-M, Julier C, Ott J. Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet. 1985;37:482–498. [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Ann Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Tang B, Seredenina T, Coppola G, et al. Gene expression profiling of R6/2 transgenic mice with different CAG repeat lenghts reveals genes associated with disease onset and progression in Huntington's disease. Neurobiol Dis. 2011;42:459–467. doi: 10.1016/j.nbd.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Yamao F. Ubiquitin system: selectivity and timing of protein destruction. J Biochem. 1999;125:223–229. doi: 10.1093/oxfordjournals.jbchem.a022277. [DOI] [PubMed] [Google Scholar]
- Amit I, Yakir L, Katz M, et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Martin-Serrano J. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol Biol Cell. 2008;19:754–763. doi: 10.1091/mbc.E07-09-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.